Abstract
Despite its relevance, protein regulation, metabolic adjustment, and the physiological status of plants under drought is not well understood in relation to the role of nitrogen fixation in nodules. In this study, nodulated alfalfa plants were exposed to drought conditions. The study determined the physiological, metabolic, and proteomic processes involved in photosynthetic inhibition in relation to the decrease in nitrogenase (Nase) activity. The deleterious effect of drought on alfalfa performance was targeted towards photosynthesis and Nase activity. At the leaf level, photosynthetic inhibition was mainly caused by the inhibition of Rubisco. The proteomic profile and physiological measurements revealed that the reduced carboxylation capacity of droughted plants was related to limitations in Rubisco protein content, activation state, and RuBP regeneration. Drought also decreased amino acid content such as asparagine, and glutamic acid, and Rubisco protein content indicating that N availability limitations were caused by Nase activity inhibition. In this context, drought induced the decrease in Rubisco binding protein content at the leaf level and proteases were up-regulated so as to degrade Rubisco protein. This degradation enabled the reallocation of the Rubisco-derived N to the synthesis of amino acids with osmoregulant capacity. Rubisco degradation under drought conditions was induced so as to remobilize Rubisco-derived N to compensate for the decrease in N associated with Nase inhibition. Metabolic analyses showed that droughted plants increased amino acid (proline, a major compound involved in osmotic regulation) and soluble sugar (D-pinitol) levels to contribute towards the decrease in osmotic potential (Ψs). At the nodule level, drought had an inhibitory effect on Nase activity. This decrease in Nase activity was not induced by substrate shortage, as reflected by an increase in total soluble sugars (TSS) in the nodules. Proline accumulation in the nodule could also be associated with an osmoregulatory response to drought and might function as a protective agent against ROS. In droughted nodules, the decrease in N2 fixation was caused by an increase in oxygen resistance that was induced in the nodule. This was a mechanism to avoid oxidative damage associated with reduced respiration activity and the consequent increase in oxygen content. This study highlighted that even though drought had a direct effect on leaves, the deleterious effects of drought on nodules also conditioned leaf responsiveness.
Keywords: Drought, Medicago sativa, N2 fixation, N remobilization, oxidative stress, photosynthesis, proteome, Rubisco
Introduction
The major environmental factor that constrains the productivity and stability of plants is water stress (Araus et al., 2002). According to the different scenarios predicted by the Intergovernmental Panel on Climate Change (Alley et al., 2007) it is expected that there will be a reduction in precipitation and rising evapotranspiration rates. The perceived need to gain further understanding of photosynthesis, so as to alleviate practical problems such as crop yield under drought conditions, has increased interest in ‘water stress physiology’ (Lawlor and Tezara, 2009).
Photosynthesis and cell growth are among the primary processes to be affected by drought (Chaves and Oliveira, 2004; Chaves et al., 2009). The photosynthetic rates of plants exposed to drought decrease due to stomatal closure and non-stomatal processes (Lawlor and Cornic, 2002; Aranjuelo et al., 2007; Lawlor and Tezara, 2009; Chaves et al., 2009). Although it is generally accepted that stomatal closure is the main factor limiting photosynthetic activity under moderate water-limiting conditions (Chaves et al., 2002, 2003), when water stress is more severe, metabolic impairment takes place (Medrano et al., 2002). Deleterious effects of drought on photosynthesis will be mediated by the responsiveness of (i) the respiration system, electron transport, and ATP synthesis in the mitochondria (Atkin and Macherel, 2009), (ii) the accumulation of stress metabolites (Zhang et al., 1999), and (iii) gene expression and protein synthesis (Lawlor and Tezara, 2009).
The balance between light capture and energy use are of great relevance to studies concerning the responsiveness of the photosynthetic apparatus under water-stress conditions (Sharp and Boyer, 1985, 1986; Cornic and Briantais, 1991; Chaves et al., 2009; Lawlor and Tezara, 2009). When photosynthesis decreases and light excitation energy is in excess, photooxidative damage may occur. The excessive excitation energy in photosystem II (PSII) will lead to an impairment of photosynthetic function, progress to an accumulation of reactive oxygen species (ROS), and thereby result in oxidative stress. Plants have developed three main mechanisms to diminish photooxidative damage: (i) preventing the production of ROS by diminishing the electron transport chain (Lawlor and Tezara, 2009); (ii) scavenging ROS formed by an integrated system of enzymatic and non-enzymatic (ascorbate–glutathione cycle) antioxidants (Asada, 1999), and (iii) diminishing photooxidation through xanthophyll cycle-dependent thermal dissipation, which is an important photoprotective process in the light-harvesting antenna of PSII (Gilmore, 1997; Verhoeven et al., 1999).
The effect of water limitation on proteins involved in C metabolism in legumes is not well understood. Analysis on this topic has been limited, and mainly focused on the characterization of the quantity and activity of Rubisco (Parry et al., 2003; Aranjuelo et al., 2007). Proteomics addresses analytical questions about the abundance and distribution of proteins in organisms, the expression profiles of different tissues and the identification and localization of individual proteins of interest (Kersten et al., 2002). This method has developed as an important approach in evaluating plant responsiveness under limited growth conditions (Desclos et al., 2008, 2009).
Although the influence of water availability on plant growth and photosynthetic activity has been studied extensively (Bushby, 1982; Fellows et al., 1987; Irigoyen et al., 1992), less attention has been given to the role of nodule activity in plant performance under drought conditions. Nodule activity depends on photosynthates supplied by the plant, which are used by the nitrogenase enzyme as a source of energy and reducing power to fix N2 (Gálvez et al., 2005; Larrainzar et al., 2009). Similarly, the products of N2 fixation, which are either amides or ureides, are exported throughout the plant via the xylem to other organs where N is required, for example, protein synthesis (Ladrera et al., 2007). This coupling results in the regulation of nitrogenase activity in plants by photosynthesis (carbon supply), nitrogen availability (N source strength), and N demand (N sink strength).
The main objective of this study was to characterize the responsiveness of the photosynthetic apparatus of nodulated alfalfa (Medicago sativa L.) plants during exposure to drought conditions. The relationship between plant and nodule metabolism was also studied to determine its possible implication in alfalfa responsiveness to drought. The combination of physiological and proteomic analyses may constitute an original contribution to understanding the drought effect on photosynthetic activity.
Materials and methods
Experimental design
Seeds of alfalfa (Medicago sativa L, Magali variety) were germinated on plates. After 3–4 d, seedlings were transplanted into 7.0 l white plastic pots filled with sand and grown in a greenhouse at 25/15 °C (day/night) with a photoperiod of 14 h under natural daylight. During the first month, plants were inoculated three times with Sinorhizobium meliloti strain 102F78. Plants were watered twice a week with Hoagland N-free full nutrient solution and once a week with deionized water to avoid salt accumulation in pots. When the plants were 91-d-old, half of the plants (randomly selected) were exposed to drought conditions (with water withholding) whereas the others were maintained in optimal water availability conditions. Over 7 d, drought plants were grown without any watering, whereas control plants were watered until pot capacity. Plant water status was evaluated by measuring the leaf water content (LWC) and osmotic potential (Ψs). Leaf water content was calculated as LWC=(FW–DW)/FW, where FW refers to fresh weight and DW refers to dry weight. Osmotic potential was determined using a Wescor 5500 osmometer (Wescor, Logan, Utah, USA) as described by Ball and Oosterhuis (2005).
Gas exchange and chlorophyll fluorescence determinations
Fully expanded apical leaves were enclosed in a Li-Cor 6400 gas exchange portable photosynthesis system (Li-Cor, Lincoln, Nebraska, USA). The gas exchange response to CO2 was measured from 0 to 1000 μmol mol−1 CO2. The light-saturated rate of CO2 assimilation (Asat) was estimated at a photosynthetic photon flux density (PPFD) of 1200 μmol m−2 s−1 using equations developed by von Caemmerer and Farquhar (1981). Stomatal conductance (gs) was determined as described by Harley et al. (1992). Estimations of the maximum carboxylation velocity of Rubisco (Vcmax) and the maximum electron transport rate contributing to RuBP regeneration (Jmax) were made using the method of Ethier and Livingston (2004). Plants were dark adapted for 30 min before dark respiration (R) measurements. Nodule respiration was studied by placing them in a respiration chamber (20×12×6×10−6 m3) connected in parallel to the sample air hose of a LI-COR-6400 according to Aranjuelo et al. (2009).
The maximal quantum efficiency of PSII (Fv/Fm) and the relative quantum efficiency of PSII photochemistry (ΦPSII) were simultaneously measured with a fluorescence chamber (LFC 6400-40; Li-COR) coupled to the Li-Cor 6400 portable photosynthesis system. For Fv/Fm determinations, leaves were dark-adapted for 30 min. Non-photochemical quenching (NPQ) was calculated as 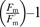 –1 as described by Bilger and Björkman (1990) Photochemical quenching (qP) was calculated according to Andrews et al. (1993). The rate of electron transport through PSII [Je(PSII)] was measured as described by Harley et al. (1992). The rate of oxygenation by Rubisco (Vo) was estimated as described by von Caemmerer and Farquhar (1981) as Vo=(Vc×pO2)/(Sr×Ci) where Vc refers to the rate of carboxylation of RuBP, pO2 refers to the ambient partial pressure of O2, Sr refers to the relative specificity of Rubisco, and Ci refers to the intercellular CO2 concentration. The rate of carboxylation by Rubisco (Vc) was estimated as Vc=(A+Rd)/[1−pO2/(2×Sr×Ci)], where Rd refers to the rate of day respiration (Miyake and Yokota, 2000). The electron fluxes in the two cycles, expressed as Je(PCR)=4×Vc and Je(PCO)= 4×Vo, respectively (Krall and Edwards, 1992), were conducted at growth conditions corresponding to each treatment.
–1 as described by Bilger and Björkman (1990) Photochemical quenching (qP) was calculated according to Andrews et al. (1993). The rate of electron transport through PSII [Je(PSII)] was measured as described by Harley et al. (1992). The rate of oxygenation by Rubisco (Vo) was estimated as described by von Caemmerer and Farquhar (1981) as Vo=(Vc×pO2)/(Sr×Ci) where Vc refers to the rate of carboxylation of RuBP, pO2 refers to the ambient partial pressure of O2, Sr refers to the relative specificity of Rubisco, and Ci refers to the intercellular CO2 concentration. The rate of carboxylation by Rubisco (Vc) was estimated as Vc=(A+Rd)/[1−pO2/(2×Sr×Ci)], where Rd refers to the rate of day respiration (Miyake and Yokota, 2000). The electron fluxes in the two cycles, expressed as Je(PCR)=4×Vc and Je(PCO)= 4×Vo, respectively (Krall and Edwards, 1992), were conducted at growth conditions corresponding to each treatment.
Nitrogenase activity
Alfalfa nodule activity was estimated by the C2H2 reduction technique (Hardy et al., 1973). Intact nodulated roots were enclosed in a 1.0 l glass flask and 100 ml of C2H2 was added. The flask was incubated at room temperature for 10 min. Afterwards, eight samples of 5 ml were withdrawn from the flask and the ethylene content in the samples was quantified using a Fractovap 4200 (Carbo Erba Strumentazione, Milan, Italy) gas chromatograph equipped with a hydrogen flame ionization detector and a column of Poropak R30/100 (2 m×1/8). Determinations were conducted at 90 °C (45 °C detector and injector) with a carrier gas flow rate of 25 ml min−1. Acetylene reduction protocol allows the meaningful analyses of growth condition effects in nitrogenase (Nase) activity (Streeter et al., 2003; King and Purcell, 2005).
Pigments and soluble sugar determinations
Extracts for pigment analysis were prepared by grinding 100 mg fresh weight in a cold mortar with 10 ml of ethanol (95%, v/v). The homogenate was centrifuged at 3165 g for 10 min at 4 °C. An aliquot of 1 ml from the supernatant was taken, 4 ml of 95% ethanol were added, and the absorbance measured at 750, 665, 649, and 470 nm. Absorbance determinations were carried out with a Spectronic 2000 (Bausch and Lomb, Rochester, USA) spectrophotometer. Extinction coefficients and equations used to calculate pigment contents were those described by Liechenthaler (1987). For sugar extraction about 50 mg of lyophilized and ground leaves were suspended with 1 ml of distilled water in an Eppendorf tube (Eppendorf Scientific, Hamburg, Germany). The solution was mixed and centrifuged at 12 000 g for 5 min at 5 °C. The supernatant was heated at 100 °C for 3 min and afterwards centrifuged at 12 000 g for 5 min at 5 °C. After centrifugation, the supernatant containing the soluble fraction was purified with a solid phase extraction column (Oasis MCX 3cc, Waters) to separate sugars from the other soluble compounds. Total soluble sugars were determined with the Spectronic 2000 spectrophotometer according to the method proposed by Yemm and Willis (1954). Sucrose, glucose, and fructose contents were analysed using a Waters 600 high performance liquid chromatograph (Waters Millipore Corp., Milford, Massachusetts, USA). The HPLC refractive index detector (Waters 2414) was set at 37 °C. Samples were eluted from the columns at 85 °C (connected in series Aminex HPX-87P and Aminex HPX-87C, 300 mm×7.8 mm, Bio-Rad) with water at a flow rate of 0.6 ml min−1 and retention time run up to 45 min.
Quantification of D-pinitol was conducted by liquid chromatography and mass spectrometry. LC–MS detection was achieved using an Applied Biosystems/PE SCIEX API 150 EX single quadrupole mass spectrometer equipped with a turbo ionspray source (PerkinElmer Series 200 Pump). Two columns, an Aminex HPX-87P and Aminex HPX-87C, were serial connected and eluted at 0.6 ml min−1 with water as the mobile phase. Column temperatures were maintained at 85 °C. A post-column addition of 18 μM of sodium acetate in acetonitrile at a flow rate of 0.6 ml min−1 was undertaken with the isocratic pump to obtain adducts of sodium. A 1:3 split was done before placing in the mass spectrometer. Typically, 50 μl of standards and samples diluted in water were injected to columns.
Free amino acid determinations by GC-MS
Frozen leaf and nodule samples were ground to a fine powder in liquid N and a sub-sample was lyophilized. Trifluoracetic Acid (TFA) 10% (v/v) was added to the sample to avoid enzymatic activity and to extract the soluble fraction. The homogenate was centrifuged at 6000 g for 15 min at 4 °C. The supernatant was purified with Ultrafree-MC 10000 NMWL (Millipore, Billerica, Massachusetts, USA) in an Eppendorf tube and centrifuged (13 000 g for 45 min at 4 °C). L-norleucine (Sigma-Aldrich, St Louis, Missouri, USA) was added as internal standard to the filtered samples and the mixture was dried with a Speed Vac desiccator overnight. Then the samples were re-suspended in 1 ml of HCl 0.1 N (v/v) and passed through a chromatographic column (Dowex 50W X8 H+, 16–40 mesh size, Sigma®) as previously described by Owen et al. (1999. The mixture of amino acids eluted from the column was completely evaporated and derivatized with N-methyl-N-(tert-butyildimethylsilyl)-trifluoroacetamide (MTBSTFA, Aldrich®) as outlined by Woo and Chang (1993) and Woo and Lee (1995). The amino acid derivatives were then injected directly into a gas chromatography-mass spectrometer (GC-MS). The concentrations of amino acids in the samples were calculated using external calibration curves for each amino acid and values were recalculated against the internal standard (L-norleucine).
Proteomic characterization
Frozen and ground leaf samples (200 mg fresh weight) were resuspended in 2 ml of cold acetone containing 10% TCA (v/v). After centrifugation at 16 000 g for 3 min at 4 °C, the supernatant was discarded and the pellet was rinsed with methanol, acetone, and phenol solutions as previously described by Wang et al. (2003). The pellet was stored at –20 °C or immediately resuspended in 200 μl of R2D2 rehydratation buffer [5 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl) dimethyl-ammonio]-1-propane-sulphonate, 2% N-decyl-N,N-dimethyl-3-ammonio-1-propane-sulphonate, 20 mM dithiothreitol, 5 mM TRIS (2-carboxyethyl) phosphine, 0.5% IPG buffer (GE Healthcare, Saclay, France), pH 4 to 7 (Mechin et al., 2003)]. The total soluble protein (TSP) concentration was determined by the method of Bradford (1976) using BSA as standard. The two-dimension electrophoresis was conducted according to what described by Desclos et al. (2008).
Image analysis of 2-DE gels
Images of the two-dimensional gels were acquired with the ProXPRESS 2D proteomic Imaging System and analysed using Phoretix 2-D Expression Software v2004 (Nonlinear Dynamics, Newcastle upon Tyne, UK). Gels from four independent biological replicates were used. An average gel, representative of each group, was automatically selected by the software with a parameter for spots to be present on more than two-thirds of the gels. The software automatically selected the average gel with the most spots as the image for the reference gel, and unmatched spots from the remaining average gel were added to the reference gel which was subsequently used for spot matching to average gels. Warping and matching were automatically performed and only adjusted on those gels where darker images led to both incorrect warping and matching. Mr and pI were calculated using Samespots software calibrated with commercial molecular mass standards (precision protein standards prestained Bio-Rad) run in a separate marker lane on the 2-DE gel. ANOVA (P <0.05) was performed using MiniTAB to compare the relative abundance of the total volume of all detected spots for each gel.
Protein identification by ESI-LC MS/MS
Excised spots were washed several times with water and dried for a few minutes. Peptide extracts were then dried and dissolved in starting buffer for chromatographic elution, which consisted of 3% CH3CN and 0.1% HCOOH in water. Peptides were enriched and separated using lab-on-a-chip technology (Agilent, Massy, France) and fragmented using an on-line XCT mass spectrometer (Agilent). The fragmentation data were interpreted using the Data Analysis program (version 3.4, Bruker Daltonic, Billerica, USA). For protein identification, tandem mass spectrometry peak lists were extracted and compared with the protein database using the MASCOT Daemon (version 2.1.3; Matrix Science, London, UK) search engine. Tandem mass spectrometry spectra were searched with a mass tolerance of 1.6 Da for precursor ions and 0.8 for MS/MS fragments.
The LC MS/MS data were converted into DTA-format files which were further searched for proteins with MASCOT Daemon. Only peptides matching an individual ion score >51 were considered. Proteins with two or more unique peptides matching the protein sequence were automatically considered as a positive identification. Among the positive matches based on one unique peptide, the fragmentation spectrum from each peptide was manually interpreted using the conventional fragmentation rules. In particular, we looked for a succession of at least five y- and/or b-ions, specific immonium ions, specific fragment ions (proline and glycine), and signatures of any modifications carried by the peptides. For protein identification, two strategies were used to mine the maximum information. Measured peptides were searched in the NCBInr-protein sequence database viridiplantae (green plants). Once the proteins were identified, we proceeded to their presumed biological function according to Bevan et al. (1998). This classification showed that up-regulated proteins belonged to the energy and protein destination and storage functional groups, whereas the down-regulated proteins belonged to metabolism, energy, protein destination and storage, transport, cell structure and disease/defence groups.
Statistical analyses
Data was processed by two-factor analysis of variance (ANOVA). Means ±standard errors (SE) were calculated, and when the F-ratio was significant, least significant differences were evaluated by Tukey's test using the statistical software package SPSS 12.0 (SPSS Inc., Chicago, IL, USA). The results were accepted as significant at P <0.05. All values shown in the figures and tables are means ±SE.
Results
Suppression of irrigation over 7 d reduced leaf water content by 33% (LWC; Table 1) and increased osmotic potential of leaves in alfalfa plants subjected to drought (Table 1). Photosynthesis (A) also decreased dramatically (58%; Table 1) in droughted plants as a consequence of stomatal closure (as reflected by stomatal conductance and transpiration decrease), a reduction in carboxylation capacity of Rubisco (Vcmax), and a reduction in the potential rate of electron transport contributing to RuBP regeneration (Jmax). Although no significant differences were observed in leaf dark respiration (Rleaf), drought decreased nodule respiration (Rnodule) as shown in Table 1. Intercellular CO2 concentration (Ci) data confirmed that drought-treated plants had lower intercellular CO2 available (Table 1). Nitrogenase (Nase) activity was markedly decreased under drought conditions (Table 1).
Table 1.
The water availability effect (control versus drought) in terms of leaf water content (LWC), leaf osmotic potential (Ψs), stomatal conductance (gs), transpiration (E), leaf temperature (Tleaf), leaf respiration (Rleaf), saturating maximum photosynthetic rate (Asat), maximum carboxylation velocity of Rubisco (Vcmax), and the maximum electron transport rate contributing to RuBP regeneration (Jmax), intercellular CO2 concentration (Ci), the maximal photochemical efficiency (Fv/Fm), relative quantum efficiency of PSII photochemistry (ΦPSII), efficiency of energy capture by open PSII reaction centres (), photochemical quenching (qP), non-photochemical quenching (NPQ), total chlorophyll (Chl a+b), electron transport through Photosystem II [Je(PSII)], electron transport through photosynthetic carbon reduction [Je(PCR)], electron transport through photorespiratory carbon oxidation [Je(PCO)], nodule respiration (Rnodule), and acetylene reduction assay (ARA) of Medicago sativa plants
Measurements were conducted at the end of the experiment, when plants were 3 months old. Each value represents the mean ±SE (n=8). The different letters indicate significant differences (P <0.05).
| Parameter | Control | Drought |
| Leaf | ||
| LWC | 76.31±0.97 a | 51.68±8.31 b |
| Ψs leaf (MPa) | –1.45±0.05 a | –3.85±0.61 b |
| gs (mmol H2O m−2 s−1) | 527.14±13.83 a | 98.14±15.81 b |
| E (mmol H2O m−2 s−1) | 0.65±0.12 a | 0.33±0.05 b |
| Tleaf (°C) | 24.04±0.31 b | 26.78±0.6 a |
| Rleaf (μmol m−2 s−1) | –2.30±0.29 a | –1.6±0.38 a |
| Asat (μmol m−2 s−1) | 25.83±3.03 a | 10.83±2.77 b |
| Vcmax(μmol m−2 s−1) | 139.15±11.7 a | 106.11±9.00 b |
| Jmax (μmol m−2 s−1) | 150.30±7.48 a | 126.84±5.12 b |
| Ci (μmol mol−1) | 297.83±27.47 a | 204.25±16.21 b |
| Fv/Fm | 0.76±0.01 a | 0.78±0.02 a |
| ΦPSII | 0.29±0.03 a | 0.20±0.02 b |
| 0.50±0.03 a | 0.41±0.03 b | |
| qP | 0.61±0.03 a | 0.44± 0.06 b |
| NPQ | 1.22±0.1 b | 2.03± 0.08 a |
| Chl a+b (mg g−1 DM) | 8.89±0.8 a | 9.87±90.05 a |
| Je(PSII) (μmol e−2 s−1) | 149.3±2.81 a | 117.45±4.54 b |
| Je(PCR) (μmol e−2 s−1) | 115.48±2.8 a | 74.00±3.31 b |
| Je(PCO) (μmol e−2 s−1) | 33.82±1.48 b | 43.44±1.67 a |
| Nodule | ||
| Ψsnodule (MPa) | –0.88±0.005 a | –1.20±0.01 b |
| Rnodule (μmol m−2 s−1) | –5.41±1.49 a | –2.11±0.62 b |
| Nase (μmol C2H2 g−1 DW h−1) | 41.80±7.84 a | 10.05±1.23 b |
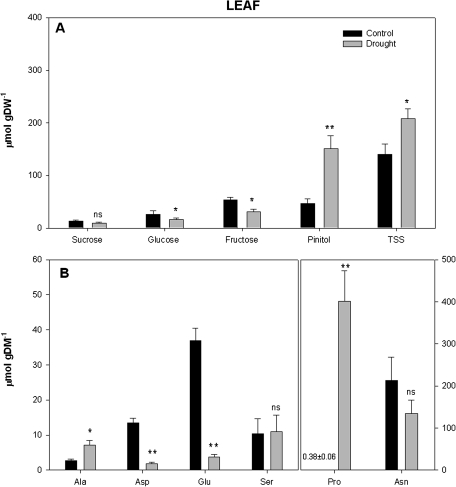
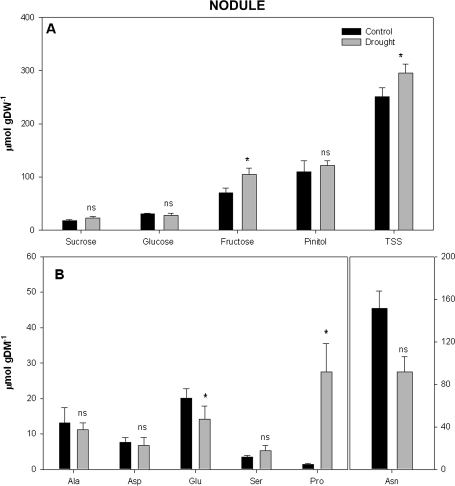
Under drought conditions ΦPSII was negatively affected (Table 1). Furthermore, the lower rate of electron transport through PSII ([Je(PSII)]) detected in droughted plants could have contributed to a reduction in the photosynthetic capacity of these plants. Electron transport through photosynthetic carbon reduction, Je(PCR), confirmed that fewer electrons were delivered to photosynthetic carboxylation processes in droughted plants. The electron flux for photorespiratory carbon oxidation, Je(PCO), was increased under drought conditions (Table 1). Photochemical quenching (qP) was lower in droughted plants, whereas non-photochemical quenching (NPQ) was stimulated by 27% in low water availability treatments (Table 1). No statistical differences were observed for total chlorophyll content associated with water availability (Table 1). Interestingly, leaf soluble sugar determinations (Fig. 1A) highlighted that glucose and fructose concentrations were diminished under low water availability conditions, even though total soluble sugar (TSS) and D-pinitol content were stimulated. No statistical differences were observed for sucrose content. The analyses of free amino acid content in leaves showed that although drought increased alanine and proline content, glutamic and aspartic acid content decreased in droughted plants (Fig. 1B). On the other hand, although no statistical differences were observed for sucrose, glucose and D-pinitol, in droughted nodules fructose and TSS increased (Fig. 2A). Concerning the nodule free amino acid content, drought increased proline content, whereas glutamic acid and asparagine (marginally significant; P=0.063) decreased. No significant differences at P <0.05 were detected in alanine, aspartic acid, and serine levels (Fig. 2B).
Fig. 1.
Water availability effect on (A) leaf soluble sugar (sucrose, glucose, fructose, pinitol, and total soluble sugar, TSS) and (B) free amino acid (Ala, alanine; Asp, aspartic acid; Glu, glutamic acid; Ser, serine; Asn, asparagine, and Pro, proline) content in Medicago sativa plants. Measurements were conducted at the end of the experiment. The y-axis scale for asparagine and proline was modified to make the text more understandable. In addition, since the proline values detected in control plants were low, the average value ±SE was added to the figure. Each value represents the mean ±SE (n= 4). Different letters indicate significant differences (P <0.05) between treatments.
Fig. 2.
Water availability effect on (A) nodule soluble sugar (sucrose, glucose, fructose, pinitol, and total soluble sugar, TSS) and (B) free amino acid (Ala, alanine; Asp, aspartic acid; Glu, glutamic acid; Ser, serine; Asn, asparagine, and Pro, proline) content in Medicago sativa plants. Measurements were conducted at the end of the experiment. Each value represents the mean ±SE (n=4). The different letters indicate significant differences (P <0.05) between treatments.
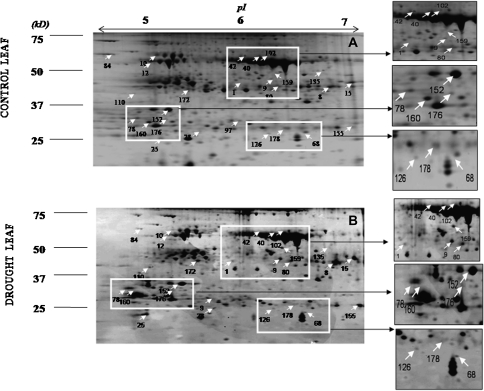
The drought effect on the leaf protein pattern in alfalfa plants was studied using 2-DE (Fig. 3). Our protocol enabled the identification of 26 proteins that differed in their expression under control and drought conditions. Twelve of those proteins were down-regulated by drought, with the remaining 16 being up-regulated (Table 2). These proteins were classified into different groups according to their presumed biological function. The down-regulated proteins were classified into two groups: energy processes (11 proteins identified) and protein destination and storage processes (1 protein identified). Among the up-regulated proteins, six groups were detected: metabolism (2 proteins identified), energy (4 proteins identified), protein destination and storage (3 proteins identified), transport (1 protein identified), cell structure (1 protein identified) and the proteins involved in disease/defence processes (5 proteins identified). The roles of these proteins are discussed in the following section with regard to changes in physiological traits in response to drought.
Fig. 3.
Silver-stained two-dimensional gel of proteins extracted from Medicago sativa leaves grown under control (A) and drought (B) conditions. In the first dimension, 125 mg of total protein was loaded on a 18 cm IEF strip with a linear gradient of pH 4–7. The second dimension was conducted in 12% polyacrylamide (w/v) gels (20×20 cm) (for details, see Materials and methods). The gel image analyses conducted with Progenesis SameSpots software v3.0 and the subsequent mass spectrometry analyses identified up to 26 proteins (marked by arrows) that, statistically, were involved in the plant response to drought.
Table 2.
Annotation of up/down-regulated identified spots following drought in silver stained two-dimensional electrophoresis gels of leaves
Spot no. represents the number of proteins assigned. Spot volume (%) is an estimation of relative protein abundance. The pI and molecular mass (Mr) values shown are the theoretical and experimental values. SC represents the protein sequence coverage (%) score, which is the Mascot score of the in-solution digestion protocol. Function, the predicted protein function is assigned according to the NCBInr-protein sequence database.
| Spot no. | Spot % volume variations | Experimental pI/Mr | Theoretical pI/Mr | PM | SC (%) | Score (P <0.05 corresponding to score >51) | Protein name/organism/NCBI accession no. | Regulation |
| 01. Metabolism | ||||||||
| 159 | 641.02 | 6.31/51.0 | 6.06/49.71 | 5 | 17 | 198 | Dihydrolipoamide dehydrogenase/Pisum sativum/gi|9955321 | Up-regulated |
| 172 | 720.36 | 5.35/40.7 | 6.29/47.08 | 14 | 19 | 466 | Glutamine synthetase/Medicago sativa /gi|17367236 | Up-regulated |
| 02. Energy | ||||||||
| 8 | 170.34 | 6.85/47.8 | 9.75/49.63 | 2 | 28 | 62 | Glyceraldehyde 3-phosphate dehydrogenase/Ficus wassa/gi|87300266 | Up-regulated |
| 10 | 16.92 | 4.85/56.5 | 4.99/61.78 | 2 | 4 | 153 | β subunit of mitochondrial ATP synthase/Chlamydomonas reinhardtii /gi|159466892 | Down-regulated |
| 12 | 16.84 | 4.77/52.8 | 5.25/52.68 | 2 | 5 | 60 | β subunit of ATP synthase/Eurya sp./gi|20269410 | Down-regulated |
| 15 | 167.44 | 7.19/46.0 | 8.93/43.411 | 5 | 20 | 218 | (NADP-dependent glyceraldehydephosphate) Glyceraldehyde-3-phosphate dehydrogenase A/Pisum sativum/gi|120658 | Up-regulated |
| 40 | 16.33 | 6.34/63.0 | 6.09/52.11 | 3 | 8 | 138 | Rubisco large subunit/Pieris phillyreifolia /gi|1352804 | Down-regulated |
| 42 | 16.29 | 6.30/62.1 | 6.0/51.00 | 4 | 10 | 159 | Rubisco large subunit/Heisteria cauliflora/gi|112408850 | Down-regulated |
| 80 | 155.37 | 6.37/41.5 | 5.92/35.47 | 5 | 21 | 191 | Cytosolic malate dehydrogenase/Cicer arietinum/gi|10334493 | Up-regulated |
| 97 | 48.84 | 5.99/35.7 | 5.83/38.63 | 5 | 18 | 258 | Fructose-bisphosphate aldolase 1/Pisum sativum/gi|399024 | Down-regulated |
| 102 | 15.01 | 6.30/62.1 | 6.22/51.85 | 6 | 14 | 233 | Rubisco large subunit/Akania bidwillii/gi|2500654 | Down-regulated |
| 135 | 57.1 | 6.70/51.0 | 6.73/29.96 | 2 | 10 | 60 | Rubisco large subunit/Echeandia sp./gi|1865802 | Down-regulated |
| 152 | 14.01 | 5.04/35.2 | 5.63/29.99 | 37 | 65 | 1057 | Rubisco activase/Medicago sativa/gi|23320705 | Down-regulated |
| 155 | 1381.95 | 7.02/34.0 | 8.80/43.31 | 14 | 24 | 607 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplast precursor/Pisum sativum/gi|120658 | Up-regulated |
| 160 | 13.37 | 4.20/30.5 | 5.83/42.21 | 10 | 19 | 306 | Sedoheptulose-1,7-bisphosphatase precursor/Oryza sativa (indica cultivar-group)/gi|27804768 | Down-regulated |
| 176 | 24.45 | 5.05/30.7 | 5.41/39.00 | 3 | 11 | 73 | Phosphoribulokinase/Pisum sativum/gi|1885326 | Down-regulated |
| 178 | 76.01 | 6.17/27.2 | 8.23/30.34 | 3 | 9 | 107 | Ribulose-phosphate 3-epimerase/Spinacia oleracea/gi|2833386 | Down-regulated |
| 06. Protein destination and storage | ||||||||
| 78 | 155.68 | 4.86/33.9 | 8.80/43.31 | 11 | 35 | 514 | 14-3-3-like protein/Cicer arietinum/gi|148612111 | Up-regulated |
| 84 | 15.47 | 4.85/62.5 | 5.20/61.18 | 21 | 35 | 985 | Putative rubisco subunit binding-protein alpha subunit/Trifolium pratense/gi|84468288 | Down-regulated |
| 110 | 148.88 | 4.61/40.0 | 4.60/38.37 | 5 | 11 | 178 | Plastoglobulin-1/Pisum sativum/gi|62900628 | Up-regulated |
| 126 | 145.18 | 6.31/27.5 | 6.30/24.60 | 1 | 8 | 69 | Proteasome subunit beta type-1 (20S proteasome alpha subunit F) (20S proteasome subunit beta-6)/Petunia×hybrida/ gi|17380185 | Up-regulated |
| 07. Transport | ||||||||
| 9 | 169.45 | 6.33/46.5 | 6.04/35.10 | 2 | 9 | 62 | Putative chloroplast inner envelope protein/Oryza sativa/gi|15341602 | Up-regulated |
| 09. Cell structure | ||||||||
| 1 | 178.63 | 5.95/40.2 | 6.01/40.82 | 3 | 11 | 149 | Reversibly glycosylated polypeptide/Gossypium hirsutum/gi|18077708 | Up-regulated |
| 11. Disease/defence | ||||||||
| 25 | 161.22 | 4.96/24.9 | 4.93/21.84 | 4 | 24 | 211 | 2-cys peroxiredoxin-like protein/Hyacinthus orientalis/gi|47027073 | Up-regulated |
| 28 | 165.27 | 5.49/25.0 | 5.47/24.04 | 2 | 8 | 111 | Dehydroascorbate reductase/Zinnia elegans/gi|50058092 | Up-regulated |
| 68 | 158.47 | 6.45/25.6 | 7.16/26.62 | 1 | 6 | 67 | Superoxide dismutase/Medicago sativa/gi|23534609 | Up-regulated |
| 78 | 155.68 | 4.86/33.9 | 8.80/43.31 | 11 | 35 | 514 | 14-3-3-like protein/Cicer arietinum/gi|148612111 | Up-regulated |
| 155 | 1381.95 | 7.02/34.0 | 8.80/43.31 | 14 | 24 | 607 | Glyceraldehyde-3-phosphate dehydrogenase A, subunit/Pisum sativum/gi|120658 | Up-regulated |
| 159 | 641.02 | 6.31/51.0 | 6.06/49.71 | 5 | 17 | 198 | Dihydrolipoamide dehydrogenase/Pisum sativum/gi|9955321 | Up-regulated |
Discussion
The inhibitory effect of drought on photosynthetic activity has been widely described and is mainly associated with stomatal and metabolic limitations (Chaves et al., 2009; Lawlor and Tezara, 2009). The decrease in leaf water content (from 76.32% to 51.68%) and the increase in leaf osmotic potential (Ψs) confirmed the deterioration of leaf water status in droughted plants.
Physiological characterization parameters revealed that diminishment of the Rubisco carboxylation (Vcmax) and the RuBP regeneration activities (Jmax), together with stomatal closure (Table 1) explained the photosynthetic decrease in droughted plants (Allen et al., 1997; Nogués and Baker, 2000; Nunes et al., 2008). Although the drought effect on Rubisco activity is a long known phenomenon, the analysis of its regulation is complex (Parry et al., 2002; Chaves et al., 2009; Lawlor and Tezara, 2009), especially in N2-fixing plants.
At the leaf level, the negative effects of drought on the large subunits of the four different Rubisco spots (Table 2) revealed that diminishment of Rubisco activity can be explained, in part, by lower Rubisco availability (Table 2). The down-regulation observed for proteins involved in Rubisco assembly (putative Rubisco, and the subunit binding-protein) confirmed this point. The reduction in Nase activity observed in droughted plants (Table 1), together with the general depletion of the main leaf amino acid content (with the exception of proline and alanine; Fig. 1B) suggests that drought negatively affected plant N availability (Gordon et al., 1997; Ramos et al., 1999; Ladrera et al., 2007; Larrainzar et al., 2009). Under these unfavourable conditions, the mobilization of N from the main leaf N reservoir (i.e. Rubisco) would contribute towards alleviating the N deficiency (Feller et al., 2008). This idea was reinforced by the up-regulation under drought conditions of enzymes with proteolytic activity, such as the proteosome β1 subunit (CCI7) (Table 2), which could have contributed to the degradation of Rubisco (Desclos et al., 2009). In accordance with this point, the up-regulation of glutamine synthetase (the enzyme involved in the GS-GOGAT cycle where assimilated NH3 is converted to glutamic acid, Glu, and glutamine, Gln) under drought conditions suggests that there was reallocation of N derived from Rubisco to other processes and organs (Ochs et al., 1999; Diaz et al., 2008; Desclos et al., 2009). The fact that, in droughted leaves, Glu levels decreased suggests that this amino acid was rapidly redirected to proline synthesis (Delauney and Verma, 1993; Fougère et al., 1991; Hare et al., 1999). The increase in free proline under water stress (Figs 1B, 2B) is associated with its role as an osmoregulant to prevent dehydration and maintaining negative water potential to avoid water loss (Irigoyen et al., 1992; Hare et al., 1998). The fact that Rubisco activase content (Table 2) diminished under droughted conditions revealed that the lower activation state could also have negatively affected the above-mentioned Rubisco activity (Chaves et al., 2002; Reddy et al., 2004; Parry et al., 2008).
The decrease in Jmax under drought conditions suggests that the diminishment of Rubisco activity and photosynthetic activity could also be associated with limitations of RuBP regeneration. The down-regulation of enzymes involved in the C regeneration processes (sedoheptulose-1,7-bisphosphatase, ribulose-phosphate 3-epimerase and phosphoribulokinase) in drought conditions (Table 2) may have caused the depletion in RuBP regeneration. The repression of genes involved in the synthesis of Calvin Cycle enzymes under drought conditions has been associated with an increase in sugar concentration in the leaf (Krapp et al., 1991; Koch, 1996; Stitt et al., 2007) suggesting that sugars may act as signalling molecules (Chaves et al., 2009). The larger leaf total soluble sugar (TSS, Fig. 1A) levels that were detected in drought conditions could have been involved in the inhibition of Calvin Cycle enzymes. The enhancement in soluble sugar content was related to an increase in pinitol content (Fig. 1A). Alongside proline (Fig. 1B), pinitol was central in maintaining water balance in alfalfa plants. As recently described by Obendorf et al. (2008), glucose is one of the precursors of pinitol synthesis and therefore the decrease in glucose content in droughted leaves could have been a consequence of glucose being directed towards pinitol synthesis. This is confirmed by the up-regulation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) which implies that more 3-phosphoglycerate was formed. 3-phosphoglycerate is a glucose (Buchanan et al., 2000) and pinitol precursor. Pinitol has been previously described in legumes as a major carbohydrate (up to the 50–60% of soluble sugars), especially under water stress conditions (Ford, 1984; McManus et al., 2000; Streeter et al., 2001) and it may act as an osmolyte (Reddy et al., 2004).
The reported inhibition of Rubisco and other enzymes involved in the Calvin cycle under drought (see above) implies a reduction in the demand for ATP and NADPH. Such a decrease was also reflected in the reduction in electron transport (Lawlor and Tezara, 2009) through photosystem II [Je(PSII); Table 1]. Drought increased the electron flux destined for photorespiratory carbon oxidation [Je(PCO); Table 1]. This enhancement was confirmed by the up-regulation of two protein isoforms of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the up-regulation of dihydrolipoamide dehydrogenase (Table 2), both involved in photorespiration (Bourguignon et al., 1996; Wingler et al., 1999). The excess of electrons was transferred to oxygen at PSI or via the Mehler reaction, with the consequent generation of reactive oxygen species (ROS; Mehta et al., 1992; Ort et al., 2002; Chaves and Oliveira, 2004; Bogeat-Triboulot et al., 2007; Moreno et al., 2008; Parry et al., 2008). In order to protect PSII against increased production of ROS, drought-stressed plants improved the mechanism of excess electron removal by dissipating part of this energy through heat emission (Ort and Baker, 2002) as observed by the enhancement of NPQ (Table 1). Moreover, the proteomic profile revealed that drought induced the up-regulation (Table 2) of well-known proteins involved in detoxification of ROS such as superoxide dismutase (SOD), dehydroascorbate reductase (DHAR), 2-cys peroxiredoxin-like protein (BAS1) (Baier et al., 2000; Dietz et al., 2002; Desclos et al., 2009) and GAPDH, which is also involved in ROS scavenging (Hancock et al., 2005).
Deleterious effects of drought on Nase activity (Table 1) have confirmed that the legume–Sinorhizobium symbiosis is very sensitive to water stress (Antolín and Sánchez-Díaz, 1992; Irigoyen et al., 1992; Streeter, 2003; Aranjuelo et al., 2007; Ladrera et al., 2007; Larrainzar et al., 2009). Under drought conditions, symbiotic nitrogen fixation (SNF) was more affected by drought than CO2-photosynthetic assimilation rates (Durand et al., 1987; Serraj et al., 1999). The deleterious effect of drought on Nase activity decreased asparagine (Asn) content in the nodules (Fig. 2B), which is the major N-transporting amino acid. Furthermore, since ammonia (another major form of N transport) content is also dependent on N2 fixation, drought might have negatively affected its availability and limited its partitioning to leaves with a consequent limitation to plant N availability. The enhancement of nodule TSS levels under drought conditions (Fig. 2A) means that sugar availability limitations were discarded as an explanation of decreased N2 fixation (Ramos et al., 1999; Gálvez et al., 2005; Naya et al., 2007). However, reduced nodule respiration rates (Table 1), in the form of lower Nase-linked respiration, reflects the possible impairment of C metabolism that could have contributed to the depletion of Nase activity (Gordon et al., 1997; Ramos et al., 1999). The lower respiration rates observed in nodules (Table 1) suggest that the TSS increase under drought conditions was associated with the decrease in carbohydrate requirements (Fellows et al., 1987; Aguirreolea and Sánchez-Díaz, 1989; Irigoyen et al., 1992). According to Gálvez et al. (2005), the down-regulation of the glycolytic pathway might provoke a shortage of substrates for bacteroid respiration, and, as a consequence, a transient accumulation of oxygen in the affected region would occur leading to an increase in the resistance of the oxygen diffusion barrier in order to avoid Nase damage.
Other studies (Hartwig, 1998; Serraj et al., 1998, 1999; King and Purcell, 2005) have suggested that N2 fixation is regulated by the N feedback mechanism. However, proline was the only amino acid that increased in droughted nodules (Fig. 2B) and its increase is associated with osmoregulatory mechanisms that maintain nodule turgor (Fougère et al., 1991; Irigoyen et al., 1992; Hare et al., 1999). Our data suggest that, as a consequence of Nase inhibition, droughted nodules invested large resources in the synthesis of proline osmoregulant. Another mechanism that could play a role in the drought-induced inhibition of N2 fixation, but has received much less attention, is oxidative stress (Marino et al., 2006; Naya et al., 2007). High proline levels in the nodules have been observed to have a protective role against ROS (Koca et al., 2007; Türkan and Demiral, 2009). The increase observed in proline levels of droughted nodules suggests that these nodules could have been subjected to oxidative stress and that proline increase played an important role in averting oxidative damage in nodules under water stress.
Conclusions
In summary, the analysis of the leaf proteome and nodule metabolism that was conducted in this work has provided new insights into the drought impairment of photosynthetic activity as revealed by physiological studies. This study showed that droughted plants invested a large quantity of C and N resources into the synthesis of osmoregulants (i.e. pinitol and proline) in order to maintain osmotic turgor in droughted leaves and nodules. Although stomatal closure initially limited the photosynthetic activity of drought-stressed alfalfa plants, proteomic analyses revealed important metabolic constraints. The deleterious effect of drought on photosynthetic activity was targeted to Rubisco and Nase activities. Drought negatively affected the availability of Rubisco binding protein content (involved in the assembly of Rubisco large and small subunits) and this could be related to the lower Rubisco availability. The lower activation state (as reflected by the lower Rubisco activase content) of this protein may have also contributed to the photosynthetic decrease. Furthermore, the proteomic characterization also revealed that the down-regulation of three Calvin cycle proteins involved in the C regeneration contributed to limiting RuBP regeneration and, consequently, Rubisco activity. The deleterious effect of drought on Nase activity was involved in the down-regulation of Rubisco protein content. The up-regulation of proteases and glutamine synthetase, together with the depletion of aspartic and glutamic acid under drought conditions, highlighted the fact that Rubisco-derived N was targeted to the synthesis of osmoregulant compounds (i.e. proline) and may be transported to the taproot. The increase in nodule TSS and the reduced respiration observed in droughted nodules suggests that Nase activity decreased due to enhancement of the oxygen diffusion barrier resistance that prevents oxidative damage to Nase. Although more research is required in this area, the increase in proline content reveals that the potential for oxidative stress damage in droughted nodules should also be considered.
Acknowledgments
This work was supported by the European Project PERMED (INCO-CT-2004-509140) and by the Spanish Science and Education Ministry (BFU-2004-05096/BFI, Juan de la Cierva research grant). The authors wish to thank Esther Miralles, David Bellido, and Regina Roca (Parc Cientific, Universtitat de Barcelona) for their technical assistance. We would also like to express our thanks for the valuable comments made by the two anonymous referees and to the editor who contributed to the improvement of the manuscript. The contribution of Laurent Coquet and Thierry Jouenne (CNRS, Université de Rouen) with the proteomic analyses is also appreciated.
Glossary
Abbreviations
- 2-CP
2-Cys peroxiredoxin BAS1
- 2-DE
two-dimensional electrophoresis
- Ala
alanine
- Asat
light-saturated rate of CO2 assimilation
- Asn
asparagine
- Asp
aspartic acid
- CCI7
proteosome β1 subunit
- Ci
intercellular CO2 concentration
- DH
dehydrogenase
- DHAR
dehydroascorbate reductase
- DW
dry weight
- E
leaf transpiration
- Fv/Fm
maximal photochemical efficiency
efficiency of energy capture by open PSII reaction centres
- FW
fresh weight
- Gln
glutamine
- G-3P
glyceraldehyde-3-phosphate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- Glu
glutamic acid
- cGAPDH
cytosolic glyceraldehyde 3-phosphate dehydrogenase
- GC-MS
gas chromatography mass spectometer
- gs
leaf stomatal conductance
- GS
glutamine synthetase
- IPCC
Intergovernmental Panel on Climate Change
- Jmax
electron transport rate contributing to RuBP regeneration
- Je(PSII)
electron transport through photosystem II
- Je(PCR)
electron transport through photorespiratory carbon oxidation
- LHCII
light-harvesting complex II
- LWC
leaf water content
- MDHAR
monodehydroascorbate reductase
- Nase
nitrogenase
- NPQ
non-photochemical quenching
- PCR
photosynthetic C reduction
- PPFD
photosynthetic photon flux
- Pro
proline
- PSII
photosystem II
- qP
photochemical quenching
- QTL
quantitative trait locus
- Rleaf
leaf respiration
- Rnodule
nodule respiration
- RuBP
ribulose bisphosphate
- ROS
reactive oxygen species
- Rubisco
ribulose 1,5 bisphosphate carboxylase oxygenase
- Ser
serine
- SOD
superoxide dismutase
- TCA
tricarboxylic acid cycle
- TFA
trifluoracetic acid
- Tleaf
leaf temperature
- TPI
triose phosphate isomerase
- TSS
total soluble sugar
- Vcmax
maximum photosynthetic rate
- ΦPSII
relative quantum efficiency of PSII photochemistry
- Ψs
osmotic potential
References
- Aguirreolea J, Sánchez-Díaz M. CO2 evolution by nodulated roots in Medicago sativa L. under water stress. Journal of Plant Physiology. 1989;134:598–602. [Google Scholar]
- Allen DJ, McKee IF, Farage PK, Baker NR. Analysis of the limitation to CO2 assimilation on exposure of leaves of two Brassica napus cultivars to UV-B. Plant, Cell and Environment. 1997;20:633–640. [Google Scholar]
- Alley R, Berntsen T, Bindoff NL, et al. Climate change 2007. The physical science basis. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL, editors. Contribution of Working Group I to the Fourth Annual Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2007. [Google Scholar]
- Andrews JR, Bredenkamp GJ, Baker NR. Evaluation of the role of state transitions in determining the efficiency of light utilization for CO2 assimilation in leaves. Photosynthesis Research. 1993;38:15–26. doi: 10.1007/BF00015057. [DOI] [PubMed] [Google Scholar]
- Antolín MC, Sánchez-Díaz M. Photosynthetic nutrient use efficiency, nodule activity and solute accumulation in drought stressed alfalfa plants. Photosynthetica. 1992;27:595–604. [Google Scholar]
- Aranjuelo I, Irigoyen JJ, Sánchez-Díaz M. Effect of elevated temperature and water availability on CO2 exchange and nitrogen fixation of nodulated alfalfa plants. Environmental and Experimental Botanny. 2007;59:99–108. [Google Scholar]
- Aranjuelo I, Pardo T, Biel C, Savé R, Azcón-Bieto J, Nogués S. Leaf carbon management in slow-growing plants exposed to elevated CO2. Global Change Biology. 2009;15:97–109. [Google Scholar]
- Araus JL, Slafer GA, Reynolds MP, Royo C. Plant breeding and water relations in C3 cereals: what to breed for? Annals of Botanny. 2002;89:925–940. doi: 10.1093/aob/mcf049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. The water–water cycle in chloroplast: scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology and Molecular Biology. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Atkin OK, Macherel D. The crucial role of plant mitochondria in orchestrating drought tolerance. Annals of Botanny. 2009;103:581–597. doi: 10.1093/aob/mcn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Noctor G, Foyer CH, Dietz KJ. Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzyme associated with ascorbate metabolism but not glutathione metabolism. Plant Physiology. 2000;120:823–832. doi: 10.1104/pp.124.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball RA, Oosterhuis DM. Measurement of root and leaf osmotic potential using the vapor-pressure osmometer. Environmental and Experimental Botany. 2005;53:77–84. [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. Analysis of 19 Mb of ontiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bilger W, Björkman O. Role of the xanthophylls cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynthesis Resesarch. 1990;25:173–185. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- Bogeat-Triboulot MB, Brosché M, Renaut J, et al. Gradual soil water depletion results in reversible changes of gene expression, protein profiles, echophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiology. 2007;143:876–892. doi: 10.1104/pp.106.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon J, Merand V, Rawsthorne S, Forest E, Douce R. Glycine decarboxylase and pyruvate dehydrogenase complexes share the same dihydrolipoamide dehydrogenase in pea leaf mitochondria: evidence from mass spectrometry and primary-structure analysis. Biochemistry Journal. 1996;313:229–234. doi: 10.1042/bj3130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annals of Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants. Rockville, MD, USA: American Society of Plant Physiologists; 2000. [Google Scholar]
- Bushby HVA. Broughton WJ, editor. Ecology. Nitrogen fixation. 1982;2 Rhizobium. Oxford, UK: Clarendon Press, 35–75. [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought: from genes to the whole plant. Functional Plant Biology. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Oliveira MM. Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. Journal of Experimental Botanny. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Pereira JS, Maroco J, Rodrigues ML, et al. How plants cope with water stress in the field: photosynthesis and growth. Annals of Botany. 2002;89:907–916. doi: 10.1093/aob/mcf105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornic G, Briantais JM. Partitioning of photosynthetic electron flow between CO2 and O2 reduction in a C3 leaf (Phaseolus vulgaris L.) at different CO2 concentrations and during drought stress. Planta. 1991;183:178–184. doi: 10.1007/BF00197786. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. The Plant Journal. 1993;4:215–223. [Google Scholar]
- Desclos M, Doubousset L, Etienne P, Le Caherec F, Satoh H, Bonnefoy J, Ourry A, Avice JC. A proteomic profiling approach to reveal a novel role of Brassica napus drought 22 kD/water-soluble chlorophyll-binding protein in young leaves during nitrogen remobilization induced by stressful conditions. Plant Physiology. 2008;147:1830–1844. doi: 10.1104/pp.108.116905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos M, Etienne P, Coquet L, Cosette P, Bonnefoy J, Segura R, Reze S, Ourry A, Avice JC. A combined 15N tracing/proteomics study in Brassica napus reveals the chronology of proteomics events associated to N remobilisation during leaf senescence induced by nitrate limitation or starvation. Proteomics. 2009;9:3580–3608. doi: 10.1002/pmic.200800984. [DOI] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Le Dily F, Masclaux-Daubresse C. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under nitrogen nutrition. Plant Physiology. 2008;147:1437–1449. doi: 10.1104/pp.108.119040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Horling F, Konig J, Baier M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. Journal of Experimental Botany. 2002;53:1321–1329. [PubMed] [Google Scholar]
- Durand JL, Sheehy JE, Minchin FR. Nitrogenase activity, photosynthesis and nodule water potential in soybean plants experiencing water-deprivation. Journal of Experimental Botany. 1987;38:311–321. [Google Scholar]
- Ethier GJ, Livingston NJ. On the need to incorporate sensitivity to CO2 transfer conductance into the Farquhar–von Caemmerer–Berry leaf photosynthesis model. Plant, Cell and Environment. 2004;27:137–153. [Google Scholar]
- Feller U, Anders I, Mae T. Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is termed. Journal of Experimental Botany. 2008;59:1615–1624. doi: 10.1093/jxb/erm242. [DOI] [PubMed] [Google Scholar]
- Fellows RJ, Patterson RP, Raper CD, Harris D. Nodule activity and allocation of photosynthate of soybean during recovery from water stress. Plant Physiology. 1987;84:456–460. doi: 10.1104/pp.84.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CW. Accumulation of low molecular weight solutes in water-stressed tropical legumes. Phytochemistry. 1984;23:1007–1015. [Google Scholar]
- Fougère F, Le Rudulier D, Streeter JG. Effects of salt stress on amino acid, organic acid and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.) Plant Physiology. 1991;96:1228–1236. doi: 10.1104/pp.96.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez L, González EM, Arrese-Igor C. Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. Journal of Experimental Botany. 2005;56:2551–2561. doi: 10.1093/jxb/eri249. [DOI] [PubMed] [Google Scholar]
- Gilmore AM. Mechanistic aspects of xanthopyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiologia Plantarum. 1997;99:197–209. [Google Scholar]
- Gordon AJ, Minchin FR, SkØt L, James CL. Stress-induces declines in soybean N2 fixation are related to nodule sucrose synthase activity. Plant Physiology. 1997;114:937–946. doi: 10.1104/pp.114.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT, Henson DH, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ. Proteomic identification of glyceraldehydes 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiology and Biochemistry. 2005;43:828–835. doi: 10.1016/j.plaphy.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Hardy RWF, Burns C, Hebert RR, Holsten RD. Applications of the acetylene–ethylene assay for measurements of nitrogen fixation. Soil Biology and Biochemistry. 1973;5:47–81. [Google Scholar]
- Hare PD, Cress WA, van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant, Cell and Environment. 1998;21:535–553. [Google Scholar]
- Hare PD, Cress WA, van Staden J. Proline synthesis and degradation: a model system for elucidating stress-related signal transduction. Journal of Experimental Botany. 1999;50:413–434. [Google Scholar]
- Harley PC, Loreto F, Marco GD, Sharkey TD. Theoretical considerations when estimating the mesophyll conductance to CO2 flux by analysis of the response of photosynthesis to CO2. Plant Physiology. 1992;98:1429–1436. doi: 10.1104/pp.98.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig UA. The regulation of symbiotic N2 fixation: a conceptual model of N feedback from ecosystem to the gene expression level. Perspectives in Plant Ecology, Evolution and Systematics. 1998;1:92–120. [Google Scholar]
- Irigoyen JJ, Emerich DW, Sánchez-Díaz M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiologia Plantarum. 1992;84:55–60. [Google Scholar]
- Kersten B, Bürkle L, Kuhn EJ, Giavalisco P, Konthur Z, Lueking A, Walter G, Eickhoff H, Schneider U. Large-scale plant proteomics. Plant Molecular Biology. 2002;48:133–141. [PubMed] [Google Scholar]
- King CA, Purcell LC. Inhibition of N2 fixation in soybean is associated with elevated ureides and amino acids. Plant Physiology. 2005;137:1389–1396. doi: 10.1104/pp.104.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koca M, Bor M, Ozdemir F, Turkan I. The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environmental and Experimental Botany. 2007;60:344–351. [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annual Review of Plant Biology. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Krall JP, Edwards GE. Relationship between photosystem II activity and CO2 fixation in leaves. Physiologia Plantarum. 1992;86:180–187. [Google Scholar]
- Krapp A, Quick WP, Stitt M. Rubisco, other Calvin cycle enzymes, and chlorophyll decrease when glucose is supplied to mature spinach leaves via the transpiration stream. Planta. 1991;186:58–69. doi: 10.1007/BF00201498. [DOI] [PubMed] [Google Scholar]
- Ladrera R, Marino D, Larrainzar E, González EM, Arrese-Igor C. Reduced carbon availability to bacteroids and elevated ureides in nodules, but not in shoots, are involved in the nitrogen fixation response to early drought in soybean. Plant Physiology. 2007;145:539–546. doi: 10.1104/pp.107.102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Scherling C, Kempa S, Ladrera R, Arrese-Igor C, Weckwerth W, González EM. Carbon metabolism and bacteroid functioning are involved in the regulation of nitrogen fixation in Medicago truncatula under drought and recovery. Molecular Plant–Microbe Interactions. 2009;22:1565–1576. doi: 10.1094/MPMI-22-12-1565. [DOI] [PubMed] [Google Scholar]
- Lawlor DW, Cornic G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant, Cell and Environment. 2002;25:275–294. doi: 10.1046/j.0016-8025.2001.00814.x. [DOI] [PubMed] [Google Scholar]
- Lawlor DW, Tezara W. Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: a critical evaluation of mechanisms and integration of processes. Annals of Botany. 2009;103:543–549. doi: 10.1093/aob/mcn244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophyll and carotenoids: pigments of photosynthetic biomembranes. In: Packer L, Douce R, editors. Methods in enzymology. Plant cell membranes. USA: Academic Press; 1987. pp. 350–382. [Google Scholar]
- Marino D, González EM, Arrese-Igor C. Drought effects on carbon and nitrogen metabolism of pea nodules can be mimicked by paraquat: evidence for the occurrence of two regulation pathways under oxidative stress. Journal of Experimental Botany. 2006;57:665–673. doi: 10.1093/jxb/erj056. [DOI] [PubMed] [Google Scholar]
- McManus MT, Bieleski RL, Caradus JR, Barker DJ. Pinitol accumulation in mature leaves of white clover in response to a water deficit. Environmental and Experimental Botany. 2000;43:11–18. [Google Scholar]
- Mechin V, Consoli L, Le Guilloux M, Damerval C. An efficient solubilization buffer for plant proteins focused in immobilized pH gradients. Proteomics. 2003;3:1299–1302. doi: 10.1002/pmic.200300450. [DOI] [PubMed] [Google Scholar]
- Medrano H, Escalona JM, Bota J, Gulias J, Flexas J. Regulation of photosynthesis of C3 plants in response to progressive drought: stomatal conductance as a reference parameter. Annals of Botany. 2002;89:895–905. doi: 10.1093/aob/mcf079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RA, Fawcett TW, Porath D, Mattoo AK. Oxidative stress causes rapid membrane translocation and in vivo degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase. Journal of Biology and Chemistry. 1992;267:2810–2816. [PubMed] [Google Scholar]
- Miyake C, Yokota A. Determination of the rate of photoreduction of O2 in the water–water cycle in watermelon leaves and enhancement of the rate by limitation of photosynthesis. Plant and Cell Physiology. 2000;41:335–343. doi: 10.1093/pcp/41.3.335. [DOI] [PubMed] [Google Scholar]
- Moreno J, García-Murria MJ, Marín-Navarro J. Redox modulation of Rubisco conformation and activity through its cysteine residues. Journal of Experimental Botany. 2008;59:1605–1614. doi: 10.1093/jxb/erm310. [DOI] [PubMed] [Google Scholar]
- Naya L, Ladrera R, Ramos J, González EM, Arrese-Igor C, Minchin FR, Becana M. The response of carbon metabolism and antioxidant defensas of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiology. 2007;144:1104–1114. doi: 10.1104/pp.107.099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogués S, Baker N. Effects of drought on photosynthesis in Mediterranean plants grown under enhanced UV-B irradiation. Journal of Experimental Botany. 2000;51:1309–1317. doi: 10.1093/jxb/51.348.1309. [DOI] [PubMed] [Google Scholar]
- Nunes C, De Sousa Araújo S, Marques da Silva J, Salema Fevereiro MP, Bernardes da Silva A. Physiological responses of the legume model Medicago truncatula cv. Jemalong to water deficit. Environmental and Experimental Botany. 2008;63:289–296. [Google Scholar]
- Obendorf RL, Sensenig EM, Wu J, Ohashi M, O'Sullivan TE, Kosina SM, Schnebly SR. Soluble carbohydrates in mature soybean seed after feeding d-chiro-inositol, myo-inositol, or d-pinitol to stem-leaf-pod explants of low-raffinose, low-stachyose lines. Plant Science. 2008;175:650–655. [Google Scholar]
- Ochs G, Shock G, Trischler M, Kosemund K, Wild A. Complexity and expression of the glutamine synthetase multigene family in the amphidiploid crop Brassica napus. Plant Molecular Biology. 1999;39:395–405. doi: 10.1023/a:1006193717093. [DOI] [PubMed] [Google Scholar]
- Ort DR, Baker NR. Photoprotective role for O2 as an alternative electron sink in photosynthesis? Current Opinion in Plant Biology. 2002;5:193–198. doi: 10.1016/s1369-5266(02)00259-5. [DOI] [PubMed] [Google Scholar]
- Owen SF, McCarthy ID, Watt PW. In vivo rates of protein synthesis in Atlantic salmon (Salmo salar L.) smolts determined using a stable isotope flooding dose technique. Fish Physiology and Biochemistry. 1999;20:87–94. [Google Scholar]
- Parry MAJ, Andralojc PJ, Khan S, Lea P, Keys AJ. Rubisco activity: effects of drought stress. Annals of Botany. 2002;89:833–839. doi: 10.1093/aob/mcf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Mitchell RAC, Madgwick PJ, Keys AJ. Manipulation of Rubisco: the amount, activity, function and regulation. Journal of Experimental Botany. 2003;54:1321–1333. doi: 10.1093/jxb/erg141. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Madgwick PJ, Carmo-Silva AE, Androlojc PJ. Rubisco regulation: a role of inhibitors. Journal of Experimental Botany. 2008;59:1569–1580. doi: 10.1093/jxb/ern084. [DOI] [PubMed] [Google Scholar]
- Ramos MLG, Gordon AJ, Minchin FR, Sprent JI, Parsons R. Effect of water stress on nodule physiology and biochemistry of a drought-tolerant cultivar of common bean (Phaseolus vulgaris L.) Annals of Botany. 1999;83:57–63. [Google Scholar]
- Reddy AR, Chaitanya KV, Vivekanandan M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. Journal of Plant Physiology. 2004;161:1189–1202. doi: 10.1016/j.jplph.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Serraj R, Sinclair TR, Allen LH. Soybean nodulation and N2 fixation response to drought under carbon dioxide enrichment. Plant, Cell and Environment. 1998;21:491–500. [Google Scholar]
- Serraj R, Sinclair TR, Purcell LC. Symbiotic N2 fixation response to drought. Journal of Experimental Botany. 1999;50:143–155. [Google Scholar]
- Sharp RE, Boyer JS. Loss in chloroplast activity at low water potentials in sunflower: the significance of photoinhibition. In: Key JL, Kosuge T, editors. Cellular and molecular biology of plant stress, UCLA Symposium on Cellular and Molecular Biology, New Series. Vol. 22. New York: AR Liss Inc; 1985. pp. 41–49. [Google Scholar]
- Sharp RE, Boyer JS. Photosynthesis at low water potentials in sunflower: lack of photoinhibitory effects. Plant Physiology. 1986;82:90–95. doi: 10.1104/pp.82.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn JE, Piques M. Multilevel genomics analysis of carbon signaling during low carbon availability: coordinating the supply and utilization of carbon in a fluctuating environment. Functional Plant Biology. 2007;34:526–549. doi: 10.1071/FP06249. [DOI] [PubMed] [Google Scholar]
- Streeter JG. Effects of drought on nitrogen fixation in soybean root nodules. Plant, Cell and Environment. 2003;26:1199–1204. [Google Scholar]
- Streeter JG, Lohnes DG, Fioritto RJ. Patterns of pinitol accumulation in soybean plants and relationships to drought tolerance. Plant, Cell and Environment. 2001;24:429–438. [Google Scholar]
- Türkan I, Demiral T. Recent developments in understanding salinity tolerance. Environmental and Experimental Botany. 2009;67:2–9. [Google Scholar]
- Verhoeven AS, Adams WW, III, Demmig-Adams B, Croce R, Bassi R. Xanthophyll cycle pigment localization and dynamics during exposure to low temperatures and light stress in Vinca major. Plant Physiology. 1999;120:727–737. doi: 10.1104/pp.120.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD. Some relationships between the biochemistry of photosynthesis and the gas-exchange of leaves. Planta. 1981;153:376–387. doi: 10.1007/BF00384257. [DOI] [PubMed] [Google Scholar]
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresto M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003;24:2369–2375. doi: 10.1002/elps.200305500. [DOI] [PubMed] [Google Scholar]
- Wingler A, Quick WP, Bungard RA, Bailey KJ, Lea PJ, Leegood RC. The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant, Cell and Environment. 1999;22:361–373. [Google Scholar]
- Woo KL, Chang DK. Determination of 22 protein amino acids as N(O)-tert-butyldimethylsilyl derivatives by gas chromatography. Journal of Chromatography. 1993;638:97–107. [Google Scholar]
- Woo KL, Lee DS. Capillary gas chromatographic determination of proteins and biological amino acids as N(O)-tert-butyldimethylsilyl derivatives. Journal of Chromatography B. 1995;665:15–25. doi: 10.1016/0378-4347(94)00515-7. [DOI] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochemistry Journal. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nguyen HT, Blum A. Genetic analysis of osmotic adjustment in crop plants. Journal of Experimental Botany. 1999;50:291–302. [Google Scholar]