Abstract
Homeodomain-leucine zipper type I (HD-Zip I) proteins are plant-specific transcription factors associated with the regulation of growth and development in response to changes in the environment. Nicotiana attenuata NaHD20 was identified as an HD-Zip I-coding gene whose expression was induced by multiple stress-associated stimuli including drought and wounding. To study the role of NaHD20 in the integration of stress responses with changes in growth and development, its expression was silenced by virus-induced gene silencing (VIGS), and control and silenced plants were metabolically and developmentally characterized. Phytohormone profiling showed that NaHD20 plays a positive role in abscisic acid (ABA) accumulation in leaves during water stress and in the expression of some dehydration-responsive genes including ABA biosynthetic genes. Moreover, consistent with the high levels of NaHD20 expression in corollas, the emission of benzylacetone from flowers was reduced in NaHD20-silenced plants. Additionally, bolting time and the opening of the inflorescence buds was decelerated in these plants in a specific developmental stage without affecting the total number of flowers produced. Water stress potentiated these effects; however, after plants recovered from this condition, the opening of the inflorescence buds was accelerated in NaHD20-silenced plants. In summary, NaHD20 plays multiple roles in N. attenuata and among these are the coordination of responses to dehydration and its integration with changes in flower transitions.
Keywords: ABA, benzylacetone, corolla, HD-Zip, Nicotiana
Introduction
The homeodomain-leucine zipper (HD-Zip) family of transcription factors (TFs) is unique to plants as it presents the combination of an HD with a Zip domain (Ariel et al., 2007). The HD is responsible for the specific binding to DNA through its helix III, and the Zip domain acts as a dimerization motif; dimerization is a prerequisite for binding to the target sequence CAAT(A/T)ATTG (Sessa et al., 1997; Palena et al., 1999). The HD-Zip family can be divided into four subfamilies, I –IV, according to the sequence similarity of the HD and Zip domains and to additional structural features outside these domains. The subfamily I (HD-Zip I) is characterized by a gene product of ∼35 kDa that has a highly conserved HD without additional conserved motifs besides the Zip domain (Ariel et al., 2007).
The function of type I HD-Zips has been associated with the regulation of development in response to changes in environmental conditions. Based on present knowledge, the expression of these genes is regulated by external factors such as drought, temperature, and osmotic stresses, and it is specific to different tissues and organs of the plant (Carabelli et al., 1993; Soderman et al., 1996, 1999; Henriksson et al., 2005; Ariel et al., 2007). For example, Arabidopsis ATHB5, 6, 7, and 12 are induced by drought and they act as growth regulators in response to water deficit conditions (Lee and Chun, 1998; Soderman et al., 1999; Hjellström et al., 2003; Johannesson et al., 2003; Olsson et al., 2004). The ectopic expression of ATHB6, 7, and 12 in Arabidopsis plants confers reduced abscisic acid (ABA) sensitivity to germinating seeds and stomata and it mimics the phenotype of wild-type plants grown under water-limiting conditions, with reduced elongation of the inflorescence stem and rosette leaves (Hjellstrom et al., 2003; Olsson et al., 2004). Plants ectopically expressing ATHB5 have a similar phenotype (Johannesson et al., 2003). Analyses of plants ectopically expressing ATHB1, 3, 13, 20, or 23 suggest that these HD-Zip I genes are involved in the regulation of cotyledon and leaf development, and ATHB13 potentially acts as a mediator of sugar signalling (Aoyama et al., 1995; Hanson et al., 2001). The sunflower (Helianthus annuus) HAHB4 is strongly induced by herbivory, wounding, methyl jasmonate (MeJA), ethylene (ET), and drought (Dezar et al., 2005; Manavella et al., 2006, 2008). Arabidopsis plants ectopically expressing HAHB4 present a strong tolerance to water stress and a particular morphological phenotype (Dezar et al., 2005) and, after wounding, they accumulate higher amounts of jasmonic acid (JA), jasmonyl-isoleucine (JA–Ile), and ET compared with wild-type plants (Manavella et al., 2008). In tomato (Solanum lycopersicum), silencing of H52 induces spontaneous cell death in leaves, activation of defence genes, overaccumulation of ET and conjugated salicylic acid (SA), and growth reduction of virulent pathogens (Mayda et al., 1999). The silencing of a second HD-Zip I in tomato, LeHB-1, reduces LeACO1 (ACC oxidase I) expression in fruits and inhibits ripening (Lin et al., 2008). Moreover, its overexpression triggers altered floral organ morphology, suggesting that this TF plays a critical role in floral organogenesis. In the resurrection plant Craterostigma plantagineum, the transcripts of CpHB6 and CpHB7 (type I HD-Zips) accumulate during dehydration whereas the transcripts of CpHB3/4/5 are reduced by dehydration in both leaves and roots (Deng et al., 2002). Transgenic tobacco and Arabidopsis plants ectopically expressing CpHB-7 also display reduced sensitivity towards ABA during seed germination and stomatal closure. Moreover, gene expression analysis performed on these plants demonstrated that some ABA-responsive genes are either induced or repressed, suggesting that this HD-Zip modifies ABA-responsive gene expression (Deng et al., 2006).
Nicotiana attenuata is an annual wild tobacco plant that germinates after fires in the Great Basin Desert in Southwestern USA (Baldwin and Morse, 1994). Because of this growth behaviour and strong intraspecific competition, N. attenuata allocates resources primarily to sustain rapid growth and seed setting. However, in its natural environment, N. attenuata is exposed to severe changing conditions as a result of the unpredictable appearance of herbivores and pathogens as well as unfavourable changes in temperature and water availability which can negatively affect its growth and reproduction. Therefore, it has to readjust its metabolism, growth, and developmental programme rapidly to cope with these unpredictable changes in the environment. For example, N. attenuata preferentially produces flowers that open at night and release benzyl acetone (4-phenyl-2-butanone; BA) to attract night-active pollinators such as moths from Manduca spp. (Kessler et al., 2008); however, when attacked by larvae of the same species, the plant increases the production of morning flowers, which do not emit BA at night and are visited preferentially by humming-birds in the morning (Kessler et al., 2010). Additional examples of the phenotypical plasticity of different plant species include (among several others): stem elongation in response to vegetation shade, root growth in response to soil moisture, leaf area and thickness in response to photon flux density, and changes in flowering time in response to unfavourable growing conditions (Callahan et al., 1997; Bell and Sultan, 1999; Sultan, 2000; Nicotra et al., 2008). Despite the wealth of studies characterizing the phenotypic plasticity of plants in response to changes in the environmental conditions, the molecular mechanisms underlying this process remain in many cases largely unknown.
Materials and methods
Plant growth, treatments, and analysis of response variables
Seeds of the 30th generation of an inbred line of N. attenuata plants were used as the wild-type genotype in all experiments. Wild-type and transformed lines of N. attenuata were germinated on Gamborg's B5 medium (Duchefa, Germany) as previously described (Krügel et al., 2002). For virus-induced gene silencing (VIGS) experiments, plants were grown in 1.0 l pots filled up with soil (Nullerde, Balster Einheitserdewerk, Fröndenberg, Germany) in chambers under 19 °C, 16 h light (1000 μmol m−2 s−1), 65% humidity, and watered daily every morning (control growth conditions).
Leaf wounding was performed by rolling a fabric pattern wheel three times on each side of the midvein. For 18:3-Glu elicitation, the wounds were supplemented with 10 μl of synthetic N-linolenoyl-glutamic acid [18:3-Glu; 0.03 nmol μl−1 in 0.02% (v/v) Tween-20/water]. Oral secretion (OS) elicitation was performed similarly but the wounds were supplemented with OS from Manduca sexta larvae (third to fifth instar) reared on N. attenuata plants. JA, SA, and ABA [solvent: 0.02% (v/v) Tween-20/water] were sprayed on leaves at 100, 300, and 300 μM, respectively. Ethephon was applied as previously described (Bonaventure et al., 2007).
To determine the response variables of reproductive phase change and flower transitions, 15 empty vector (EV) and NaHD20-silenced plants were used for each treatment and they were randomly distributed in the chamber. Data on bolting time, stalk length, and buds and flower numbers were obtained every 2 d from the beginning of the treatments. Rosette stage plants were elicited by wounding and OS elicitation, every day for a period of 11 d, starting 3 d before the initiation of bolting. Pseudomonas syringae pv. tomato DC3000 infection was performed by spraying rosette stage plants (3 d before the initiation of bolting) with a bacterial solution as previously described (Katagiri et al., 2002). The same treatment was repeated 4 d later. For the water stress experiment, the soil of rosette stage plants (3 d before bolting) was water saturated, the excess water drained, and the plants were left without additional water for 18 d (and re-watered afterwards). For root analysis, rosette stage hydroponically grown wild-type plants were removed from the growing pots and the roots were exposed to ambient air for different times.
Cloning of NaHD20
Specific primers (Supplementary Table S1 available at JXB online) were designed based on the Nicotianan tabacum HD20, HD48, HD47, HD49, and HDkl sequences (see Accession numbers) (Rushton et al., 2008) to PCR-amplify their respective homologues in N. attenuata. PCR products were obtained using N. attenuata leaf cDNA as template, cloned into pGEM®-T Easy vector (Promega, Madison, WI, USA), and sequenced (Supplementary Table S2 and Accession numbers). The 3′ and 5′ HD20 cDNA segments were obtained using, respectively, the RACE system for rapid amplification of cDNA ends (Invitrogen, Karlsruhe, Germany) and the SMARTer RACE cDNA amplification kit (Clontech, Mountain View, CA, USA) following the manufacturer's instructions and using gene-specific primers (Supplementary Table S1).
Virus-induced gene silencing
VIGS based on the tobacco rattle virus (TRV) was used to silence the NaHD20 gene transiently in N. attenuata. A cDNA fragment of 287 bp was amplified by PCR with the specific primers listed in Supplementary Table S1 at JXB online. PCR products were digested with BamHI and SalI, and inserted into the plasmid pTV00 in the antisense orientation (Saedler and Baldwin, 2004). Agrobacterium tumefaciens-mediated transformation was performed as previously described (Saedler and Baldwin, 2004). Plants transformed with the EV were used as control. The efficiency of gene silencing was evaluated by quantification of NaHD20 mRNA levels by qPCR (see below) using the primers listed in Supplementary Table S1.
Phytohormone extraction and quantification
For phytohormone analysis, 0.1 g of frozen leaf material was extracted with 1 ml of ethyl acetate spiked with 0.1 μg of [9,10-2H2]dihydro-JA, [13C6]JA–Ile, [2H4]SA, or [2H6]ABA. Homogenates were centrifuged for 10 min at 4 °C, the upper organic phase was collected, and the plant material was re-extracted with 0.5 ml of ethyl acetate. The organic phases were combined and the samples were evaporated to dryness. The residue was reconstituted in 0.2 ml of 70% (v/v) methanol/water for analysis on a Varian 1200 Triple-Quadrupole-LC-MS system (Varian) as previously described (Kallenbach et al., 2010).
Ethylene measurements
ET levels were analysed by photoacoustic spectrometry (INVIVO; https://www.invivo-gmbh.de) as previously described (Korner et al., 2009). Leaves of rosette stage plants were either wounded or OS elicited and immediately placed in 100 ml sealed glass vessels for 5 h before ET quantification (one leaf per vessel). For the analysis of ET emitted by flowers, flowers were emasculated before anther dehiscence and either hand-pollinated or not (control) and immediately placed in 100 ml sealed glass vessels (five flowers per vessel) for 5 h before ET quantification. Three biological replicates for EV and NaHD20-silenced plants and treatment were performed.
Quantification of benzylacetone emission from flowers
Flower volatiles were collected by enclosing individual night flowers in plastic cups connected to Super-Q traps (ARS, Philadelphia, PA) under an air flow of 30 ml min−1. Volatiles were collected from 14:00 h to 9:00 h. Traps were spiked with 400 ng of tetraline as an internal standard and eluted with 250 μl of dichloromethane. Volatile analysis was performed by gas chromatography–mass spectrometry (GC-MS) with a CP-3800 GC instrument (Varian, Palo Alto, CA, USA) on a DB-Wax column (Agilent, Waldbronn, Germany). BA was identified by comparing retention times and mass spectra with a commercial standard. Peak areas were integrated and normalized to the area of the internal standard.
Secondary metabolite extraction and quantification
Control and 18:3-Glu-elicited leaves of rosette stage plants were harvested after 0, 36, and 72 h. Approximately 0.2 g of leaf material was extracted with 1 ml of extraction buffer [40% (v/v) methanol/water; 0.1% (v/v) acetic acid/water] and samples centrifuged (16 100 g) for 20 min at 4 °C. The supernatant was transferred into a new tube and centrifuged again to remove insoluble particles. Samples were analysed on an HPLC-1100 series instrument (Agilent) using a Chromolith FastGradient RP18e column (50×2 mm; Merck, Darmstadt, Germany) attached to a Gemini NX RP18 pre-column (2×4.6 mm). The solvents were: A [0.1% (v/v) formic acid/water; 0.1% (v/v) ammonium hydroxide/water pH 3.5] and B (methanol). The solvent gradient was 0% solvent B for 0.5 min, to 80% solvent B in 6.5 min and isocratic for 3 min. The flow rate was 0.8 ml min−1, the injection volume was 1 μl, and the column oven was set at 50 °C. The recorded detector channels were set at 260 nm (nicotine), 210 nm [diterpene glucosides (DTGs)], and 320 nm (caffeoylputrescine, caffeoylspermidine, and chlorogenic acid).
RNA isolation, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from different tissues by the TRIzol® reagent (Invitrogen) and DNase I treated (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions. A 5 μg aliquot of total RNA was reverse transcribed using oligo(dT)18 and SuperScript reverse transcriptase II (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR (qPCR) was performed with a Mx3005P Multiplex qPCR system (Stratagene, La Jolla, CA, USA) and the qPCR Core kit for SYBR® Green I (Eurogentec, Liege, Belgium) using gene-specific primers (Supplementary Table S1 at JXB online). Quantification of NaHD20 mRNA levels was performed either by normalization with the eukaryotic elongation factor 1A (NaeEF1A-α) mRNA according to the ΔCt method or per μg of total cDNA after digestion with RNase A and RNase H and quantification by UV absorbance after purification. The two methods gave similar kinetics and relative amounts between tissues of NaHD20 transcripts. The NaeEF1A-α mRNA was selected as a standard for normalization because its levels showed minimal variations after the treatments tested and in the different tissues.
Sequence analysis and statistics
ClustalW was used for multiple and pairwise alignments using the Mega4 software (Tamura et al., 2007). Statistical analysis was performed with SPSS 17.0 (SPSS Inc., Chicago, IL, USA) or with Excel (Microsoft, Washington, USA).
GenBank accession numbers
NaHD20 (HM107874); NaNCED1 (HM068892); NaOSM1 (HM068893); NaLTP1 (HM068895); NtHD20 (ET048311); NtHD49 (ET044131); NtHD48 (ET042012); NtHD47 (ET051066); NtHDkl (EB680839).
Results
Identification of NaHD20 as a multiple-stress-inducible HD-Zip gene in N. attenuata leaves
With the purpose of identifying putative HD-Zip TFs involved in the regulation of changes in growth and development induced by biotic and abiotic stimuli in N. attenuata, the publicly available sequences corresponding to the N. tabacum HD TFs (Rushton et al., 2008) were used as queries to identify HD-Zips in N. attenuata. Specific primers were designed based on the sequences from N. tabacum HD-Zips I and II, and five cDNAs encoding putative HD-Zips (two type I and three type II) were retrieved from leaves of N. attenuata plants (Supplementary Tables S1, S2 at JXB online). The mRNA levels corresponding to one of these HD-Zips (type I) were found to be inducible by multiple biotic- and abiotic-related stresses (see below). In contrast, the transcript levels for the remaining four HD-Zips were not affected by the stimuli tested (data not shown and see below). The multiple-stress-inducible HD-Zip had >95% amino acid sequence identity to the putative N. tabacum NtHD20 (Rushton et al., 2008) (Supplementary Fig. S1 at JXB online) and, accordingly, it was named NaHD20 and it was further characterized in this study.
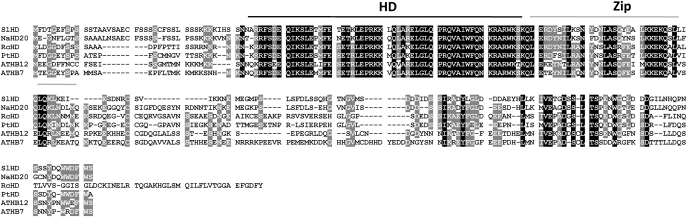
Analysis of sequence similarity by basic local alignment (BLAST) using the complete predicted amino acid sequence of NaHD20 as a query against non-redundant GenBank databases showed that, in addition to NtHD20, the closest homologues of NaHD20 were three uncharacterized HD-Zips from tomato, castorbean (Ricinus communis), and black cottonwood (Populus trichocarpa) and two characterized Arabidopsis HD-Zips, ATHB12 and ATHB7. The amino acid sequence alignment of these HD-Zips showed that the highest degree of homology between them was restricted to the HD and Zip domains; however, sequence conservation was also present outside these two domains (Fig. 1).
Fig. 1.
Alignment of NaHD20 amino acid sequences with HD-Zip I proteins from different plant species. Multiple alignment of the NaHD20 amino acid sequence with its closest homologues from other plant species. ATHB7, A. thaliana homeobox 7 (At2g46680); ATHB12, A. thaliana homeobox 12 (At3g61890); SlHD, S. lycopersicum homeodomain (AK323400); RcHD, Ricinus communis homeodomain (XM_002529914); PtHD, Populus trichocarpa homeodomain (XM_002302623); HD, homeodomain; Zip, leucine-zipper domain.
NaHD20 transcript levels are induced by different stress-associated treatments and are abundant in corollas
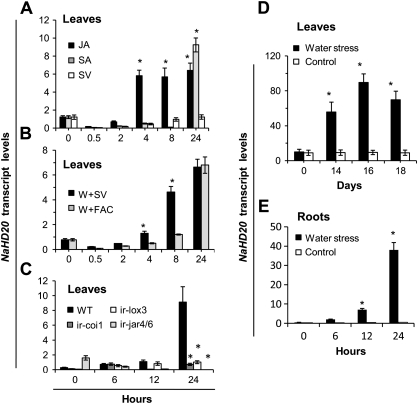
NaHD20 mRNA levels were induced by wounding, exogenous JA, SA, ABA, and ET, and by dehydration. The accumulation of NaHD20 transcript levels in leaves after exogenous JA application was slow, starting to increase only after 2 h of the treatment to reach a maximum at 4 h (6- to 7-fold compared with basal levels) and remained constant for up to 24 h (Fig. 2A). Application of exogenous SA also stimulated a slow accumulation of NaHD20 mRNA (6- to 7-fold compared with basal levels; Fig. 2A). After wounding, the induction of NaHD20 mRNA levels was slower compared with JA application and it was further decelerated by supplementing the wounds with the insect-derived elicitor 18:3-Glu; however, the levels of induction were similar between treatments at 24 h (Fig. 2B). Ethephon (an ET-releasing agent) stimulated a more rapid (within 1 h) but lower (2.5-fold) and transient accumulation (Supplementary Fig. S2 at JXB online).
Fig. 2.
Analysis of NaHD20 transcript levels in N. attenuata leaves and roots after different stress treatments. Total RNA was extracted from leaves or roots of rosette stage wild-type (WT) or transgenic N. attenuata plants after different times and treatments, reverse transcribed, and NaHD20 mRNA levels were quantified by qPCR. (A) Leaf tissue from WT plants treated with exogenous JA (100 μM), SA (300 μM), or solvent (SV). (B) Leaf tissue from wounded WT plants with the addition of either solvent (W+SV) or 18:3-Glu (W+FAC). (C) Leaf tissue from wounded WT, ir-lox3, ir-coi1, and ir-jar4/6 silenced plants. (D) Leaf tissue from WT plants subjected to water stress (by withholding soil irrigation) or control conditions for several days. (E) Roots of WT plants subjected to water stress (by air drying) or control conditions for up to 24 h. *Student t-test, P <0.05 [control versus treatment (A, D, E); wounding versus FAC elicitation (B); WT versus RNA interference-silenced plants (C)]; (n=3, bars denote ±SE).
To study the dependence of NaHD20 on the endogenous biogenesis and signalling of JA, N. attenuata plants silenced in the expression of LOX3 (ir-lox3), JAR4/6 (ir-jar4/6), and COI1 (ir-coi1) were analysed. The ir-lox3 and ir-jar4/6 plants are deficient in the synthesis of JA and JA–Ile, respectively (Halitschke and Baldwin, 2003; Wang et al., 2007), and ir-coi1 plants in jasmonate perception (Paschold et al., 2008). Consistent with the induction by wounding and exogenous JA application, the induction of NaHD20 transcripts was abolished in ir-lox3, ir-jar4/6, and ir-coi1 (Fig. 2C), indicating that expression of the NaHD20 gene depends on the endogenous production of these two jasmonates and signalling through COI1.
When wild-type plants were grown under water stress conditions by withholding irrigation for >2 weeks, NaHD20 mRNA levels increased in leaves (>10-fold induction after 16 d from the beginning of the treatment compared with basal levels; Fig. 2D) and treatment of leaves with exogenous ABA also induced the expression levels of this transcript (Supplementary Fig. S2 at JXB online). Finally, roots exposed to the air for several hours also induced a fast and strong accumulation of NaHD20 mRNA (150-fold compared with root basal levels; Fig. 2E).
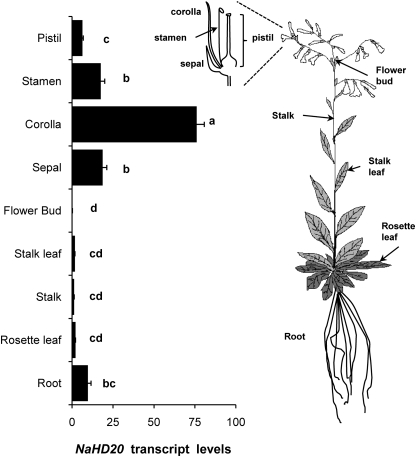
The expression of NaHD20 transcripts was detected in all the different tissues of wild-type plants tested, with high levels of accumulation in floral parts compared with vegetative tissue and with the highest levels in the corolla, sepals, and stamens (Fig. 3).
Fig. 3.
Tissue-specific expression of NaHD20 mRNA. Total RNA was extracted from different tissues of rosette stage and mature N. attenuata wild-type (WT) plants, reverse transcribed, and used for quantification of NaHD20 mRNA levels by qPCR (n=3, bars denote ±SE). Different letters indicate significant differences (univariate ANOVA, F8,18=228.8, P <0.001, followed by a Scheffé post-hoc test, P <0.05).
NaHD20 affects ABA accumulation and expression of drought-responsive transcripts during water stress
To investigate the role of NaHD20 in N. attenuata plants, its expression was silenced by VIGS. Plants transformed with the EV were used as controls. Gene silencing efficiency was analysed by qPCR and the NaHD20 mRNA levels were reduced by 86±7% in NaHD20-silenced plants compared with EV plants (Supplementary Fig. S4 at JXB online). The rosette size and rosette morphology of NaHD20-silenced plants were indistinguishable from those of EV plants.
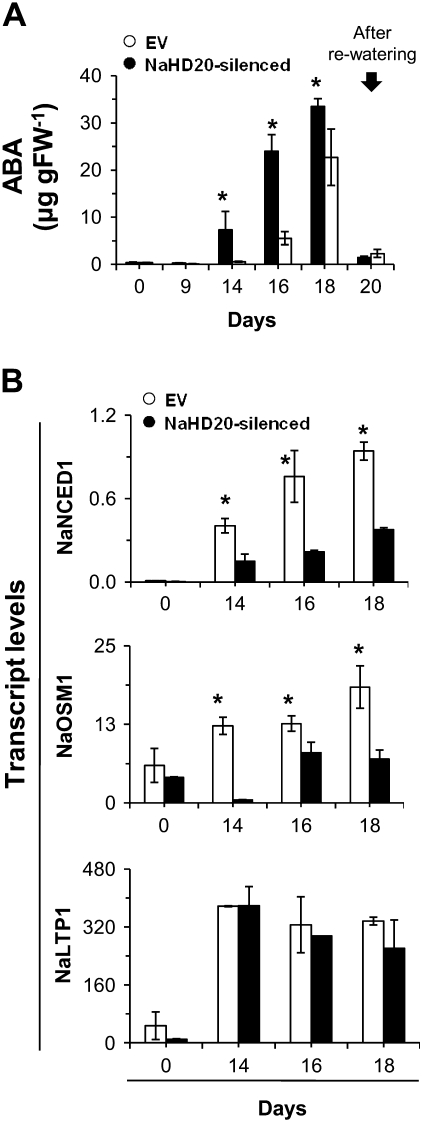
Based on its similarity to Arabidopsis ATHB7 and 12, a potential role for NaHD20 in water stress responses was first investigated. For this experiment, the soil of rosette stage EV and NaHD20-silenced plants was allowed to take up water to its field capacity and plants were left without additional water for 18 d. Under these conditions, rosette leaf wilting became evident after 13 d in both EV and NaHD20-silenced plants and, when plants were re-watered (day 18 after the beginning of the treatment), the rosette leaves from both plant types regained turgidity within 24 h. During the experiment, no obvious differences in leaf number and morphology (e.g. leaf area) between plant types were detected (data not shown). The levels of ABA in leaves of EV plants reached 33 μg gFW−1 (per gram of fresh weight) at day 18 and after re-watering they declined to 1.5 μg gFW−1 (Fig. 4A). In NaHD20-silenced plants, ABA accumulated more slowly than in EV plants and reached 22 μg gFW−1 at day 18 (65% of EV levels), and it declined to 2.2 μg gFW−1 after re-watering (Fig. 4A; see legend for statistical analysis). Expression of ABA levels per gDW (per gram of dry weight) gave similar results (Supplementary Fig. S5 at JXB online).
Fig. 4.
Analysis of ABA accumulation and gene expression during water stress conditions. (A) Rosette stage EV and NaHD20-silenced plants were water stressed for several days and leaves (one leaf per plant for each time point) were harvested at different times, extracted, and ABA levels quantified by LC-MS/MS. The arrow marks the day of re-watering (day 18 after the beginning of the experiment). *One-way ANOVA with Fisher's PLDS, P <0.05 (n=5). (B) Analysis of NaNCED1, NaOSM11, and NaLTP1 expression levels in EV and NaHD20-silenced plants during conditions of water deficit. Total RNA was extracted from leaves at different times, reverse transcribed, and used for quantification of NaNCED1, NaOSM1, and NaLTP1 mRNA levels by qPCR. *Student t-test, P <0.05 (EV versus NaHD20-silenced plants) (n=3, bars denote ±SE).
To investigate if the silencing of NaHD20 affected the expression of drought-responsive genes, the transcript levels of NaNCED1 (9-cis-epoxycarotenoid dioxygenase 1), NaLTP1 (lipid transfer protein 1), and NaOSM1 (osmotin 1) were evaluated by qPCR during water stress in EV and NaHD20-silenced plants. The transcript levels for NaNCED1 constantly increased in water-stressed EV plants, to reach a >200-fold increase compared with basal levels after 18 d (Fig. 4B). The levels of this transcript were also induced in NaHD20-silenced plants, however to 2- to 3-fold lower levels than in EV plants. Similarly, the transcript levels for NaOSM1 were induced 2- to 4-fold by water deficiency in leaves of EV plants whereas they were not induced or even repressed (day 14) in NaHD20-silenced plants (Fig. 4B). In contrast, even though NaLTP1 transcript levels were strongly up-regulated by water stress, the induction was not affected by the silencing of NaHD20 (Fig. 4B). Treatment of leaves with exogenous ABA induced the accumulation of these three transcripts (Supplementary Fig. S3 at JXB online).
Ectopic expression of the sunflower HAHB4 in Arabidopsis induces the enhanced accumulation of JA and ET after wounding and reduces the accumulation of SA after infection with a virulent strain of P. syringae (Manavella et al., 2008). Therefore, the levels of JA, JA–Ile, and ET were quantified in EV and NaHD20-silenced plants after wounding, and 18:3-Glu elicitation and the levels of SA were quantified after 24 h and 48 h of P. syringae infection. All these phytohormones accumulated to similar levels in EV and NaHD20-silenced plants after the treatments (data not shown). Consistently, the accumulation of several defence-associated secondary metabolites which depend on JA and JA–Ile (nicotine and DTGs) and phenolics (caffeoylputrescine, caffeoylspermidine, and chlorogenic acid) was also similar between EV and NaHD20-silenced plants (data not shown).
NaHD20 is a positive regulator of the emission of benzylacetone from N. attenuata flowers but not of ethylene production after pollination
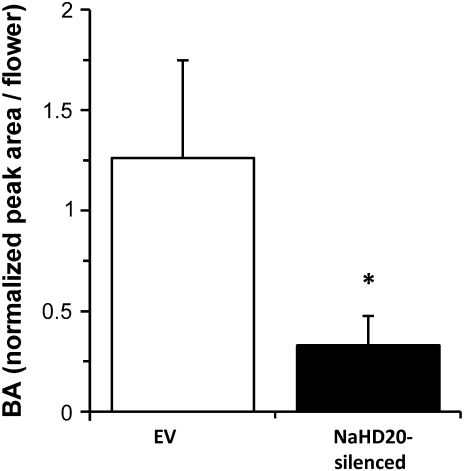
Based on the high levels of expression of NaHD20 mRNA in the corolla and other floral parts, experiments were conducted to assess whether NaHD20 was associated with processes regulating the production of either corolla-derived volatiles or ET after pollination. The night flowers of N. attenuata emit volatile compounds at night, and BA is one of the major constituents of the blend (Euler and Baldwin, 1996). Corolla-emitted BA from night flowers of EV and NaHD20-silenced plants was trapped during the night period and analysed by GC-MS. Emission of BA was reduced by 75% in NaHD20-silenced plants compared with EV plants (Fig. 5; see legend for statistical analysis).
Fig. 5.
Analysis of benzylacetone (BA) emitted by night flowers. Volatiles emitted by night flowers of EV and NaHD20-silenced plants were trapped from 14:00 h to 9:00 h and analysed by GC-MS. BA levels correspond to 10 biological replicates (n=10) for EV and NaHD20-silenced plants and they are expressed as the normalized peak area to the internal standard. *Mann–Whitney U-test; P <0.05; NaHD20-silenced plants versus EV plants.
Similar to many plant species, the primary pollination event in N. attenuata flowers is accompanied by an increase in ET production within the first hours after pollination. Emission of ET in flowers of EV and NaHD20 plants was evaluated after controlled pollination by emasculation of flowers before anther dehiscence and hand pollination of the stigma. Control flowers were emasculated but not pollinated. Flowers were placed immediately into glass vessels for quantification of ET. The results showed that total ET emission was not affected by the silencing of NaHD20 (data not shown).
NaHD20 is a positive regulator of the transition from the vegetative to the reproductive stage and of flower differentiation in control growth conditions
Based on the role of several HD-Zip I genes in processes controlling the growth of the inflorescence stem and flower organogenesis in plants and on the high level of NaHD20 expression in floral parts in N. attenuata, the participation of this TF in processes controlling the reproductive phase change and the regulation of flower differentiation was evaluated under different growth conditions.
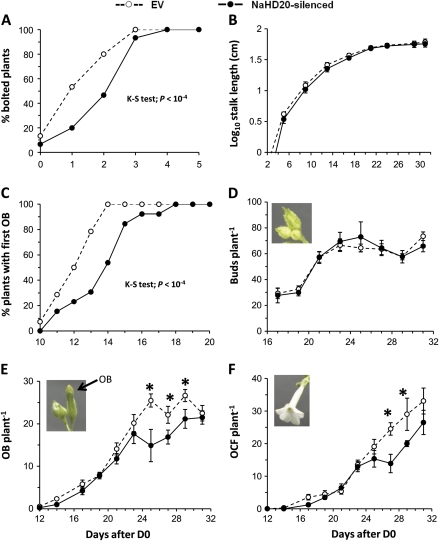
Under control growth conditions, NaHD20-silenced plants presented a delay in bolting (defined as the time when the flowering stalk emerged) compared with EV plants (Fig. 6A, see legend for statistical analysis). For the subsequent experiments, day 0 (D0) was defined as the time when the first EV plant bolted (Fig. 6A). Thus, by days 1 and 2, 50% and 75% of the EV plants have bolted; however, only 20% and 45% of NaHD20-silenced plants, respectively, had bolted (Fig. 6A, see legend for statistical analysis). By day 4, 100% of both EV and NaHD20-silenced plants had bolted. The rate of stalk elongation and the final stalk length were similar between the plant types (Fig. 6B).
Fig. 6.
Analysis of reproductive phase change and flower transitions in NaHD20-silenced plants grown under control conditions. EV and NaHD20-silenced plants were grown under control conditions in ambient growth chambers. For all experiments, day 0 (D0) was defined as the time in which the first EV plants bolted and time therefore refers to days after D0. (A) Bolting time expressed as the percentage of bolted plants relative to D0 [two-sample Kolmogorov–Smirnov (K–S) test; P <10−4]. (B) Rate of stalk elongation. (C) Percentage of plants that had at least one opened bud (OB; two-sample K–S test; P <10−4). (D) Number of closed buds per plant (buds per plant). (E) Number of opened buds (OBs) per plant. (F) Number of opened corolla flowers (OCFs) per plant. *Mann–Whitney U-test; P <0.05; NaHD20-silenced plants versus EV plants. Each data point corresponds to data collected for 15 plants per plant type. (This figure is available in colour at JXB online.)
As a response variable of the differentiation of flowers: (i) the number of buds produced per plant; (ii) the time when the first bud opened; (iii) the number of opened buds (OBs) per plant; and (iv) the number of opened corolla flowers (OCFs) per plant were analysed in EV and NaHD20-silenced plants. Consistent with the differences in bolting time between EV and NaHD20-silenced plants (Fig. 6A), the production of the first OB was delayed in NaHD20-silenced plants (Fig. 6C, see legend for statistical analysis). All EV plants had a first OB by day 14, whereas this was the case for only 50% of NaHD20-silenced plants. By day 18, all NaHD20-silenced plants have at least one OB (Fig. 6C). The number of buds per plant produced by EV and NaHD20-silenced plants was similar (Fig. 6C). Likewise, the number of OBs per plant was similar between the plant types during days 12–23; however, the number was significantly reduced for NaHD20-silenced plants after day 23, reaching similar values at day 31 (Fig. 6E, Mann–Whitney U-test, P <0.05 for days 25, 27, and 29). For example, at day 25, EV plants had on average 25 OBs per plant, whereas NaHD20-silenced plants had 15 (40% reduction; Fig. 6E). Consistently, the number of OCFs per plant was similar between plant types between days 12 and 25, whereas the number was reduced in NaHD20-silenced plants after this period (Fig. 6F, Mann–Whitney U-test, P <0.05 for days 27 and 29). For example, at day 27, EV plants had on average 24 OCFs whereas NaHD20-silenced plants had 14 (40% reduction; Fig. 6F). The macroscopic morphology of the flowers and flower parts was similar between EV and NaHD20-silenced plants (data not shown).
NaHD20 is a negative regulator of the production of OB and OCF after recovery from water deficiency
The role of NaHD20 in the transition to the reproductive stage and flower differentiation was also studied during three stress conditions: drought, wounding plus M. sexta OS elicitation, and infection with a virulent strain of P. syringae.
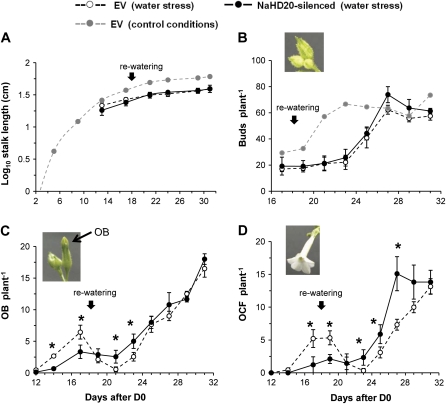
For the water stress experiment, EV and NaHD20-silenced plants at the early rosette stage (3 d before bolting) were used and the treatment was identical to that described above for ABA quantification. The results are presented in Fig. 7 and the data corresponding to stalk length and number of buds for EV plants under control growth conditions (Fig. 6B, C) are also included in Fig. 7 to help with the comparisons. The bolting time and the time when the first OB appeared in both EV and NaHD20-silenced plants was similar to that of plants grown under control conditions, since they occurred before the plants were under water deficit (the soil was still wet). Under water limitation, the rates of stalk elongation from EV and NaHD20-silenced plants were slowed down by day 13, when the plants started to show evident macroscopic signs of dehydration (leaf wilting), and they were similar in both plant types (Fig. 7A). Likewise, the number of buds per plant produced by EV and NaHD20-silenced plants was reduced during water stress compared with the control treatment; however, the reductions were again similar between the types of plants (Fig. 7B). After re-watering, the number of buds per plant increased in both EV and NaHD20-silenced plants to reach numbers similar to those of plants grown under control conditions after 2 weeks (Fig. 7B). The number of OBs per plant was significantly reduced in NaHD20-silenced plants during water deficiency (Fig. 7C; Mann–Whitney U-test, P <0.05 for days 12–17) consistent with the data presented in Fig. 6E; however, after re-watering, the number significantly increased (Fig. 7C; Mann–Whitney U-test, P <0.05 for days 21–23) in NaHD20-silenced plants compared with EV plants, to reach similar levels thereafter (Fig. 7C, days 25–31). Consistent with these differences, the number of OCFs per plant was also reduced in NaHD20-silenced plants compared with EV before re-watering (Fig. 7D, Mann–Whitney U-test, P <0.05 for days 12–19); however, it was increased to levels significantly higher than in EV plants after re-watering (Fig. 7D; Mann–Whitney U-test, P <0.05 for days 23–27). For example, at day 27, EV plants had on average seven OCFs whereas NaHD20-silenced plants had 15 OCFs (Fig. 7D). At day 31, the number of OCFs per plant was similar between plant types (Fig. 7D).
Fig. 7.
Analysis of reproductive phase change and flower transitions in NaHD20-silenced plants during water stress and after recovery. Rosette stage EV and NaHD20-silenced plants were subjected to conditions of water deficit for 18 d and afterwards re-watered (black arrows: day 18). Day 0 (D0) was defined as the time in which the first EV plants bolted and time therefore refers to days after D0. The data corresponding to stalk length and number of buds for EV plants under control growth conditions (Fig. 6) are also presented (grey dotted lines) to help with the comparisons between treatments. (A) Rate of stalk elongation. (B) Number of closed buds per plant. (C) Number of opened buds (OBs) per plant. (D) Number of opened corolla flowers (OCFs) per plant. *Mann–Whitney U-test; P <0.05; NaHD20-silenced plants versus EV plants. Each data point corresponds to data collected for 15 plants per plant type. (This figure is available in colour at JXB online.)
The results obtained after analysis of EV plants elicited by wounding and OS and infected by a virulent strain of P. syringae (see Materials and methods for the experimental design) showed a negative effect of the treatments on bolting time, opening of the first bud, and number of OBs and OCFs per plant in EV plants (Supplementary Figs S6, S7 at JXB online). These treatment-associated negative effects were also observed in NaHD20-silenced plants; however, they were proportional to the levels observed in EV plants under the same conditions, indicating that they were independent of NaHD20 expression (data not shown).
Discussion
NaHD20 plays a positive role in leaf ABA accumulation during water stress
The levels of NaHD20 mRNA were induced several fold in leaves and roots of wild-type plants under conditions of water stress and exogenous ABA treatment. This up-regulation was consistent with the drought-dependent regulation of other members of the HD-Zip I family in other plant species, including Arabidopsis, the resurrection plant C. plantagineum, and sunflower (Soderman et al., 1996, 1999; Olsson et al., 2004; Dezar et al., 2005; Deng et al., 2006). Consistent with a potential role for NaHD20 in the regulation of drought-associated responses, the rate of ABA accumulation in leaves of water-stressed NaHD20-silenced plants was decelerated and reduced compared with control EV plants, and ABA levels reached on average 65% of those in EV plants at day 18 of the treatment (Fig. 4A and Supplementary Fig. S5 at JXB online). Although speculative at this point, NaHD20 could be involved in a positive feedback loop controlling ABA biosynthesis in leaves.
In plants, increased ABA levels under water-deficit conditions result mainly from increased de novo biosynthesis through the transcriptional activation of ABA biosynthetic genes; the inhibition of transcription or translation impairs stress-induced ABA biosynthesis (Milborrow, 2001; Wasilewska et al., 2008). The transcription of 9-cis-epoxycarotenoid dioxygenase (NCED) genes (catalysing the synthesis of xanthonin from 9-cis-epoxycarotenoids) is rapidly induced in leaves by drought stress in most plant species (Tan et al., 1997; Burbidge et al., 1999; Qin and Zeevaart, 1999; Chernys and Zeevaart, 2000) and the step catalysed by this enzyme is considered the rate-limiting step for ABA biosynthesis in this tissue. Consistent with these previous results, the levels of NaNCED1 mRNA were strongly up-regulated by water stress in N. attenuata leaves (Fig. 4B). Moreover, consistent with the reduced leaf ABA levels in NaHD20-silenced plants, the induction of this transcript was reduced by 2- to 3-fold in these plants (Fig. 4B). A reduced induction of the ABA- and dehydration-responsive NaOSM1 gene was also observed in NaHD20-silenced plants compared with EV plants, whereas the induction of the NaLTP1 gene was similar between these plant types (Fig. 4B). Whether NaHD20 can directly bind to the promoter region of NaOSM1 and NaNCED1 or the effect on their expression is indirect (based on lower ABA levels or the regulation of secondary transcription factors) is at present unknown and it remains the focus of future investigations. Moreover, the reduction in ABA and NaNCED1 mRNA accumulation due to NaHD20 silencing could be due to developmental, physiological, or biochemical changes (e.g. reduced leaf area, stomatal closure, or osmotic adjustment) that could delay loss of turgor after withdrawal of watering. More detailed experiments are needed to demonstrate the actual NaHD20-dependent mechanisms underlying the regulation of ABA levels.
NaHD20 does not affect the biosynthesis of JA, SA, and ET in N. attenuata plants
Similar to the sunflower HAHB4, the expression of NaHD20 was induced by wounding, insect elicitation, and exogenous applications of JA. The ectopic expression of HAHB4 in Arabidopsis stimulates an increased accumulation of JA and ET after wounding but a reduced accumulation of SA after pathogen infection compared with control plants (Manavella et al., 2008). In NaHD20-silenced plants, the accumulation of JA, JA–Ile, and ET was similar to that in control EV plants after wounding, and 18:3-Glu elicitation and the accumulation of SA were also similar between these plant types after P. syringae infection, suggesting that NaHD20 does not affect the production of these phytohormones in leaves of N. attenuata.
The analysis of plants deficient in JA and JA–Ile biosynthesis and perception showed that both the endogenous production and perception of these jasmonates are required for the induction of NaHD20 mRNA levels. Since the kinetics of NaHD20 mRNA induction by wounding and exogenous JA application were slow, this HD-Zip may have an effect in late responses to these stimuli. The accumulation of secondary metabolites known to be at least partially dependent on JA biosynthesis and perception was not altered in NaHD20-silenced plants compared with EV plants. Thus, the role of NaHD20 in responses associated with wounding remains unknown.
NaHD20 is a positive regulator of BA emission from night flowers
Expression of NaHD20 transcripts was detected in all tissues analysed, with the highest level of expression in corollas (Fig. 3). Consistently, previous studies have shown that HD-Zip I genes are expressed in most plant tissues including flowers (Lee and Chun, 1998; Henriksson et al., 2005; Deng et al., 2006). The emission of BA from corollas of night flowers was reduced by 75% in NaHD20-silenced plants, indicating that NaHD20 positively regulates the emission or production of this volatile molecule. What mechanisms underlie this effect is at present unknown. BA is emitted from the outer corolla lip, and possible roles for NaHD20 in the control of BA emission involve the control of the expression of BA biosynthetic genes in flowers or of the mechanisms that regulate the release of this volatile compound.
NaHD20 has a dual role as a positive and negative regulator of inflorescence bud opening depending on the external growth conditions
The transition from the vegetative to the reproductive stage is controlled by multiple environmental cues, such as the photoperiod, temperature, and water availability. Drought often delays developmental events because of the inhibition of growth by water deficit (Trewavas and Jones, 1991); drought inhibits or reduces the new assimilation and translocation of carbohydrates, the translocation of nutrients, and protein synthesis, whereas it increases proteolysis (Fox, 1990).
Arabidopsis ATHB7 and ATHB12 function as regulators of cell elongation, expansion, and differentiation during water stress responses. The ectopic expression of these transcription factors delays bolting, reduces the rates of elongation of the primary inflorescence stem, and induces alterations in leaf shape; however, flowering time is unaffected (Hjellström et al., 2003; Olsson et al., 2004). The silencing of NaHD20 in N. attenuata also affected developmental processes both in control and in a context of water-deficit conditions. The silencing of NaHD20 induced a delay in bolting time but it did not affect the rate of stalk elongation and the final stem length under control growth conditions. Water stress decelerated the elongation of the stem; however, the elongation rates and length remained similar between EV and NaHD20-silenced plants (Fig. 7A). Likewise, the similar number of inflorescence buds produced per plant between these two plant types indicated that NaHD20 did not affect the development of the floral meristems. Moreover, the similar reduction in the number of buds between EV and NaHD20-silenced plants during water deficit and their similar increase after recovery supported this conclusion (Fig. 7B). The opening of the inflorescence buds was delayed in NaHD20-silenced plants by a specific time during development, when plants reached the maximum number of OBs per plant (Fig. 6E). This delay was reflected in the number of OCFs produced per plant (Fig. 6F). The delay in the formation of OBs in NaHD20-silenced plants also occurred during water deficit conditions, and, strikingly, ∼5 d after recovery from this treatment, a differential acceleration in the production of OBs and OCFs occurred in NaHD20-silenced plants relative to EV plants (Fig. 7C, D). This acceleration resulted in double numbers of OBs and OCFs in NaHD20-silenced plants compared with EV plants between days 23 and 29 (Fig. 7C, D). The silencing of NaHD20 did not however affect the final number of flowers produced (Figs. 6E, F, 7C, D), consistent with the similar number of inflorescence buds produced per plant. These differences in the production of OBs and OCFs between EV and NaHD20-silenced plants could be the result of a direct effect of NaHD20 on the regulation of flower transition processes or an indirect effect via the control of additional developmental regulatory factors or phytohormones (e.g. ABA). At this point, however, the possibility that a differential sensibility of developing flowers to water stress (due, for example, to the more advanced flowering stage of EV plants) could affect the re-activation of flowering after re-watering cannot be ruled out.
The effect of insect-derived chemical cues and bacterial infection on flowering-associated processes was independent of the silencing of NaHD20, suggesting a degree of specificity in the function of this gene as a regulator of flower transitions during water stress.
Conclusion
It was demonstrated that NaHD20 has multiple functions in N. attenuata as a positive effector of ABA accumulation in leaves, of BA emission from night flowers, and as a positive or negative effector of processes associated with the opening of inflorescence buds and bolting time depending on the growth conditions. Whether these mechanisms are connected to changes in ABA levels is at present unknown. The effect of ABA on reproductive-associated processes is controversial; previous studies have shown that ABA may delay or advance flowering time depending on the plant species and the developmental stage of the plant (Trewavas and Jones, 1991). Future experiments using N. attenuata plants stably silenced in NaHD20 expression will focus on understanding the mechanisms affecting flower transitions and their potential relationships to changes in ABA production.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Alignment of HD20 amino acid sequences from N. tabacum and N. attenuata.
Figure S2. Analysis of NaHD20 transcript levels in leaves after Ethephon and ABA application.
Figure S3. Leaf levels of NaNCED1, NaOSM1, and NaLTP1 after ABA treatment.
Figure S4. Silencing efficiency of NaHD20 expression in NaHD20-silenced plants.
Figure S5. ABA accumulation in EV and NaHD20-silenced plants (g dry weight−1).
Figure S6. Analysis of reproductive phase change and flower transition in EV plants after OS elicitation.
Figure S7. Analysis of reproductive phase change and flower transition in EV plants after Pseudomonas syringae infection.
Table S1. Primer sequences used for cloning and real-time qPCR.
Table S2. Sequences of Nicotiana attenuata cDNAs encoding HD-Zip proteins.
Supplementary Material
Acknowledgments
The DFG (BO3260/3-1), the DAAD, and the Max Planck Society are acknowledged for funding. DR, CAD, and RLC are members of Conicet-Argentina and funded by Foncyt (PICT 38103 and PAE-PICT 37100-022).
References
- Aoyama T, Dong CH, Wu Y, Carabelli M, Sessa G, Ruberti I, Morelli G, Chua NH. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. The Plant Cell. 1995;7:1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends in Plant Sciences. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Morse L. Up in Smoke 2. Germination of Nicotiana attenuata in response to smoke-derived cues and nutrients in burned and unburned soils. Journal of Chemical Ecology. 1994;20:2373–2391. doi: 10.1007/BF02033208. [DOI] [PubMed] [Google Scholar]
- Bell DL, Sultan SE. Dynamic phenotypic plasticity for root growth in Polygonum: a comparative study. American Journal of Botany. 1999;86:807–819. [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Rodriguez VM, Armand F, Farmer EE. The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant and Cell Physiology. 2007;48:1775–1789. doi: 10.1093/pcp/pcm151. [DOI] [PubMed] [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. The Plant Journal. 1999;17:427–431. doi: 10.1046/j.1365-313x.1999.00386.x. [DOI] [PubMed] [Google Scholar]
- Callahan HS, Pigliucci M, Schlichting CD. Developmental phenotypic plasticity: where ecology and evolution meet molecular biology. Bioessays. 1997;19:519–525. doi: 10.1002/bies.950190611. [DOI] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. The Plant Journal. 1993;4:469–479. doi: 10.1046/j.1365-313x.1993.04030469.x. [DOI] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiology. 2000;124:343–353. doi: 10.1104/pp.124.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Phillips J, Brautigam A, et al. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Molecular Biology. 2006;61:469–489. doi: 10.1007/s11103-006-0023-x. [DOI] [PubMed] [Google Scholar]
- Deng X, Phillips J, Meijer AH, Salamini F, Bartels D. Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 2002;49:601–610. doi: 10.1023/a:1015501205303. [DOI] [PubMed] [Google Scholar]
- Dezar CA, Gago GM, Gonzalez DH, Chan RL. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Research. 2005;14:429–440. doi: 10.1007/s11248-005-5076-0. [DOI] [PubMed] [Google Scholar]
- Euler M, Baldwin IT. The chemistry of defense and apparency in the corollas of Nicotiana attenuata. Oecologia. 1996;107:102–112. doi: 10.1007/BF00582240. [DOI] [PubMed] [Google Scholar]
- Fox G. Drought and the evolution of flowering time in desert annuals. American Journal of Botany. 1990;77:1508–1518. [Google Scholar]
- Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. The Plant Journal. 2003;36:794–807. doi: 10.1046/j.1365-313x.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- Hanson J, Johannesson H, Engstrom P. Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Molecular Biology. 2001;45:247–262. doi: 10.1023/a:1006464907710. [DOI] [PubMed] [Google Scholar]
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engstrom P, Soderman E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiology. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjellström M, Olsson A, Engström P, Söderman E. Constitutive expression of the water deficit-inducible homeobox gene ATHB7 in transgenic Arabidopsis causes a suppression of stem elongation growth. Plant, Cell and Environment. 2003;26:1127–1136. [Google Scholar]
- Johannesson H, Wang Y, Hanson J, Engstrom P. The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Molecular Biology. 2003;51:719–729. doi: 10.1023/a:1022567625228. [DOI] [PubMed] [Google Scholar]
- Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid–amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiology. 2010;152:96–106. doi: 10.1104/pp.109.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He S. The Arabidopsis thaliana– Pseudomonas syringae interaction. In: Somerville C, Meyerowitz E, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. pp. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Diezel C, Baldwin IT. Changing pollinators as a means of escaping herbivores. Current Biology. 2010;20:237–242. doi: 10.1016/j.cub.2009.11.071. [DOI] [PubMed] [Google Scholar]
- Kessler D, Gase K, Baldwin IT. Field experiments with transformed plants reveal the sense of floral scents. Science. 2008;321:1200–1202. doi: 10.1126/science.1160072. [DOI] [PubMed] [Google Scholar]
- Korner E, von Dahl CC, Bonaventure G, Baldwin IT. Pectin methylesterase NaPME1 contributes to the emission of methanol during insect herbivory and to the elicitation of defence responses in Nicotiana attenuata. Journal of Experimental Botany. 2009;60:2631–2640. doi: 10.1093/jxb/erp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology. 2002;12:177–183. [Google Scholar]
- Lee YH, Chun JY. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Molecular Biology. 1998;37:377–384. doi: 10.1023/a:1006084305012. [DOI] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. The Plant Journal. 2008;55:301–310. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Arce AL, Dezar CA, Bitton F, Renou JP, Crespi M, Chan RL. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. The Plant Journal. 2006;48:125–137. doi: 10.1111/j.1365-313X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Manavella PA, Dezar CA, Bonaventure G, Baldwin IT, Chan RL. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. The Plant Journal. 2008;56:376–388. doi: 10.1111/j.1365-313X.2008.03604.x. [DOI] [PubMed] [Google Scholar]
- Mayda E, Tornero P, Conejero V, Vera P. A tomato homeobox gene (HD-zip) is involved in limiting the spread of programmed cell death. The Plant Journal. 1999;20:591–600. doi: 10.1046/j.1365-313x.1999.00633.x. [DOI] [PubMed] [Google Scholar]
- Milborrow BV. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. Journal of Experimental Botany. 2001;52:1145–1164. [PubMed] [Google Scholar]
- Nicotra AB, Cosgrove MJ, Cowling A, Schlichting CD, Jones CS. Leaf shape linked to photosynthetic rates and temperature optima in South African Pelargonium species. Oecologia. 2008;154:625–635. doi: 10.1007/s00442-007-0865-1. [DOI] [PubMed] [Google Scholar]
- Olsson AS, Engstrom P, Soderman E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Molecular Biology. 2004;55:663–677. doi: 10.1007/s11103-004-1581-4. [DOI] [PubMed] [Google Scholar]
- Palena CM, Gonzalez DH, Chan RL. A monomer–dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochemical Journal. 1999;341:81–87. [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Bonaventure G, Kant MR, Baldwin IT. Jasmonate perception regulates jasmonate biosynthesis and JA–Ile metabolism: the case of COI1 in Nicotiana attenuata. Plant Cell and Physiology. 2008;49:1165–1175. doi: 10.1093/pcp/pcn091. [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proceedings of the National Academy of Sciences, USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Bokowiec MT, Laudeman TW, Brannock JF, Chen X, Timko MP. TOBFAC: the database of tobacco transcription factors. BMC Bioinformatics. 2008;9:53. doi: 10.1186/1471-2105-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. Journal of Experimental Botany. 2004;55:151–157. doi: 10.1093/jxb/erh004. [DOI] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. DNA-binding specificity of the homeodomain-leucine zipper domain. Journal of Molecular Biology. 1997;274:303–309. doi: 10.1006/jmbi.1997.1408. [DOI] [PubMed] [Google Scholar]
- Soderman E, Hjellstrom M, Fahleson J, Engstrom P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Molecular Biology. 1999;40:1073–1083. doi: 10.1023/a:1006267013170. [DOI] [PubMed] [Google Scholar]
- Soderman E, Mattsson J, Engstrom P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. The Plant Journal. 1996;10:375–381. doi: 10.1046/j.1365-313x.1996.10020375.x. [DOI] [PubMed] [Google Scholar]
- Sultan SE. Phenotypic plasticity for plant development, function and life history. Trends in Plant Sciences. 2000;5:537–542. doi: 10.1016/s1360-1385(00)01797-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proceedings of the National Academy of Sciences, USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A, Jones H. Abscisic acid: physiology and biochemistry. In: Davies W, Jones H, editors. An assessment of the role of ABA in plant development. Oxford: BIOS; 1991. pp. 169–188. [Google Scholar]
- Wang L, Halitschke R, Kang JH, Berg A, Harnisch F, Baldwin IT. Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta. 2007;226:159–167. doi: 10.1007/s00425-007-0477-3. [DOI] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. An update on abscisic acid signaling in plants and more. Molecular Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.