Abstract
The LATERAL ORGAN BOUNDARIES DOMAIN (LBD) gene family encodes plant-specific transcription factors. In this report, the LBD gene DOWN IN DARK AND AUXIN1 (DDA1), which is closely related to LATERAL ORGAN BOUNDARIES (LOB) and ASYMMETRIC LEAVES2 (AS2), was characterized. DDA1 is expressed primarily in vascular tissues and its transcript levels were reduced by exposure to exogenous indole-3-acetic acid (IAA or auxin) and in response to dark exposure. Analysis of a T-DNA insertion line, dda1-1, in which the insertion resulted in misregulation of DDA1 transcripts in the presence of IAA and in the dark revealed possible functions in auxin response and photomorphogenesis. dda1-1 plants exhibited reduced sensitivity to auxin, produced fewer lateral roots, and displayed aberrant hypocotyl elongation in the dark. Phenotypes resulting from fusion of a transcriptional repression domain to DDA1 suggest that DDA1 may act as both a transcriptional activator and a transcriptional repressor depending on the context. These results indicate that DDA1 may function in both the auxin signalling and photomorphogenesis pathways.
Keywords: Auxin, Aux/IAA, HY5, LBD genes, photomorphogenesis
Introduction
The plant-specific LATERAL ORGAN BOUNDARIES DOMAIN (LBD) gene family comprises 43 members in Arabidopsis (Shuai et al., 2002). Members of this family share the conserved LOB domain, which has recently been shown to have DNA-binding activity (Husbands et al., 2007). While the functions of the majority of LBD genes are unknown, members of this family have been implicated in a number of developmental processes including leaf polarity establishment (Lin et al., 2003; Xu et al., 2003), lateral root formation (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007), tracheary element development (Soyano et al., 2008), boundary delimitation (Shuai et al., 2002; Borghi et al., 2007; Lin et al., unpublished results), cytokinin signalling (Naito, 2007), inflorescence branch formation (Bortiri et al., 2006), female gametophyte development (Evans, 2007), and KNOX gene regulation (Ori et al., 2000; Semiarti et al., 2001; Chalfun-Junior et al., 2005; Borghi et al., 2007). The founding member of this family, LATERAL ORGAN BOUNDARIES (LOB) was isolated from an enhancer-trap screen, based on its expression on the adaxial side of lateral organ boundaries (Shuai et al., 2002). In Arabidopsis, LOB defines a subgroup of LBD genes that also includes LBD10/ASL2, LBD25/ASL3, LBD36/ASL1, and AS2/LBD6 (Iwakawa et al., 2002). Among this subgroup, AS2 (ASYMMETRIC LEAVES2) is the only gene with clearly defined functions. AS2 is required to prevent expression of the class I KNOX homeobox genes BREVIPEDICELLUS (BP), KNAT2, and KNAT6 in the leaf (Ori et al., 2000; Semiarti et al., 2001; Lin et al., 2003). AS2 is expressed on the adaxial side of lateral organs (Iwakawa et al., 2002, 2007; Wu et al., 2008) and misexpression leads to the formation of adaxialized leaves (Lin et al., 2003), implicating AS2 in adaxial cell fate specification. LBD36/ASL1 is expressed primarily in the vasculature and, when misexpressed, also results in repression of BP (Chalfun-Junior et al., 2005). LBD36/ASL1 may have limited redundancy with AS2 to control cell fate determination in petals (Chalfun-Junior et al., 2005). Functions have not been ascribed to the two remaining members of this subgroup, LBD10/ASL2 and LBD25/ASL3.
The hormone auxin has been implicated in multiple developmental responses in plants (reviewed in Vanneste and Friml, 2009). Auxin signalling is regulated through proteolysis of the Aux/IAA proteins, which act as transcriptional repressors (Worley et al., 2000; Gray et al., 2001; Reed, 2001; Dharmasiri and Estelle, 2002). Aux/IAA proteins form heterodimers with auxin response factors (ARFs), negatively regulating their activity to repress downstream auxin responses (Dharmasiri and Estelle, 2002, 2004; Liscum and Reed, 2002). The F-box protein TIR1, which acts as part of the SCF complex, is an auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005) that, upon interaction with auxin, targets Aux/IAA proteins for degradation. Degradation of Aux/IAA proteins frees the ARFs to regulate gene expression through auxin response elements (AuxREs) present in the promoters of auxin-regulated genes (Worley et al., 2000; Dharmasiri and Estelle, 2002, 2004; Liscum and Reed, 2002). Despite the extensive body of knowledge about auxin signalling that has been amassed, several components of this pathway await characterization.
Recent data have implicated several LBD genes in various aspects of auxin signalling. Microarray experiments identified a number of Arabidopsis LBD genes that are regulated by auxin (Nemhauser et al., 2004; Paponov et al., 2008). Crown rootless1 (Crl1)/Adventitious rootless1 (Arl1), which is required for formation of crown and lateral roots in rice, is a direct target of the ARF protein OsARF1 (Inukai et al., 2005; Liu et al., 2005). In Arabidopsis, the three genes most closely related to rice Crl1, LBD16, LBD18, and LBD29, are also regulated by auxin (Okushima et al., 2005; Lee et al., 2009). All three of these genes function in lateral root formation downstream of ARF7 and ARF19 (Okushima et al., 2007; Lee et al., 2009). Furthermore, LBD16 and LBD29 are directly regulated by ARF7 and ARF19 (Okushima et al., 2005, 2007). The maize gene rootless concerning crown and seminal roots (RTCS) is also a presumptive Crl1 orthologue involved in lateral root formation (Taramino et al., 2007). Additionally, the Arabidopsis LBD gene JAGGED LATERAL ORGANS (JLO) regulates the expression of the auxin efflux carrier PIN, suggesting a role in auxin signalling (Borghi et al., 2007).
The phenotypes observed in some auxin mutants suggest that there is an interaction between the auxin and light signalling pathways (Reed, 2001; Liscum and Reed, 2002). In fact, both pathways involve protein degradation via the proteasome (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). In the dark, the light-inactivatable repressor of photomorphogenesis COP1 is translocated to the nucleus (von Arnim and Deng, 1994). In the nucleus, COP1 binds directly and specifically to HY5 (Ang et al., 1998), a bZIP transcription factor that promotes photomorphogenesis by mediating light-controlled gene expression (Chattopadhyay et al., 1998). The interaction of HY5 and COP1 targets HY5 for proteasome-mediated proteolysis (Osterlund et al., 2000), resulting in the inhibition of light-regulated gene expression in the dark (Yadav et al., 2002). Analyses of the hy5 mutant indicate that HY5 might also be involved in auxin signalling, further supporting the idea that the auxin and light pathways intersect (Cluis et al., 2004).
In this study, it was shown that the Arabidopsis LBD gene DOWN IN DARK AND AUXIN1 (DDA1), formerly LBD25/ASL3, functions in both auxin signalling and aspects of photomorphogenesis. DDA1 transcript levels were reduced following treatment with exogenous indole-3-acetic acid (IAA) or exposure to dark conditions. The dda1-1 mutant, which behaves as a conditional gain-of-function semi-dominant allele, had a diminished auxin response and displayed aberrant hypocotyl elongation in the dark, indicative of defects in some aspects of auxin response and photomorphogenesis, respectively.
Material and methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown in soil or on 1× Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) as described previously (Shuai et al., 2002). All genotypes were in the Col-0 ecotype, with the exception of hy5-1, which is in Ler. The dda1-1 T-DNA mutant, SALK_033840, was isolated from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). Homozygous mutants were isolated by genomic DNA gel blot analysis and PCR-based genotyping using gene-specific primers DDA1-H (5′-CTTGGGAAATTGAGAATAATCCATAC-3′) and DDA1-F (5′-CCAACCCATGTCTCCTCTTTATCTC-3′) in combination with the T-DNA primer LBA1 (5′-TGGTTCACGTAGTGGGCCATCG-3′).
Plasmid constructs
The DDA1 promoter region (from –3201 bp upstream of the ATG to +18) was amplified from Col genomic DNA using Ex-Taq Polymerase (Takara, Shiga, Japan) with the primers pDDA1-F (5′-TCTAGAGATTCGGGTTGATATCTGAT-3′) and pDDA1-R (5′-GGATCCTGTTTCTCTCTTGGGCATTA-3′), which contained introduced XbaI and BamHI sites. PCR products were cloned into pCR-II TOPO (Invitrogen, Carlsbad, CA, USA) and sequenced to confirm their integrity, then subcloned into the XbaI and BamHI sites of pCB308 (Xiang et al., 1999) to create an in-frame translational fusion of the first six amino acids of DDA1 to β-glucuronidase (GUS).
To generate fusions to the hormone-binding domain of the rat glucocorticoid receptor (GR) (Picard et al., 1988), a Gateway destination vector was constructed. pBI-ΔGR (Lloyd et al., 1994) was digested with BamHI and the overhangs filled in using Klenow polymerase. The resulting DNA was ligated to Gateway conversion Cassette C (Invitrogen), to create the destination vector pBI-ΔGR-GW, which allows the generation of in-frame fusions to the hormone-binding domain of GR. The DDA1-GR construct was generated using a Gateway recombination with pBI-ΔGR-GW and PYAT3G27650, which contains the DDA1 coding sequence in a Gateway entry vector (Gong et al., 2004), according to the manufacturer's instructions (Invitrogen).
The DDA1-EAR construct was generated using a Gateway recombination reaction between entry clone PYAT3G27650 (Gong et al., 2004) and destination vector pDNG, kindly provided by Rüdiger Simon. pDNG contains the alcA promoter (Roslan et al., 2001) and a synthetic EAR (ERF-associated amphiphilic repression) domain (Hiratsu et al., 2003) flanking the ccdB cassette. The resulting DDA1-EAR construct contained the AlcA promoter driving an in-frame fusion of DDA1 to the EAR domain. The 35S:AlcR construct, pJH0022, was kindly provided by Syngenta.
All binary vectors were transformed into Agrobacterium tumefaciens strain GV3101 and subsequently transformed into Col wild-type plants using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998).
GUS expression analyses
Single-copy homozygous pDDA1:GUS plants were grown on MS medium with or without supplementation with 10 μM IAA or 85 nM 2,4-D for 7 d under a 16 h light/8 h dark photoperiod or in total darkness. Histochemical analyses and microscopy were performed as previously described (Shuai et al., 2002).
Phenotypic characterization
To determine lateral root numbers, seedlings were grown vertically for 4 d on unsupplemented MS medium, then transferred to medium supplemented with 85 nM 2,4-D, or to unsupplemented control medium, and grown for an additional 4 d. Visible lateral roots formed on the primary root were counted. Hypocotyl measurements were determined for 7-day-old seedlings grown on MS medium in total darkness or under a 16 h light/8 h dark photoperiod. To increase the level of endogenous auxin, seedlings were grown at 28 °C as previously described (Gray et al., 1998). Root growth sensitivity to auxin was determined as previously described (Lincoln et al., 1990). A standard table-top scanner was used to obtain images of seedlings on plates, and measurements were obtained using MCID Elite 7.0 software (Imaging Research Inc., Ontario, Canada).
Ethanol induction
F1 plants derived from a cross between a homozygous single-copy pAlcA:DDA1-EAR plant and a homozygous single-copy 35S:AlcR plant were termed 35S>>DDA1-EAR and were used in all ethanol induction experiments. Seedlings were grown on MS medium in closed transparent containers. Seedlings were induced by exposure to ethanol vapour—two 1.5 ml tubes containing 1 ml of 50% ethanol each were placed inside the containers for 2 h d−1 for 4 d. Control-treated plants were maintained in a closed container in a separate growth chamber.
Expression analyses
For expression analyses, seedlings were grown for 6 d on MS solid medium (Murashige and Skoog, 1962), then transferred to MS liquid medium (Murashige and Skoog, 1962), and maintained overnight to equilibrate. Auxin or dark exposure treatments were done the following day by the addition of 20 μM IAA or by wrapping the plates in aluminium foil. RNA extraction and cDNA syntheses were performed as previously described (Lin et al., 2003). PCR conditions for DDA1 and ACTIN2 (ACT2) amplification were: denaturation at 94 °C for 10 min, followed by 15 cycles (DDA1) or 10 cycles (ACT2) of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 2 min, and one final cycle of 72 °C for 10 min using the primers DDA1-F (5′-GAATTCATGCCCAAGAGAGAAAC-3′) and DDA1-R (5′-GCGGCCGCACCCCTCCGACCACC-3′) for DDA1, and ACT2-N (5′-AAAATGGCCGATGGTGAGG-3′) and ACT-C2 (5′-ACTCACCACCACGAACCAG-3′) for ACT2. The blotting and hybridization were performed as previously described (Lin et al., 2003). RT-PCR analyses of DDA1 and ACT2 transcript levels using different amounts of cDNA template demonstrated that the PCRs were quantitative under these conditions (see Supplementary Fig. S1 available at JXB online).
Results
LBD25 (At3g27650, also known as ASL3) is a member of the LBD gene family and belongs to a subclade of LBD genes that includes LOB, AS2, LBD36/ASL1, and LBD10/ASL2 (Iwakawa et al., 2002; Shuai et al., 2002). lob loss-of-function mutants did not display conspicuous phenotypes, therefore it was suspected that other LBD genes might have functions overlapping those of LOB. Phylogenetic analyses indicated that LBD25 was a likely candidate, as it is more closely related to LOB than any other LBD gene (Iwakawa et al., 2002; Shuai et al., 2002). Based on the observed down-regulation of LBD25 expression by auxin and dark conditions (see below), LBD25 was named DOWN IN DARK AND AUXIN1 (DDA1).
DDA1 is transcriptionally regulated in response to auxin and dark
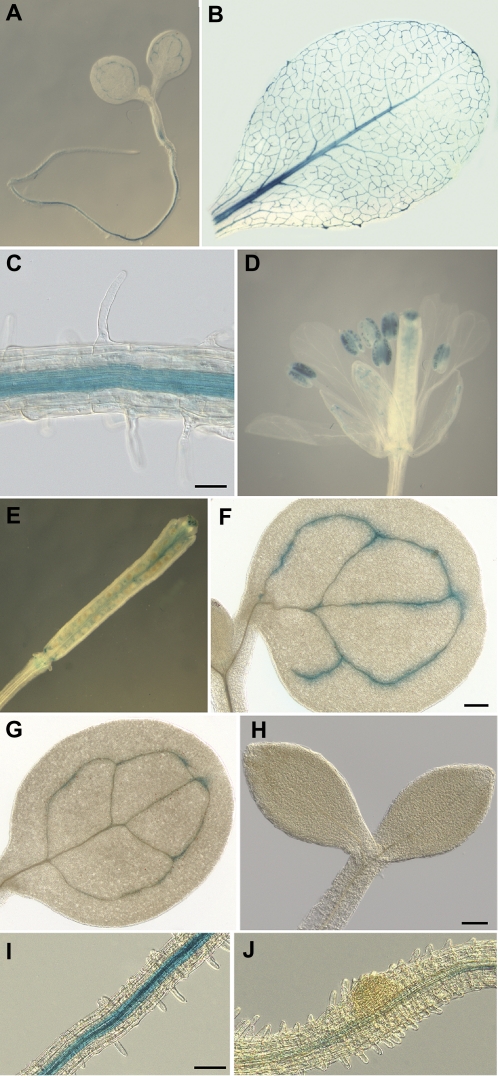
Previous expression analyses using RT-PCR showed that DDA1 was expressed more broadly than LOB (Shuai et al., 2002), but a detailed characterization of the DDA1 expression pattern has not previously been reported. To examine DDA1 expression, a promoter–reporter gene construct containing a 3.2 kb region upstream of the translation start site and including the first six DDA1 codons fused, in-frame, to uidA (GUS) was introduced into Arabidopsis. More than 30 independent transgenic plants were analysed and all showed a similar GUS expression pattern (data not shown). Several single-copy pDDA1:GUS transgenic lines were identified, one of which was used for detailed expression analyses. In pDDA1:GUS seedlings, GUS expression was detected in the vasculature of cotyledons, at the base of the hypocotyl, and in the root, but was excluded from the root tip (Fig. 1A, C). GUS expression was also observed in the vasculature of rosette leaves (Fig. 1B) and cauline leaves, although GUS activity was weaker in the latter (data not shown). In the flower, GUS expression was detectable in the vasculature of sepals but not petals, in the stigma, in the placenta, in pollen grains, and at the base of floral organs (Fig. 1D). As some promoter:GUS fusions have been reported to result in artefactual GUS activity in pollen (Mascarenhas and Hamilton, 1992), it was confirmed that DDA1 transcripts were detectable in anthers using RT-PCR (data not shown). After pollination, GUS activity was observed at the base of the silique, in the placenta, and in the degenerating stigma (Fig. 1E), similar to the pattern observed in flowers.
Fig. 1.
DDA1 is expressed in the vasculature and is transcriptionally regulated by auxin and dark exposure. Histochemical GUS analysis of pDDA1:GUS transgenic plants. (A) Seven-day-old seedling. (B) Mature rosette leaf. (C) Root of 7-day-old seedling. (D) Open flower. (E) Silique. (F) Cotyledon of 7-day-old seedling grown under standard conditions. (G) Cotyledon of 7-day-old seedling grown in 10 μM IAA. (H) Seven-day-old seedling grown in constant dark. (I) Root of 7-day-old seedling grown on unsupplemented medium. (J) Seven-day-old seedling root grown on medium supplemented with 85 nM 2,4-D. Size bar in (C) = 50 μm, in (F) = 200 μm, and in (H) and (I) = 100 μm. The magnification in (F) and (G) is the same; the magnification in (I) and (J) is the same.
Examination of publicly available microarray data revealed that DDA1 transcript levels were reduced by treatment with auxin (Nemhauser et al., 2004) and exposure to dark (www.arabidopsis.org). To investigate DDA1 regulation further, GUS activity was compared in 7-day-old pDDA1:GUS seedlings grown in the presence or absence of exogenous auxin and in seedlings grown under a long-day light–dark cycle or in complete darkness. Growth on 10 μM IAA resulted in a substantial decrease in GUS activity in cotyledon vasculature (Fig. 1G) compared with seedlings grown on unsupplemented medium (Fig. 1F), while GUS activity was nearly abolished in dark-grown seedlings (Fig. 1H). GUS activity was also reduced in the roots of pDDA1:GUS plants that were grown on 85 nM 2,4-D (compare Fig. 1I and J). These observations indicate that the regulation of DDA1 in response to auxin and dark exposure is likely to be, at least in part, at the transcriptional level.
dda1-1 mutants exhibit reduced auxin responses
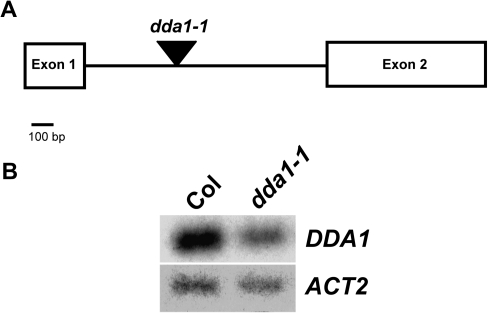
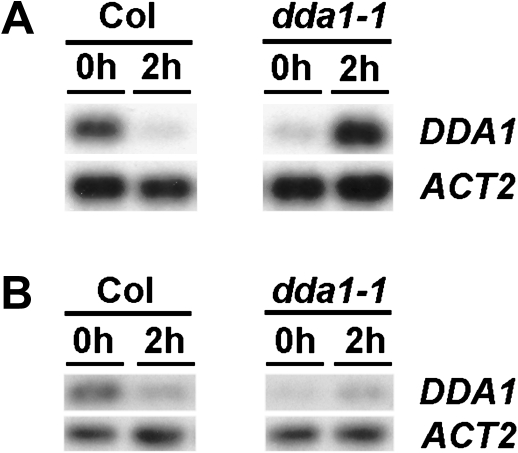
To better understand the function of DDA1, a T-DNA insertion line, SALK_033840 (Alonso et al., 2003), was identified which was designated dda1-1. This line contained an insertion in the sole intron of the DDA1 gene (Fig. 2A). To determine whether the T-DNA insertion affected DDA1 transcript accumulation, RT-PCR was used to amplify the coding region of DDA1 transcripts in homozygous dda1-1 seedlings. Reduced transcript levels were detected in dda1-1 homozygotes compared with the Col wild type, suggesting it is a hypomorphic allele (Fig. 2B). Sequencing of RT-PCR products demonstrated that transcripts produced in dda1-1 were accurately spliced and therefore apparently functional (data not shown).
Fig. 2.
Location and consequences of T-DNA insertion in the dda1-1 mutant. (A) The genomic structure of DDA1 indicating the position of the T-DNA insertion in dda1-1 (triangle). (B) RT-PCR analysis of DDA1 transcript levels in 7-day-old seedlings of Col and dda1-1. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 cycles (DDA1) or 10 cycles (ACT2) of amplification. The primers used for DDA1 amplification span the entire coding region.
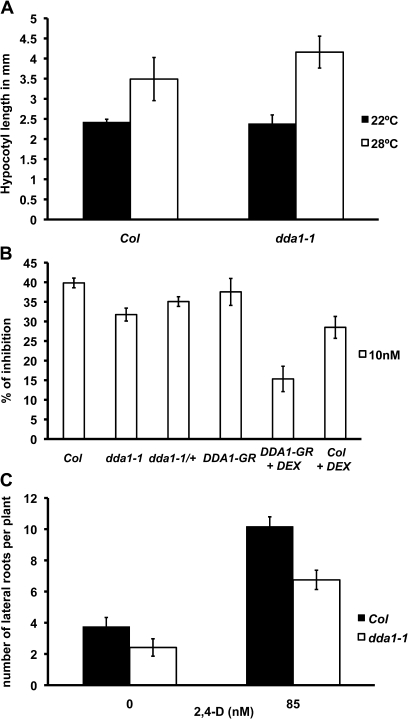
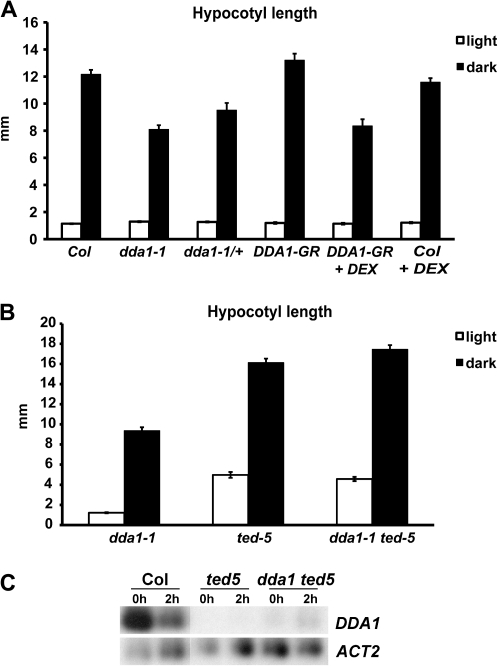
Based on the observed regulation of DDA1 in the presence of auxin, the dda1-1 mutant was examined for auxin responses. Whether the hypocotyl of dda1-1 responded normally to increased auxin concentrations was first examined. dda1-1 plants were grown at 28 °C, a condition that increases endogenous auxin levels (Gray et al., 1998). dda1-1 seedlings did not show a significant difference in hypocotyl length compared with the Col wild type when grown at either 22 °C or 28 °C (Fig. 3A), indicating that auxin signalling is not perturbed in the hypocotyl of the dda1-1 mutant.
Fig. 3.
dda1-1 mutant seedlings show reduced sensitivity to auxin. (A) Hypocotyl measurements of 7-day-old Col and dda1-1 seedlings grown at 22 °C or 28 °C. A minimum of 10 seedlings was assayed for each background and temperature. Error bars represent the standard error. t-test indicates that the values between genotypes are not significantly different. (B) Reduction in root growth resulting from 2,4-D exposure. Seedlings were grown on unsupplemented medium for 4 d, then transferred to 2,4-D-supplemented medium. After 3 d, root length was measured. Inhibition of root growth is calculated from growth on 2,4-D relative to growth on unsupplemented medium. A minimum of 10 seedlings was assayed for each background. Error bars represent the standard error. t-test P <0.01 (Col×ddal1-1); P< 0.05 (Col×dda1-1/+); P <0.01 (Col+DEX×DDA1-GR+DEX). (C) The number of lateral roots per 8-day-old seedling following transfer to unsupplemented or 2,4-D-supplemented medium after 4 d growth. A minimum of 12 seedlings was assayed for each background and treatment. Error bars represent the standard error. t-test for 0 nM indicates that the values are not significantly different and for 85 nM, P <0.001.
Auxin sensitivity assays were performed to determine whether auxin responses were affected in dda1-1 roots. Four-day-old seedlings were transferred to medium containing 2,4-D or to unsupplemented control medium, and root growth in a 3 d period was determined. Sensitivity to a range of 2,4-D concentrations was examined. The most significant difference between dda1-1 and wild-type Col plants was observed using 10 nM 2,4-D (see Supplementary Fig. S2 at JXB online), therefore subsequent experiments used 10 nM 2,4-D. Wild-type Col plants exhibited an ∼40% inhibition in root growth in response to auxin treatment. dda1-1 mutants displayed reduced sensitivity to auxin compared with Col, showing ∼32% inhibition (Fig. 3B).
Lateral root formation in dda1-1 seedlings was also examined as an additional indicator of auxin responsiveness. Compared with the wild type, dda1-1 mutants did not show a significant difference in lateral root number when grown on unsupplemented medium (Fig. 3C). To examine auxin-induced lateral root production, 4-day-old plants were transferred to medium containing 85 nM 2,4-D and lateral root numbers were determined after 4 d of growth. dda1-1 mutants produced ∼35% fewer lateral roots than the wild type. These data are consistent with reduced auxin sensitivity in dda1-1 roots.
The axr3-1 mutant disrupts DDA1 regulation by auxin
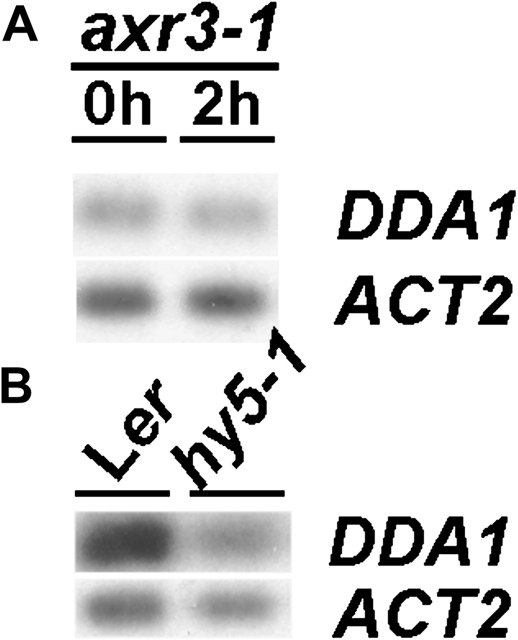
In order to gain insight into the regulation of DDA1, DDA1 transcript levels were examined in the axr3-1 mutant background. AXR3 encodes the Aux/IAA protein IAA17 (Leyser et al., 1996), a repressor of auxin responses that is targeted to the proteasome for degradation in the presence of auxin. The axr3-1 mutation results in protein stabilization and a resulting alteration in auxin responses (Leyser et al., 1996; Rouse et al., 1998). In axr3-1 seedlings, DDA1 transcript abundance was not altered by treatment with IAA (Fig. 4A), indicating that AXR3 degradation is required for the reduction of DDA1 transcripts following exposure to exogenous auxin. This places down-regulation of DDA1 downstream of auxin-mediated proteolysis of Aux/IAA proteins. Given that there are many Aux/IAA proteins in Arabidopsis and axr3-1 is a gain-of-function mutant, it is possible that AXR3 does not normally participate in DDA1 regulation in wild-type plants, where this role might be performed by other related Aux/IAA proteins.
Fig. 4.
DDA1 transcript levels are regulated by AXR3 and HY5. (A) RT-PCR analysis of DDA1 transcript levels in 7-day-old axr3-1 mutant seedlings following 0 h or 2 h exposure to 20 μM IAA. (B) RT-PCR analysis of DDA1 transcript levels in 7-day-old Ler and hy5-1 seedlings. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 cycles (DDA1) or 10 cycles (ACT2) of amplification.
dda1-1 displays aberrant response to dark growth conditions
As DDA1 transcript levels were modulated by exposure to dark conditions, dark-grown dda1-1 seedlings were examined for etiolation characteristics such as hypocotyl elongation, apical hook formation, and closed cotyledons (McNellis et al., 1994). Dark-grown dda1-1 seedlings exhibited lack of chlorophyll pigmentation, an apical hook, and closed cotyledons, similar to the wild type (data not shown). However, dark-grown dda1-1 hypocotyls were significantly shorter than those of the wild type (Fig. 5A). In contrast, dda1-1 hypocotyls were slightly longer than those of the wild type when grown under a 16 h light/8 h dark photoperiod (Fig. 5A), indicating that they exhibited an aberrant response to dark growth conditions. As other etiolation responses were normal, DDA1 appears to function in only one aspect of etiolation—hypocotyl elongation. dda1-1 mutant hypocotyls responded normally to auxin (Fig. 3A), therefore the aberrant hypocotyl elongation observed in dark-grown plants does not appear to be the result of disturbed auxin responses.
Fig. 5.
dda1-1 mutants display shorter hypocotyls in the dark. (A) Hypocotyl length of 7-day-old Col, dda1-1, and DDA1-GR seedlings grown under a 16 h light/8 h dark photoperiod (white columns) or in the dark (black columns). A minimum of 12 seedlings was assayed for each background and growth condition. Error bars represent the standard error. t-test for dark treatment, P <0.0001 (Col×dda1-1); P <0.001 (Col×dda1-1/+); P <0.0001 (Col+DEX×DDA1-GR+DEX). (B) The same experiment as in (A) but using dda1-1, ted5-1, and dda1-1 ted5-1 seedlings. t-test for light treatment, P <0.0001 (dda1-1×ted5-1); P <0.0001 (dda1-1×dda1-1 ted5-1). ted5-1 and dda1-1 ted5-1 were not significantly different (P <0.2). t-test for dark treatment, P <0.0001 (dda1-1×ted5-1); P <0.0001 (dda1-1×dda1-1 ted5-1); P <0.05 (ted5-1×dda1-1 ted5-1). (C) RT-PCR analysis of DDA1 transcript levels in 7-day-old Col, ted5-1, and dda1-1 ted5-1 seedlings following 0 h or 2 h exposure to dark conditions. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 cycles (DDA1) or 10 cycles (ACT2) of amplification.
A major factor in the promotion of photomorphogenesis is the bZIP transcription factor HY5, which is targeted to the proteasome for degradation in dark conditions (Ang et al., 1998; Chattopadhyay et al., 1998; Osterlund et al., 2000; Yadav et al., 2002). To investigate the relationship between DDA1 and HY5, steady-state levels of DDA1 transcripts were examined in the hy5-1 mutant background. DDA1 transcript levels were significantly reduced in hy5-1 seedlings compared with the wild type (Fig. 4B), indicating that HY5 activity contributes to DDA1 regulation. To investigate this relationship further, double mutants were generated between dda1-1 and the HY5 mutant allele ted5-1 (Pepper and Chory, 1997). In both light- and dark-grown conditions, ted5-1 mutant hypocotyls were longer than dda1-1 hypocotyls (Fig. 5B). dda1-1 ted5-1 double-mutant hypocotyls were similar to those of ted5-1 single mutants (Fig. 5B). The restoration of dark-induced hypocotyl elongation in the double mutant, relative to the dda1-1 single mutant, indicates that ted5-1 is epistatic to dda1-1. To investigate the molecular nature of this epistasis, DDA1 transcript abundance was examined in dda1-1 ted5-1 double mutants. DDA1 transcript levels were reduced in dda1-1 ted5-1 seedlings, similar to the levels observed in ted5-1 (Fig. 5C). Further, there was no apparent dark-induced transcript regulation in the double mutants (Fig. 5C). These data are consistent with the hypothesis that DDA1 negatively regulates hypocotyl elongation during photomorphogenesis.
The dda1-1 mutation affects DDA1 transcript accumulation in the presence of auxin and in the dark
As DDA1 transcript levels were reduced in response to auxin or dark exposure, the phenotypes observed in the dda1-1 mutant—reduced sensitivity to auxin and aberrant response to dark—were inconsistent with its apparent hypomorphic nature. Because of this contradiction, DDA1 steady-state transcript levels were analysed in both wild-type and dda1-1 seedlings following treatment with auxin or exposure to dark conditions. Exposure to 20 μM IAA for 2 h resulted in a reduction in the abundance of DDA1 transcripts in wild-type seedlings (Fig. 6A), in agreement with the behaviour of the pDDA1:GUS reporter line. However, while dda1-1 seedlings showed reduced steady-state transcript levels prior to auxin treatment, an increase in transcript abundance was observed following the induction (Fig. 6A). Exposure to total darkness for 2 h resulted in a small increase in transcript abundance in dda1-1 seedlings (Fig. 6B), in contrast to the reduction observed in wild-type seedlings. Thus, although dda1-1 seedlings had reduced transcript levels under standard growth conditions, transcript accumulation was not regulated appropriately in response to auxin or dark exposure.
Fig. 6.
Transcript levels are aberrantly regulated by auxin and dark exposure in dda1-1 mutants. (A) RT-PCR analysis of DDA1 transcript levels in 7-day-old Col and dda1-1 seedlings following 0 h or 2 h exposure to 20 μM IAA. (B) RT-PCR analysis of DDA1 transcript levels in 7-day-old Col and dda1-1 seedlings transferred to the dark. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 cycles (DDA1) or 10 cycles (ACT2) of amplification.
Since the T-DNA insertion in dda1-1 is in the sole intron, it was speculated that the differential transcript accumulation compared with Col might be due to effects on splicing efficiency. If this were the case, then different regions of the transcript might differ in abundance in mutant plants. To test this possibility, dda1-1 cDNA was amplified using primers spanning the first exon of DDA1, which is upstream of the insertion site. These primers produced an RT-PCR product similar in abundance to that obtained with primers spanning the entire coding region (data not shown), indicating that the increase in transcript levels is not likely to be due to changes in splicing efficiency. The nature of the altered DDA1 regulation in dda1-1 is not clear.
dda1-1 is a hypermorphic allele in the presence of auxin and in the dark
If dda1-1 plants exhibited increased DDA1 activity in the presence of auxin, as would be predicted for a hypermorphic allele, then it is expected to behave in a semi-dominant manner. To test this hypothesis, dda1-1/+ heterozygotes were analysed for auxin responses. dda1-1/+ seedlings exhibited moderate auxin resistance, showing an intermediate level of growth inhibition between that seen in dda1-1 homozygotes and wild-type seedlings. This result is consistent with the conclusion that dda1-1 is a hypermorphic allele in the presence of exogenous auxin (Fig. 3B).
To investigate further the role of DDA1 in plant development, transgenic plants were generated expressing a dexamethasone (DEX)-inducible form of DDA1, a translational fusion to the hormone-binding domain of the rat GR, under control of the ubiquitously expressed cauliflower mosaic virus 35S promoter. In the absence of DEX, the response of 35S:DDA1-GR seedlings to auxin was not significantly different from that of the wild type (Fig. 3B). When grown in the presence of DEX, however, DDA1-GR seedlings showed reduced auxin sensitivity compared with wild-type plants grown on DEX (Fig. 3B). It is noted that wild-type plants grown on DEX also exhibited a diminished response to auxin. However, DEX-grown DDA1-GR seedlings showed a mild but significant reduction in auxin sensitivity compared with wild-type seedlings grown on DEX. DDA1-GR plants on DEX showed ∼15% root growth inhibition due to auxin, while wild-type plants on DEX showed ∼30% inhibition. Reduced sensitivity to auxin in plants that had increased levels of DDA1 transcript, in auxin-treated dda1-1 mutants and DDA1-GR plants, indicates that DDA1 acts as a negative regulator of the auxin signalling pathway.
As dda1-1 mutants had higher transcript levels following dark exposure, in contrast to wild-type plants, which had reduced transcript accumulation (Fig. 6B), this allele also appears to be hypermorphic under dark-grown conditions. dda1-1/+ heterozygotes and DDA1-GR plants were therefore examined for aberrant hypocotyl elongation in the dark. Dark-grown dda1-1/+ seedlings produced hypocotyls that were intermediate in length between Col wild type and dda1-1 homozygotes, consistent with dda1-1 being a semi-dominant allele (Fig. 5A). DDA1-GR plants grown on DEX showed a reduction in hypocotyl length of ∼33% compared with the wild type on DEX, while DDA1-GR grown on medium without DEX exhibited normal hypocotyl elongation (Fig. 5A). These data support the hypothesis that dda1-1 behaves as a hypermorphic allele in the dark and suggest that DDA1 is involved in suppressing hypocotyl elongation during photomorphogenesis.
Overexpression of DDA1 fused to a transcriptional repression domain reveals differences in DDA1 function in the auxin and photomorphogenesis pathways
Several LBD proteins have been shown to bind DNA, and the closely related LOB protein has transcriptional activation activity (Husbands et al., 2007). DDA1 is therefore likely to function as a transcriptional regulator. To examine the role of this protein further in processes related to photomorphogenesis and auxin signalling, transgenic plants were generated expressing a fusion of DDA1 to an EAR domain, which has strong transcriptional repression activity (Ohta et al., 2001; Hiratsu et al., 2003). This fusion protein is expected to function as a strong transcriptional repressor, which should provide insights into the function of DDA1. Given that it was not possible to obtain plants with significantly elevated levels of DDA1 when using a constitutive promoter (data not shown), transgenic plants with inducible DDA1-EAR expression were generated using the two-component alc system (Deveaux et al., 2003). Transgenic plants expressing DDA1-EAR under the control of the AlcA promoter (AlcA:DDA1-EAR) were crossed to plants expressing the AlcR transcription factor under control of the 35S promoter (35S:AlcR). AlcR is active only in the presence of ethanol (Lockington, 1987), allowing DDA1-EAR expression to be induced by ethanol vapour. F1 plants, designated 35S>>DDA1-EAR, were examined for ethanol-dependent phenotypes.
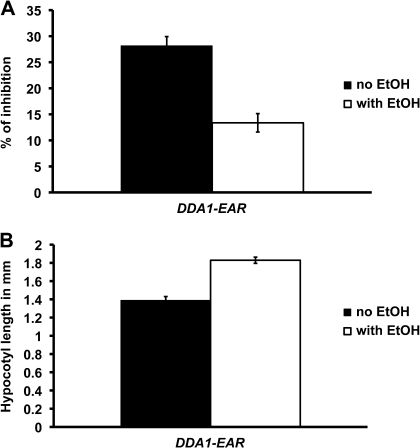
In the absence of ethanol, 35S>>DDA1-EAR transgenic plants were phenotypically normal, indistinguishable from wild-type plants grown in either the presence or absence of ethanol (data not shown). The phenotypes of dda1-1 and DDA1-GR plants, both of which have increased DDA1 activity in the presence of exogenous auxin, indicated that DDA1 is a negative regulator of auxin signalling. In order to investigate the nature of this negative regulation, 35S>>DDA1-EAR plants were examined for auxin-response phenotypes. Root growth inhibition assays were conducted on 35S>>DDA1-EAR and wild-type plants that were either induced with ethanol vapour or grown in control conditions without exposure to ethanol. Relative to uninduced plants, ethanol-induced 35S>>DDA1-EAR plants exhibited reduced growth inhibition in response to auxin exposure (Fig. 7A), while the response of wild-type plants to auxin was unaffected by ethanol treatment (data not shown). Following treatment with ethanol vapour, auxin treatment of 35S>>DDA1-EAR roots resulted in ∼14% growth inhibition, while roots of uninduced control plants showed ∼28% growth inhibition (Fig. 7A). The reduced auxin sensitivity of 35S>>DDA1-EAR plants is similar to that observed in DDA1-GR plants, suggesting that the addition of a repressor domain to DDA1 did not alter its function. These data are consistent with DDA1 functioning as a transcriptional repressor to suppress some aspects of the auxin response.
Fig. 7.
Phenotypes observed in DDA1-EAR seedlings. (A) Reduction in root growth resulting from 2,4-D exposure. Seedlings were grown as described in Fig. 3. On the third day, ethanol induction was initiated and maintained for 4 d. The percentage growth inhibition was calculated from growth on 2,4-D relative to growth on unsupplemented medium. A minimum of 10 seedlings was assayed for each condition. Error bars represent the standard error. t-test P <0.001 (no ethanol×with ethanol). (B) Hypocotyl length of 7-day-old seedlings grown under a 16 h light/8 h dark photoperiod. A minimum of 12 seedlings was assayed for each condition. Error bars represent the standard error. t-test P <0.000001 (no ethanol×with ethanol).
Based on the phenotypes of dda1-1 and DDA1-GR plants, which have increased levels of DDA1 activity, DDA1 appears to contribute to the repression of hypocotyl elongation during photomorphogenesis. To gain further insights into this aspect of DDA1 function, light-grown 35S>>DDA1-EAR plants were examined for ethanol-dependent changes in hypocotyl length. Following treatment with ethanol vapour, the hypocotyls of 35S>>DDA1-EAR plants grown in standard growth conditions were slightly longer than those of uninduced control plants, while exposure to ethanol vapour did not affect hypocotyl length in wild-type plants (data not shown). Hypocotyls of induced 35S>>DDA1-EAR plants were ∼30% longer than those of uninduced controls (Fig. 7B). No difference in hypocotyl length was observed between induced and control dark-grown 35S>>DDA1-EAR plants (data not shown). The phenotype of induced 35S>>DDA1-EAR plants resembled that of hy5 mutants, which also have longer hypocotyls in the light (Oyama et al., 1997), although the hy5 phenotype is more dramatic. The observation that 35S>>DDA1-EAR plants exhibited a longer hypocotyl in the light while the gain-of-function DDA1-GR and dda1-1 plants exhibited a shorter hypocotyl in the dark suggests that the activity of the DDA1-EAR protein is different from that of the native DDA1 protein, consistent with the fusion protein functioning as a dominant negative. It is worth noting that when grown under a long-day photoperiod, in which the transcript levels of DDA1 were reduced (Fig. 2B), dda1-1 plants had slightly longer hypocotyls than Col plants (Fig. 4A). These data are consistent with a model in which DDA1 functions as a transcriptional activator to repress hypocotyl elongation in the light. Collectively, the results suggest that DDA1 acts as a transcriptional repressor during auxin response while it acts as a transcriptional activator in the photomorphogenesis pathway.
Discussion
Nature of the dda1-1 allele
Although the T-DNA insertion in dda1-1 produced a hypomorphic allele under standard growth conditions, the dda1-1 mutant behaves as a gain-of-function allele in the presence of exogenous IAA or in the dark. The reason for this discrepancy is not yet clear. The T-DNA insertion in dda1-1 does not appear to alter splicing efficiency, as RT-PCR using primers annealing to the first exon, which is upstream of the insertion in dda1-1, also revealed elevated transcript levels. It is possible that the T-DNA insertion, which is in the sole DDA1 intron, disrupts a cis-acting element required for the transcriptional down-regulation of DDA1 in response to auxin or growth in the dark. However, the intron sequences are not essential for this regulation, as a DDA1 promoter:GUS construct lacking the intron conferred regulation by auxin and dark. Another possibility is that transcript accumulation is due to reduced post-transcriptional degradation of the DDA1 transcript. As the transcript produced by the dda1-1 mutant is predicted to be identical to the wild-type DDA1 transcript, this explanation seems implausible.
dda1-1 mutants do not present severe phenotypes
dda1-1 mutant plants exhibited a diminished response to both auxin and dark growth conditions. Although consistent, the phenotypes were quite subtle compared with those of other auxin and light signalling mutants (Lincoln et al., 1990; Wei et al., 1994; Leyser et al., 1996). Because both gain-of-function DDA1 and 35S>>DDA1-EAR phenotypes were fairly subtle, it is likely that DDA1 would not have been identified in conventional mutagenesis screens. The subtle nature of the phenotypes may result from the fact that DDA1 functions in both the auxin and light perception pathways, perhaps contributing quantitatively to both responses.
In recent years, a large body of data relating to the auxin and light signal transduction pathways has been amassed, leading to a dramatic increase in our understanding of these important responses. Several mutant screens led to the identification of major players in both pathways. Most of those screens identified components that act very early in the respective pathway (Wei and Deng, 1996, 1999; Holm and Deng, 1999; Hardtke and Deng, 2000; Dharmasiri and Estelle, 2002, 2004; Liscum and Reed, 2002; Dharmasiri et al., 2005; Kepinski and Leyser, 2005). To gain a complete understanding of the auxin and dark responses, it will be crucial also to identify and characterize late-acting genes. DDA1 appears to be one such gene, participating in both light and auxin pathway responses.
LBD genes involved in auxin-related processes
DDA1 is one of a number of LBD genes that play a role in plant responses to auxin. The expression of several Arabidopsis LBD genes has been shown to be regulated by auxin (Nemhauser et al., 2004; Paponov et al., 2008). Although biological functions for most of the auxin-regulated LBD genes have not been reported, the rice gene Crl1/Arl1, which is a direct target of OsARF1 (Inukai et al., 2005; Liu et al., 2005), is required for crown root formation. The Arabidopsis genes LBD16, LBD18, and LBD29, which are closely related to Crl1/Arl1, also function in lateral root formation and are regulated by ARF7 and ARF19 (Okushima et al., 2007; Lee et al., 2009), indicating that function within this LBD subfamily is conserved across monocots and dicots. The Arabidopsis LBD gene JLO is also involved in auxin responses. JLO activity negatively regulates the expression of members of the PIN family of auxin efflux factors (Borghi et al., 2007), although it remains to be shown if this regulation is direct.
Cross-talk between the auxin and light pathways
Several pieces of evidence support the idea that there is communication between the auxin and light signal transduction pathways. HY5, a bZIP transcription factor involved in the light response pathway, promotes the expression of the Aux/IAA genes AUXIN RESISTANT2 and SOLITARY ROOT, which function as negative regulators of auxin signalling (Cluis et al., 2004). HY5 also seems to promote the expression of DDA1, which has been shown to be a negative regulator of auxin responses. HY5 regulation of DDA1 transcription is probably indirect, as HY5-binding sites were not found in the DDA1 promoter (data not shown) and DDA1 was not identified as a HY5 target in ChIP-chip experiments (Lee et al., 2007). Other evidence of cross-talk between the light and auxin pathways comes from the observation that some gain-of-function Aux/IAA mutants are also constitutively photomorphogenic (Reed, 2001; Liscum and Reed, 2002). DDA1 also functions in both pathways, contributing to negative regulation of auxin responses and to repression of hypocotyl elongation in the light. One mutant involved in both auxin and light responses is axr3-1, and it was shown that DDA1 levels were stably maintained in this background even in the presence of auxin. The present data are consistent with the idea that DDA1 is a negative regulator of the auxin signalling pathway and promotes hypocotyl elongation in the light.
DDA1 is involved in auxin signalling and promotion of photomorphogenesis
Taken together, the data shown here have led to a model that would explain the regulation of DDA1 and its function. IAA negatively regulates Aux/IAA proteins such as AXR3 by inducing their proteolysis (Dharmasiri and Estelle, 2002, 2004). It was found that in the dominant axr3-1 mutant, the level of DDA1 transcripts was stabilized in the presence of IAA, which normally causes a decrease in DDA1 transcript accumulation. Therefore, the IAA-induced reduction of DDA1 transcription may act through the degradation of AXR3 or related Aux/IAA proteins. In the presence of auxin, the levels of DDA1 transcripts were increased in the dda1-1 background. Hence, the reduced auxin responses in dda1-1 are due to enhanced levels of DDA1 transcripts, leading to the conclusion that DDA1 is a negative regulator of auxin signalling. This is in agreement with the fact that AXR3, which is a positive regulator of DDA1, is also a negative regulator of this same pathway (Reed, 2001; Dharmasiri and Estelle, 2002, 2004; Liscum and Reed, 2002).
It is known that the photomorphogenesis-promoting transcription factor HY5 is targeted for degradation in the dark (von Arnim and Deng, 1994; von Arnim et al., 1997; Ang et al., 1998; Osterlund et al., 2000). It has been shown here that HY5 positively regulates the expression of DDA1 and, in the dark, when HY5 is absent, the transcript levels of DDA1 are decreased. Based on these results, a model is proposed in which a mechanism for down-regulation of DDA1 in the dark is through the degradation of its positive regulator, HY5. In the dda1-1 mutant, the levels of DDA1 transcripts are increased in the dark. Therefore, the aberrant dark responses in dda1-1 are due to elevated levels of DDA1, leading to the conclusion that DDA1 is involved in promotion of photomorphogenesis. The fact that HY5, a key player in the promotion of photomorphogenesis (Chattopadhyay et al., 1998; Yadav et al., 2002), is a positive regulator of DDA1 corroborates this conclusion.
Based on the phenotypes observed in 35S>>DDA1-EAR plants, DDA1 appears to function as both a transcriptional activator and a transcriptional repressor depending on the pathway. Transcription factors in a number of different families have been reported to have both transcriptional activation and transcriptional repression activities depending on interactions with other factors or protein modifications (Hoecker et al., 1995; Ammanamanchi et al., 2003; Canon and Banerjee, 2003; Kesarwani et al., 2007; Ikeda et al., 2009). The ability of a transcription factor both to activate and to repress transcription depending on context contributes substantially to the overall complexity of the transcriptional response.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Dose–response curve comparing auxin sensitivity in dda1-1, Col, and axr1-3.
Figure S2. RT-PCR using different amounts of cDNA template to demonstrate that PCRs are quantitative.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing dda1-1 (SALK_033840), axr3-1, and hy5-1 seed, A. Pepper for providing ted5-1 seeds, A. Lloyd for providing pBI-ΔGR, D. Oliver for providing pCB308, R. Simon for providing pDNG, Syngenta for use of the alc system, V. Jaganatha for generating pAlcA:DDA1-EAR lines, and A. Husbands for generating 35S:AlcR lines. We especially thank David Carter for assistance with the MCID-generated data, Bahman Ehdaie for help with statistical analyses, Mark Estelle for helpful discussions about auxin signalling, Mercedes Schroeder for technical help, and Thomas Eulgem and Harley Smith for critically reviewing the manuscript. This work was supported by the National Science Foundation (IBN-0318822 and IBN-0420202 to PSS). AM was funded by a PhD Fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (BEX 1213/02-4).
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Ammanamanchi S, Freeman JW, Brattain MG. Acetylated Sp3 is a transcriptional activator. Journal of Biological Chemistry. 2003;278:35775–35780. doi: 10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. The Plant Cell. 2007;19:1795–1808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. The Plant Cell. 2006;18:574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Banerjee U. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes and Development. 2003;17:838–843. doi: 10.1101/gad.1064803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Molecular Biology. 2005;57:559–575. doi: 10.1007/s11103-005-0698-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. The Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AJ. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. The Plant Journal. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JA, Doonan JH, Laufs P. The ethanol switch: a tool for tissue-specific gene induction during plant development. The Plant Journal. 2003;36:918–930. doi: 10.1046/j.1365-313x.2003.01922.x. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M. Auxin signaling and regulated protein degradation. Trends in Plant Science. 2004;9:302–308. doi: 10.1016/j.tplants.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Estelle M. The role of regulated protein degradation in auxin response. Plant Molecular Biology. 2002;49:401–408. [PubMed] [Google Scholar]
- Evans MMS. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. The Plant Cell. 2007;19:46–62. doi: 10.1105/tpc.106.047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Shen YP, Ma LG, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiology. 2004;135:773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiology. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes and Development. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Holm M, Deng XW. Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Molecular Biology. 1999;41:151–158. doi: 10.1023/a:1006324115086. [DOI] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Research. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. The Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. The Plant Journal. 2007;51:173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiology. 2007;144:336–346. doi: 10.1104/pp.106.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiology. 2009;151:1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. The Plant Journal. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. The Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology. 2002;49:387–400. [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. ARL1, a LOB-domain protein required for adventitious root formation in rice. The Plant Journal. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Lockington R, Scazzocchio C, Sequeval D, Mathieu M, Felenbok B. Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans. Molecular Microbiology. 1987;1:275–281. doi: 10.1111/j.1365-2958.1987.tb01933.x. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP, Hamilton DA. Artifacts in the localization of GUS activity in anthers of petunia transformed with a CaMV 35S–GUS construct. The Plant Journal. 1992;2:405–408. [Google Scholar]
- McNellis TW, von Arnim AG, Deng XW. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. The Plant Cell. 1994;6:1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/ LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Bioscience, Biotechnology, and Biochemistry. 2007;71:1269–1278. doi: 10.1271/bbb.60681. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology. 2004;2:1460–1471. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7and ARF19. The Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes and Development. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, Palme K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Molecular Plant. 2008;1:321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- Pepper AE, Chory J. Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends in Plant Science. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. The Plant Journal. 2001;28:225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiology. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2-LIKE19/ LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/ LBD18 regulate tracheary element differentiation in Arabidopsis. The Plant Cell. 2008;20:3359–3373. doi: 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G, Sauer M, Stauffer JL, Multani D, Niu X, Sakai H, Hochholdinger F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. The Plant Journal. 2007;50:649–659. doi: 10.1111/j.1365-313X.2007.03075.x. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiology. 1997;114:779–788. doi: 10.1104/pp.114.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/ DET/ FUS genes in light control of Arabidopsis seedling development. Plant Physiology. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends in Genetics. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis TW, Piekos B, Deng XW. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. The Plant Cell. 1994;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. The Plant Journal. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proceedings of the National Academy of Sciences, USA. 2008;105:16392–16397. doi: 10.1073/pnas.0803997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Molecular Biology. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- Yadav V, Kundu S, Chattopadhyay D, Negi P, Wei N, Deng X-W, Chattopadhyay S. Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. The Plant Journal. 2002;31:741–753. doi: 10.1046/j.1365-313x.2002.01395.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.