Abstract
The impact of ectomycorrhiza formation on the secretion of exoenzymes by the host plant and the symbiont is unknown. Thirty-eight F1 individuals from an interspecific Populus deltoides (Bartr.)×Populus trichocarpa (Torr. & A. Gray) controlled cross were inoculated with the ectomycorrhizal fungus Laccaria bicolor. The colonization of poplar roots by L. bicolor dramatically modified their ability to secrete enzymes involved in organic matter breakdown or organic phosphorus mobilization, such as N-acetylglucosaminidase, β-glucuronidase, cellobiohydrolase, β-glucosidase, β-xylosidase, laccase, and acid phosphatase. The expression of genes coding for laccase, N-acetylglucosaminidase, and acid phosphatase was studied in mycorrhizal and non-mycorrhizal root tips. Depending on the genes, their expression was regulated upon symbiosis development. Moreover, it appears that poplar laccases or phosphatases contribute poorly to ectomycorrhiza metabolic activity. Enzymes secreted by poplar roots were added to or substituted by enzymes secreted by L. bicolor. The enzymatic activities expressed in mycorrhizal roots differed significantly between the two parents, while it did not differ in non-mycorrhizal roots. Significant differences were found between poplar genotypes for all enzymatic activities measured on ectomycorrhizas except for laccases activity. In contrast, no significant differences were found between poplar genotypes for enzymatic activities of non-mycorrhizal root tips except for acid phosphatase activity. The level of enzymes secreted by the ectomycorrhizal root tips is under the genetic control of the host. Moreover, poplar heterosis was expressed through the enzymatic activities of the fungal partner.
Keywords: Heterosis, heritability, host genetic control, Laccaria bicolor, poplar, secreted enzymes
Introduction
The fine roots of tree species in temperate and boreal forests are symbiotically associated with fungi, forming a composite organ called ectomycorrhiza (ECM) (Smith and Read, 2008). The establishment and the functioning of ECM lead to complex morphological and physiological changes in both the plant and the fungus (Martin and Nehls, 2009; Courty et al., 2010a). The ECM symbiosis has been described as a mutualistic association where the autotrophic plant supplies photosynthates to the heterotrophic fungus, which in turn supplies water and nutrients to the host (Smith and Read, 2008). Several studies also have shown that ectomycorrhizal fungi (ECMf) are able to produce extracellular enzymes, such as proteases, involved in the direct mobilization of nutrients from organic substrates (Courty et al., 2005, 2006, 2010b; Lindahl et al., 2005; Koide et al., 2008). In addition, a given species may contribute to significant functional variations through metabolic activities (Buée et al., 2007; Courty et al., 2010b).
The ecological fitness and the metabolic activity of ECMf depend on their genotypes, environmental factors (van der Heijden and Sanders, 2002; Smith and Read, 2008), host plant genotypes (Barker et al., 2002; Linderman and Davis, 2004), and the interactions between all these factors (Khasa et al., 2002; Gehring et al., 2006; Karst et al., 2009). Recent studies also suggest that the host plant genome may play a role in determining the dominant mycorrhizal type in dually colonized hosts (van der Heijden and Kuyper, 2001; Khasa et al., 2002). However, no studies have simultaneously examined the effect of host plant genotypes and the metabolic activity of one ECMf species in controlled conditions. In the Laccaria bicolor/poplar ECM symbiosis, Tagu et al. (2005) have shown that the host genotype impacts on root colonization by the fungus. The heritability of mycorrhizal colonization of poplar was also studied (Tagu et al., 2001, 2005). However, the metabolic activity of one ECM fungal genotype colonizing different genotypes of the same host species was never studied. In the present study, the use of poplar as the host tree model was motivated by the availability of large genetic and genomic resources for this species (Brunner et al., 2004; Tuskan et al., 2006). Moreover, heritability and variability of physiological parameters [i.e. water use efficiency, dry weight and leaf maximum area (LMA)] at the family level have been intensively studied in poplar (Marron et al., 2005; Dillen et al., 2007).
In this study, a standard set of seven enzymatic activities routinely used in field studies were selected as relevant functional traits (Courty et al., 2005). The enzyme activity of a secreted laccase, an oxidative enzyme involved in the degradation of recalcitrant plant residues, such as lignin, five secreted glycosyl hydrolases (cellobiohydrolase, β-glucosidase, β-xylosidase, β-glucuronidase, and N-acetylglucosaminidase) acting on polysaccharides, and a phosphomonoesterase involved in the mobilization of phosphorus from soil organic matter were assessed. Laccaria bicolor has a small set of glycosyl hydrolases able to hydrolyse plant cell wall polysaccharides (Martin et al., 2008). However, its genome encodes several carbohydrate-active enzymes able to degrade bacterial, fungal, and animal polysaccharides (Martin et al., 2008).
The impact of the host genotype on the ECM metabolic activity is unknown. Here, the responding functional trait in focus is the capacity to produce secreted or cell wall-bound enzymes. The first objective was to determine whether the enzymatic activities expressed in mycorrhizal roots differed significantly between two parents, Populus deltoides and P. trichocarpa, and different poplar hybrid genotypes (P. deltoides×P. trichocarpa). The second objective was to determine the effect of host genotypes on fungal traits by measuring the heritability of enzymatic activities in mycorrhizal and non-mycorrhizal root tips and by assessing a possible heterosis for these traits among the progeny.
Materials and methods
Plant material, strain, and culture conditions
Poplar material consisted of 38 F1 individuals from an interspecific P. deltoides (female clone from Illinois, no. 73028-62) and P. trichocarpa (male clone from Washington, no. 101-74) controlled cross (family 54B) (Tagu et al., 2001, 2005). The ability of the two parents and the 38 breeds to form mycorrhizas was tested by inoculating them with L. bicolor S238N (Di Battista et al., 1996; Tagu et al., 2001). The 38 F1 genotypes were chosen at random among the 336 genotypes used for the construction of a genetic map (Cervera et al., 2001; Jorge et al., 2005). The L. bicolor S238N fungal strain, coming from the INRA-Nancy collection of ECMf, was maintained on Pachlewski's. This model fungal strain was chosen for its ability to form ECMs with poplar and for the avaibility of genomic resources (Tagu et al., 2001; Martin et al., 2008). The inoculum of L. bicolor S238N was prepared by aseptically growing the mycelium in a peat–vermiculite nutrient mix in glass jars for 2 months in the dark at 25 °C, and it was kept at 4 °C for 2 months before use (Le Tacon and Bouchard, 1986).
Inoculation
Cuttings of one internode of each of the 38 poplar progeny and the two parents were rooted and individually inoculated at the same time, in 1.0 l pots containing a mixture of fungal inoculum (1:9 v/v) and calcinated attapulgite (Oil Dri US Special) for 12 weeks, in a greenhouse during spring with day–night temperatures of 28 °C and 15 °C, respectively. Plants were watered during the whole experiment until measurements were taken. From the second month, a low N, low P nutrient solution was applied weekly (Frey-Klett et al., 1997). In order to control environmental heterogeneity of the greenhouse, eight replicates were examined for each poplar genotype and were randomly distributed in eight blocks. Each block contained one pot of each of the 38 progeny and the two parents.
Root colonization
Entire root systems, except roots present at 1 cm depth from the collar, were carefully washed under tap water and cut into ∼1 cm pieces. For each root system, 100 randomly selected root tips were examined and assessed as mycorrhizal or non-mycorrhizal under a stereomicroscope (magnification ×40) for calculation of ECM percentages.
Chlorophyll content, leaf morphological measurement, and dry weight
Before harvesting plants, the chlorophyll a and b content was measured with a Minolta SPAD chlorophyll meter (Minolta Corp., Ramsey, NJ, USA). Three SPAD measurements were done on three leaves of each plant and then averaged (Monje and Bugbee, 1992). To convert SPAD measures into chlorophyll content, a standard curve was built by extracting chlorophylls with the dimethylsulphoxide (DMSO) extraction technique (Monje and Bugbee, 1992; Richardson et al., 2002). Total leaf chlorophyll concentration (mg cm−2) of the extracts was calculated from the equation: 0.0202A645+0.00802A663. SPAD measurements were then converted to chlorophyll content using a third-order polynomial equation: –0.0064SPAD3+0.5895SPAD2+2.0891SPAD+10.024.
Once mycorrhizal infection had been determined, leaves, stems, and roots were separated. The leaves were placed in plastic bags and kept at 4 °C until leaf morphological measurements were completed. The leaf area (cm2) of all leaves of each plantlet was measured by using a LI-COR 3100 (Li-Cor Inc., Lincoln, NE, USA). Leaves, stems, and roots were then dried at 70 °C for 1 week. The LMA was calculated for each clone using the relationship between the area of each leaf and its corresponding dry weight.
Enzymatic activity profiling of ectomycorrhizal and non-mycorrhizal root tips
One mycorrhizal root tip and one non-mycorrhizal root tip were collected from each of the 320 cuttings in order to determine their potential enzymatic activities, using the high-throughput photometric and fluorimetric microplate assays described and detailed in Courty et al. (2005), and applied in previous studies (Buée et al., 2007; Courty et al., 2010b). As the variability of enzyme activities among ECM tips within a root system is low, one tip is sufficient to obtain a representative value (Courty et al., 2005). Each well of a 96-well microtitration plate contained either one ectomycorrhizal root tip or one non-ectomycorrhizal root tip. Seven activities were successively measured on root tips: β-xylosidase (EC 3.2.1.37), β-glucuronidase (EC 3.2.1.31), cellobiohydrolase (EC 3.2.1.91), N-acetylglucosaminidase (EC 3.2.1.14), β-glucosidase (EC 3.2.1.3), acid phosphatase (EC 3.1.3.2), and laccase (EC 1.10.3.2). The enzymes activities were expressed as pmol mm−2 min−1 of developed surface area of root tips. The developed surface area of the root tips was measured after scanning and image analysis using the Mac/Win Rhizo software (Regent Instruments, Quebec City, Canada). They correspond to the activities of enzymes present on the surface of the roots or mycorrhiza mantles and released into the medium during the incubation.
Whole-genome expression oligoarray analyses
Genes coding for laccase (Lac), N-acetylglucosaminidase (Nag), and acid phosphatase (Pap) were already known and had been characterized in the genome of L. bicolor and P. trichocarpa. As the genes involved in β-xylosidase, β-glucuronidase, cellobiohydrolase, and β-glucosidase activity had not been characterized, it was not possible to measure the corresponding transcript expression. Accumulation of predicted Lac, Nag, and Pap transcripts was detected in free-living mycelium of L. bicolor S238N, and in ectomycorrhizal and non-mycorrhizal root tips of poplar using the NimbleGen L. bicolor whole-genome expression oligoarray v2 (Martin et al., 2008) and the NimbleGen P. trichocarpa whole-genome expression oligoarray (Tuskan et al., 2006). Data are available at the GEO platform GPL2699. The L. bicolor 4-plex whole genome expression array contained 18 653 gene models with three oligonucleotide probes for each gene model. For 4702 gene models, technical duplicates were included on the oligoarray (A Kohler and F Martin, unpublished results). Average expression levels were calculated for each gene from the independent probes and were used for further analysis. To estimate the signal background and the resulting threshold value for significant expression, the mean intensity of 2032 random probes present on the microarray was calculated. Gene models with expression exceeding the threshold by ≥3 were considered to be transcribed. Raw array data were filtered for non-specific probes and renormalized using ARRAYSTAR software (DNASTAR). Three biological replicates were used. Therefore, the reported gene expression values corresponded to the mean intensity of hybridization signals obtained for the specific oligonucleotide probes. A Student t-test with false discovery rate (FDR; Benjamini–Hochberg) multiple testing corrections was applied on the data (P <0.05), using ARRAYSTAR sofware (DNASTAR).
Statistical analysis
The percentage of mycorrhizal colonization was transformed by arcsin √X/100 function prior to analysis of variance (ANOVA). β-xylosidase, β-glucuronidase, cellobiohydrolase, N-acetylglucosaminidase β-glucosidase, acid phosphatase, and laccase activities, root, shoot, and stem dry weight, and LMA were also submitted to ANOVA. The following mixed linear model was applied on an individual basis to detect significant differences among the clones:
where μ is the overall mean, B is the block effect (fixed), G is the genotype effect (random), and ϵ is the random residual error.
Restricted maximum likelihood estimates of genetic, block, and residual variance components (σ2G, σ2B, and σ2ϵ) were computed, and, for each trait, individual broad sense heritability (h2) was estimated as follows:
where n is the average number of replicates per genotype. Standard deviations (SD) were derived from classic estimation of SD for a ratio x/y where x=σ2G and y=σ2G+σ2ϵ/n.
All analyses were performed with the statistical programs JMP 5.0 (SAS Institute Inc., Cary, NC, USA) and R version 1.8.0 (R Development Core Team, 2006, www.R-project.org).
The genetic coefficient of variation (CVG) was used (Cornelius, 1994) to compare the relative amounts of genetic variation of traits with different means:
Relationships between the different traits were also analysed by Pearson linear correlations.
The developed projected area of mycorrhizal and mycorrhizal root tips were compared between genotypes by ANOVA.
Results
A total of 320 plants were harvested and studied in this experiment. Three dead plants were not used in the analysis. No significant block effect was found for any measured traits.
Poplar ecophysiological traits
Significant differences (P <0.001) were found between plant genotypes for all measured traits (chlorophyll content, LMA, and stem and root dry weight). Significant differences were found between the parents for all measured traits except for LMA.
Effect of poplar genotype on root colonization by L. bicolor
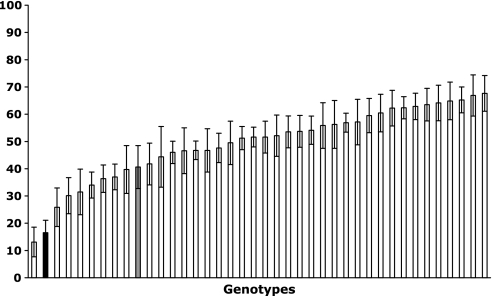
Twelve weeks after inoculation, progeny and parental clones were only colonized by L. bicolor. No other contaminant ECMf were found on roots. The two parental genotypes differed significantly in their mycorrhizal development, P. trichocarpa exhibiting a rate of colonization of 40±8%, and P. deltoides a rate of 16±4%. The percentage root colonization of the different genotypes (progeny) varied from 12±8% to 64±6%, with an average of 31% (Fig. 1). The ANOVA showed a significant genotype effect and no block effect. The developed projected area of mycorrhizal or non-mycorrhizal root tips was not significantly different between genotypes.
Fig. 1.
Percentage root colonization of the different poplar clones (n=40). Genotypes are ranked in mean percentage of root colonization. Bars represent the SE (n=8). Grey corresponds to Populus trichocarpa (male) and black to Populus deltoides (female).
Enzymatic activity patterns of the parental and the hybrid root system
For each plant, the seven enzymatic activities were measured successively on one mycorrhizal and one non-mycorrhizal root tip (40 poplar genotypes×8 plant replicates×2 root tips). Mycorrhizal root tips never lost their ability to secrete the seven enzymes under the test conditions, even if sometimes this was at a very low level (e.g. for β-xylosidase, β-glucuronidase, and laccase).
The seven enzymatic activities expressed in non-mycorrhizal roots did not differ significantly between the two parents (Table 1a). The seven enzymatic activities measured on ECM root tips differed significantly between the two parents (Table 1a): five activities (β-xylosidase, cellobiohydrolase, β-glucosidase, acid phosphatase, and laccase) had a higher level in P. trichocarpa and two (N-acetylglucosaminidase and β-glucuronidase) had a higher level in P. deltoides.
Table 1.
Average enzymatic activities of ectomycorrhizal and non-Mycorrhizal root tips of poplar
| (a) Comparison of the parents: P. trichocarpa (Pt) and P. deltoides (Pd) | ||||
| Mycorrhizal root tips |
Non-mycorrhizal root tips |
|||
| Pt | Pd | Pt | Pd | |
| β-xylosidase | 25.43±1.67 | 14.04±1.17* | 0.94±0.52 | 0.46±0.14 |
| β-glucuronidase | 23.29±1.82 | 35.47±1.47* | 0.69±0.41 | 0.81±0.51 |
| N-acetylglucosaminidase | 297.83±13.86 | 412.81±17.72* | 3.02±0.70 | 4.1±1.42 |
| Cellobiohydrolase | 86.51±2.21 | 22.42±1.61* | 0.57±0.17 | 0.62±0.24 |
| β-glucosidase | 358.94±16.96 | 119.77±9.77* | 4.13±1.47 | 2.78±1.45 |
| Acid phosphatase | 368.46±19.47 | 155.94±12.84* | 25.33±5.15 | 25.77±4.47 |
| Laccase | 5.64±0.95 | 4.76±0.72* | 0 | 0 |
| (b) Comparison of the 40 plant genotypes (the two parents and the 38 progeny). Significant differences in enzymatic activities between mycorrhizal and non-mycorrhizal root tips are also reported | ||||
| n | Mycorrhizal root tips | Non-mycorrhizal root tips | Mycorrhizal versus non-mycorrhizal ratio | |
| β-xylosidase | 317 | 31.32±1.76* | 1.23±0.07 | 25.5* |
| β-glucuronidase | 317 | 41.41±2.90* | 0.82±0.06 | 50.5* |
| N-acetylglucosaminidase | 317 | 418.30±17.26* | 3.14±0.16 | 133.2* |
| Cellobiohydrolase | 317 | 85.07±6.48* | 0.87±0.06 | 97.8* |
| β-glucosidase | 317 | 306.78±16.93* | 4.68±0.52 | 65.5* |
| Acid phosphatase | 317 | 396.65±26.42* | 24.81±1.35* | 16* |
| Laccase | 317 | 4.8±0.2 | 0 | ND |
Enzyme activities are expressed as pmol mm−2 min−1 (Courty et al., 2005). Mean and SE are given for each activity. An asterisk indicates a significant difference (P <0.001). The effect of plant genotype on enzyme activities was assessed for mycorrhizal root tips and non-mycorrhizal root tips.
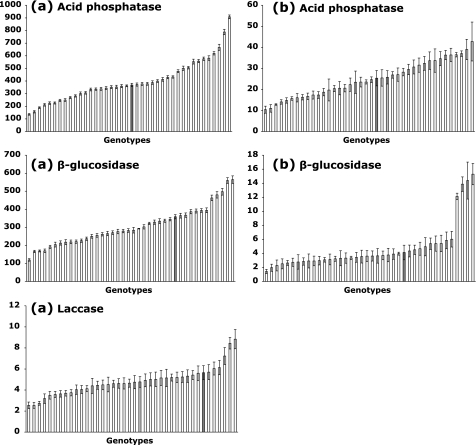
Enzyme activity patterns of mycorrhizal and non-mycorrhizal root tips of the parents and of their progeny were different (Fig. 2). Significant differences were found between plant genotypes for all activities measured on ECM root tips, except for laccase activity (Table 1b). No significant differences were found between plant genotypes for any activities measured on non-mycorrhizal root tips, except for acid phosphatase activity (Table 1b).
Fig. 2.
Values of the seven enzymatic activities successively measured on one root tip of the different poplar cuttings (40 clones, eight replicates per clone) either colonized (a) or non-colonized (b) by L. bicolor. No laccase activity was detected on non-mycorrhizal root tips. Genotypes are ranked by mean activity. Enzyme activities are expressed as pmol mm−2 min−1 (Courty et al., 2005). Bars represent the SE (n=8). Grey corresponds to Populus trichocarpa (male) and black to Populus deltoides (female).
Six of the enzymatic activities differed significantly between mycorrhizal and non-mycorrhizal roots, while no laccase activity could be detected in non-mycorrhizal roots (Table 1b). Compared with non-mycorrhizal root tips, N-acetylglucosaminidase activity was increased by >100-fold in mycorrhizas, while β-glucuronidase, cellobiohydrolase, and β-glucosidase activities were multiplied by a factor ranging between 50 and 100, and β-xylosidase and acid phosphatase between 15 and 50 (Table 1b).
Heritability
Heritability values (h2) of plant phenotypic characters ranged from 0.21 to 0.48. The highest values of heritability were found for LMA (0.48±0.01), chlorophyll content (0.45±0.01), and stem and leaf dry weight (0.43±0.01 and 0.50±0.01, respectively). The lowest value was found for root dry weight (0.21±0.01). A value of 0.45±0.02 was found for the percentage of mycorrhizal colonization. Heritability values of enzymatic activities were similar for ectomycorrhizal and non-mycorrhizal root tips, except for laccase activity, which was not detected on non-mycorrhizal root tips (0.29±0.01 in mycorrhizal root tips; 0 in non-mycorrhizal root tips). The highest heritabilities were found for N-acetylglucosaminidase (mycorrhizal root tips, 0.42±0.01; non-mycorrhizal root tips, 0.40±0.01), acid phosphatase (mycorrhizal root tips, 0.41±0.01; non-mycorrhizal root tips, 0.40±0.01), β-glucosidase (mycorrhizal root tips, 0.36±0.01; non-mycorrhizal root tips, 0.34±0.01), and cellobiohydrolase (mycorrhizal root tips, 0.33±0.02; non-mycorrhizal root tips, 0.31±0.02) activity. The lowest value was found for β-glucuronidase activity (mycorrhizal root tips 0.04±0.01; non-mycorrhizal root tips, 0.04±0.01). A medium value was found for β-xylosidase activity (mycorrhizal root tips, 0.16±0.01; non-mycorrhizal root tips, 0.19±0.01).
Heterosis
For each trait, the ratio between the average of the hybrids and the best parent (a) or between the average of the hybrids and the average of the two parents (b) was calculated (Supplementary Table S1 available at JXB online). The LMA exhibited a high positive heterosis (a= +46, b= +51), while the leaf dry weight exhibited a negative heterosis (a= – 41, b= –36). The percentage mycorrhizal colonization also exhibited a positive heterosis (a= +25, b= +75). In mycorrhizal and non-mycorrhizal root tips, all the enzymatic activities displayed a positive heterosis at least for the b values, with the exception of laccase activity in mycorrhizal root tips (a= –15, b= –8) and acid phosphatase activity in non-mycorrhizal root tips (a= –4, b= –3).
Gene expression
Expression of the genes Lcc, Nag, and Pap was assessed by whole-genome expression oligoarray analyses in poplar and L. bicolor (Table 2). In poplar, gene expression was compared between non-mycorrhizal and mycorrhizal roots. Thirty-two laccases (Lcc1–Lcc32) were detected in poplar. Three of them are mitochondrial and 27 have a signal peptide meaning that they belong to secreted pathways. Of the 32 genes coding for laccases in poplar, the expression of 11 could be assessed. Of the 11 expressed and also putatively expressed, one (Lcc6) was significantly up-regulated in mycorrhizas and two were down-regulated in mycorrhizas (Lcc16 and Lcc31). Two N-acetylglucosaminidase (Nag1 and Nag2) genes were expressed and a signal peptide was found for both of them. The expression of Nag1 and Nag2 was not modified by mycorrhizal establishment. Seven acid phosphatase genes (Pap1–Pap7) were expressed and a signal peptide was found for five of them (Pap2, Pap3, Pap5, Pap6, and Pap7). Two (Pap5 and Pap7) were significantly up-regulated in mycorrhizas.
Table 2.
Quantification by exon expression array of the transcript levels of laccase (Lcc), N-acetylglucosaminidase (Nag), and acid phosphatase (Pap) genes under different conditions
| (a) Poplar genes. The NimbleGen array analysis was carried out using P. trichocarpa root tips with or without mycorrhizal infection by L. bicolor. Transcript levels in non-mycorrhizal root tips were used as the control values. NT, not transcribed; –, gene not on the array or no reliable probe left. | ||||
| Protein ID | Signal-P | Target-P | Ratio | |
| Lcc1 | 820390 | 24 | S | 1.0 |
| Lcc2 | 557962 | 24 | S | – |
| Lcc3 | 797646 | 22 | S | NT |
| Lcc4 | 576931 | 23 | S | – |
| Lcc5 | 762473 | 25 | S | – |
| Lcc6 | 767563 | 28 | S | 4.3* |
| Lcc7 | 219290 | 29 | S | 1.4 |
| Lcc8 | 759686 | – | 1.2 | |
| Lcc9 | 653089 | 24 | S | – |
| Lcc10 | 819177 | 26 | S | NT |
| Lcc11 | 797888 | 31 | S | NT |
| Lcc12 | 579478 | 32 | S | – |
| Lcc13 | 235935 | 32 | S | – |
| Lcc14 | 831900 | 34 | S | NT |
| Lcc15 | 235930 | 32 | S | – |
| Lcc16 | 768177 | 26 | S | 0.4* |
| Lcc17 | 548008 | 32 | S | – |
| Lcc18 | 783559 | 32 | S | NT |
| Lcc19 | 822366 | 32 | S | 0.4 |
| Lcc20 | 560853 | 23 | S | NT |
| Lcc21 | 592533 | 23 | S | – |
| Lcc22 | 832603 | 31 | S | 0.3 |
| Lcc23 | 738903 | M | – | |
| Lcc24 | 574533 | 30 | S | – |
| Lcc25 | 777748 | 23 | S | 0.8 |
| Lcc26 | 738893 | M | – | |
| Lcc27 | 571858 | 33 | S | – |
| Lcc28 | 574985 | 28 | S | – |
| Lcc29 | 205176 | M | 1.6 | |
| Lcc30 | 569758 | 28 | S | – |
| Lcc31 | 420672 | 17 | S | 0.4* |
| Lcc32 | 774519 | – | 0.2 | |
| Nag1 | 772972 | 27 | S | 1.4 |
| Nag2 | 202916 | 25 | S | 1.5 |
| Pap1 | 821155 | – | 1.4 | |
| Pap2 | 831269 | 28 | S | 0.6 |
| Pap3 | 818768 | 27 | S | 0.5 |
| Pap4 | 816041 | – | 1.4 | |
| Pap5 | 272725 | 22 | S | 2.1* |
| Pap6 | 259486 | 23 | S | 1.9 |
| Pap7 | 825753 | 32 | S | 1.8* |
| (b) L. bicolor genes. The NimbleGen array analysis was carried out using P. trichocarpa (Pt) and P. deltoides (Pd) with mycorrhizal infection by L. bicolor. Transcript levels in the mycelium grown in pure culture were used as the control values. NE, not expressed in mycorrhizas (signal below the background); –, gene not on the array or no reliable probe left. | |||||
| Protein ID | Signal-P | Target-P | Ratio Pt | Ratio Pd | |
| Lcc1 | 399743 | 17 | S | 0.1 | 0.2 |
| Lcc2 | 399744 | 17 | S | – | – |
| Lcc3 | 399745 | 17 | S | 4.4 | 2.0 |
| Lcc4 | 399746 | 18 | S | 0.8 | 0.6 |
| Lcc5 | 399747 | 19 | M | NE | NE |
| Lcc6 | 399748 | 20 | S | 0.4 | 0.3* |
| Lcc7 | 399750 | 19 | S | – | – |
| Lcc8 | 399749 | 22 | S | 2.0* | 1.4 |
| Lcc9 | 399751 | 16 | S | 0.3* | 0.3* |
| Pap1 | 310810 | 21 | S | 0.6 | 0.9 |
| Nag1 | 309753 | 0.6 | 0.6* | ||
| Nag2 | 182604 | 18 | S | 0.4* | 0.4* |
The length (bp) of the signal peptide (Signal-P) was predicted with Signal P 3.0 (http://www.cbs.dtu.dk/services/SignalP/). The prediction of the subcellular location of the proteins (Target-P) was performed with TargetP 1.1 available on the webpage (http://www.cbs.dtu.dk/services/TargetP/); M, mitochondrial; S, secreted; –, unknown.
Three biological replicates were used for each treatment with NimbleGen oligoarrays (v.2.0; NG2). A Cyber-T test was performed on the mean for each transcript (*P <0.05).
For L. bicolor, gene expression was compared between mycorrhizal roots of P. trichocarpa and P. deltoides, and mycelium growing in pure culture. On the nine laccases previously described (Courty et al., 2009), one was mitochondrial (Lcc5) and eight had a signal peptide meaning that they belong to secreted pathways. Seven of these laccase genes were expressed, while two were not (Lcc2 and Lcc7). Lcc5 was only expressed in the free-living mycelium. Lcc9 was significantly down-regulated in P. deltoides and P. trichocarpa mycorrhizas, while Lcc6 was significantly down-regulated only in P. deltoides mycorrhizas and Lcc8 was up-regulated in P. trichocarpa mycorrhizas. The other laccases were not significantly regulated. The only acid phosphatase (Pap1) expressed displayed a peptide signal. Its expression was not significantly different between mycorrhizas and free-living mycelium. Among the two N-acetylglucosaminidase (Nag1 and Nag2) expressed, only Nag2 exhibited a signal peptide. The expression of Nag2 was higher in the free-living mycelium than in P. trichocarpa–L. bicolor and P. deltoides–L. bicolor mycorrhizas.
Correlations between the different traits
No poplar trait was correlated with enzymatic activities of non-mycorrhizal root tips (Table 3). LMA, and stem and root dry weight were not correlated with any activities from either ectomycorrhizal or non-mycorrhizal root tips (Table 3). Chlorophyll content was significantly negatively correlated with β-xylosidase, cellobiohydrolase, and β-glucosidase activities, three enzymes involved in cellulose and hemicellulose catabolism.
Table 3.
Correlation matrix (Pearson correlation r) between poplar traits, enzymatic activities, and percentage mycorrhizal colonization
| % | Chl | DW stem | DW roots | LMA | M Xyl | M Glr | M Nag | M Cel | M Gls | M Pho | M Lac | NM Xyl | NM Glr | NM Nag | NM Cel | NM Gls | NM Pho | |
| 1 | −0.09 | 0.20 | 0.20 | 0.06 | 0.22 | 0.14 | 0.13 | 0.07 | 0.13 | 0.04 | 0.05 | 0.05 | 0.13 | 0.02 | −0.02 | 0.06 | 0.13 | |
| Chl | 1 | −0.12 | −0.12 | −0.01 | –0.19 | −0.01 | −0.07 | –0.18 | –0.18 | −0.05 | 0.01 | −0.00 | 0.02 | 0.08 | 0.08 | 0.00 | 0.11 | |
| DW stem | 1 | 0.42 | –0.28 | −0.05 | −0.04 | 0.04 | −0.07 | −0.13 | −0.02 | −0.01 | −0.01 | 0.09 | 0.12 | 0.00 | −0.02 | 0.01 | ||
| DW roots | 1 | –0.20 | 0.05 | 0.04 | 0.06 | −0.01 | 0.02 | −0.03 | 0.06 | −0.07 | 0.00 | −0.09 | −0.09 | −0.10 | −0.02 | |||
| LMA | 1 | 0.14 | 0.09 | 0.14 | 0.06 | 0.10 | 0.12 | 0.13 | −0.08 | −0.13 | −0.04 | −0.07 | −0.06 | 0.04 | ||||
| M Xyl | 1 | 0.12 | 0.41 | 0.64 | 0.56 | 0.25 | 0.25 | 0.07 | 0.02 | −0.03 | 0.11 | −0.03 | 0.09 | |||||
| M Glr | 1 | 0.27 | 0.18 | 0.15 | 0.30 | 0.15 | 0.01 | 0.07 | −0.12 | −0.13 | −0.07 | 0.15 | ||||||
| M Nag | 1 | 0.35 | 0.33 | 0.47 | 0.31 | −0.11 | −0.08 | −0.04 | −0.01 | –0.19 | 0.32 | |||||||
| M Cel | 1 | 0.69 | 0.22 | 0.16 | 0.06 | −0.04 | 0.02 | 0.05 | −0.06 | 0.05 | ||||||||
| M Gls | 1 | 0.25 | 0.27 | 0.01 | −0.05 | −0.07 | 0.06 | −0.05 | 0.09 | |||||||||
| M Pho | 1 | 0.29 | 0.01 | 0.08 | −0.01 | 0.02 | 0.01 | 0.36 | ||||||||||
| M Lac | 1 | −0.12 | −0.05 | −0.14 | −0.06 | −0.10 | 0.21 | |||||||||||
| NM Xyl | 1 | 0.47 | 0.27 | 0.43 | 0.35 | −0.03 | ||||||||||||
| NM Glr | 1 | 0.25 | 0.39 | 0.29 | 0.04 | |||||||||||||
| NM Nag | 1 | 0.24 | 0.31 | −0.03 | ||||||||||||||
| NM Cel | 1 | 0.28 | −0.03 | |||||||||||||||
| NM Gls | 1 | 0.07 | ||||||||||||||||
| NM Pho | 1 |
Abbreviations: %, percentage mycorrhizal colonization; Chl, chlorophyll (g m−2); DW, dry weight (g); LMA, keaf maximum area (m2); M, mycorrhizal root tips; NM, non-mycorrhizal root tips; Pho, acid phosphatase; Nag, N-acetylglucosaminidase; Gls, β-glucosidase; Cel, cellobiohydrolase; Xyl, β-xylosidase; Lac, laccase; Glr, β-glucuronidase.
Correlation is significant for P <0.01 (values in bold).
Enzymatic activities of ectomycorrhizal root tips were not correlated with those of non-mycorrhizal root tips. All the enzymatic activities of mycorrhizal root tips were correlated with each other, except for laccase activity. Similarly, except for acid phosphatase, all the enzymatic activities of non-mycorrhizal root tips were correlated with each other. β-xylosidase activity of mycorrhizal roots was the only activity positively correlated with the percentage of mycorhizal infection (Table 3). Stem and root dry weights were also significantly correlated with the percentage of root colonization.
Discussion
Enzymatic activities of non-mycorrhizal root tips and mycorrhizas
The potential activities of enzymes involved in organic matter breakdown or organic phosphorus mobilization measured on poplar root tips colonized or not by L. bicolor were significantly different. Here, it was found that the ectomycorrhizal complex adds to or substitutes for enzymes secreted from poplar roots. Compared with non-mycorrhizal root tips, N-acetylglucosaminidase activity is 100-fold greater in mycorrhizas, while β-glucuronidase, cellobiohydrolase, and β-glucosidase activities were between 50- and 100-fold greater, and β-xylosidase and acid phosphatase were between 15- and 50-fold greater. Moreover, laccase activity could not be detected on non-mycorrhizal roots. By degrading organic compounds, including those from their own mycelia, and channelling nutrients directly to the host tree, ECMf have the capacity to shorten mineralization pathways in which free-living decomposers are involved. It is well known that ECMf are able to secrete enzymes which allow the release of nutrients from soil organic matter (Cullings and Courty, 2009; Courty et al., 2010a). However, the aim of this study was not to understand the role of ECMf in the release of nutrients. The enzymes measured should be considered as functional traits to study the effects of soil or host tree parameters on ECMf. It is the first time that the breadth of the modifications induced by the symbiotic association on the potential enzymatic secretion by the root system has been measured.
Expression of genes involved in enzymatic activities
Although the complete sequences of the P. trichocarpa and L. bicolor genome are available, all of the genes putatively encoding the proteins responsible for the measured activities were not identified. The correspondence between enzymatic assay and gene expression could be determined for three of them: laccase (Lcc), N-acetylglucosaminidase (Nag), and acid phosphatase (Pho).
Two genes code for a secreted N-acetylglucosaminidase (Nag1 and Nag2) involved in chitin catabolism in poplar and also two in L. bicolor. In this work, poplar Nag1 and Nag2 were expressed in both root tips and mycorrhizas but were not regulated by the symbiosis. In contrast, the expression of L. bicolor Nag1 and Nag2 was down-regulated in mycorrhizas. Nevertheless, the activity of N-acetylglucosaminidase was 130-fold greater in the mycorrhizas compared with non-mycorrhizal root tips. The following assumptions can be made: (i) L. bicolor N-acetylglucosaminidases are secreted outside the mycelium of the sheath in mycorrhizas, while the poplar N-acetylglucosaminidases are not secreted outside the root tips; (ii) L. bicolor N-acetylglucosaminidases can be involved in nitrogen mobilization from chitin by degrading its own mycelia and in defence against soil pathogenic fungi; and (iii) L. bicolor can have the same ability as Trichoderma asperellum (Ramot et al., 2004) to store a high amount of this enzyme in an active form and secrete it when the mycelium senses the substrate.
In the L. bicolor genome, nine genes coding for laccases were characterized (Courty et al., 2009). In this experiment, six putatively secreted genes were expressed and three (Lcc1, Lcc3, and Lcc4) were not significantly regulated by symbiosis. One (Lcc8) was significantly overexpressed in mycorrhizas and two (Lcc6 and Lcc9) were underexpressed. In the P. trichocarpa genome, 32 genes code for laccases, 27 display a signal peptide meaning that they belong to secreted pathways, and, in this work, 20 were expressed in root tips. Despite the large number of laccase genes which were expressed, no laccase activity was detected by the ABTS test on non-mycorrhizal root tips. This means that poplar laccases are not secreted in the rhizosphere. In Arabidopsis thaliana, only a few of the laccase genes were expressed in a pattern that could be considered consistent with a major role for these enzymes in lignin deposition (McCaig et al., 2005). Poplar laccases seems to be not cell wall bound, or secreted outside the cells. They are probably involved in the polymerization of lignin precursors or in other functions.
Acid phosphatases, able to free phosphate groups from complex organic compounds, are widespread in living organisms. Both ECMf and plants secrete acid phosphatases in the rhizosphere. In most of the studies, mycorrhizas secrete more phosphatases than non-colonized roots (Colpaert et al., 1997; Conn and Dighton, 2000). Nevertheless, there are some exceptions (Cumming, 1996). ECMf exhibit high phosphatase release in their environment, particularly under mineral phosphorus deficiency (Dighton, 1983; Nygren and Rosling, 2009). The L. bicolor genome comprises only one gene coding for a putative secreted acid phosphatase, while the P. trichocarpa genome contains five. The phosphatase Pap1 from L. bicolor was not regulated by symbiosis. Pap5 and Pap7 from poplar were significantly highly expressed under mycorrhizal conditions, whereas the three others (Pap2, Pap3, and Pap6) were not significantly expressed.
Ezawa et al. (2005) have shown, on Tagetes petala in symbiosis with Archaeospora leptoticha, that the level of transcripts of the T. petala acid phosphatase (TpPAP1) was increased 8-fold by A. leptoticha colonization. The present results support the hypothesis of Ezawa et al. (2005) on the fungal activation of the low-phosphate adaptation system of the plant partner and seem to show that the same mechanism of plant phosphatase activation exists in both arbuscular mycorrhizas and ECMs.
Another hypothesis could be involved in the explanation of the differences in enzyme secretion between non-mycorrhizal and mycorrhizal roots. Inside the mycorrhizas, the root tissues, being isolated from the external medium by the fungal sheath, probably contribute poorly to enzyme secretion. The ability to degrade cellulose, hemicelluloses, and lignin is widespread among fungi and soil bacteria (i.e. Streptomyces sp., Bacillus sp., and Cellulomonas sp.; Lynd et al., 2002). However, it is assumed that most of the cellulose degradation in soil is performed by fungi (de Boer et al., 2005). Even if it has been shown that laccase genes were present in bacteria (Kellner et al., 2008), bacterial lignin degradation appears to be negligible in terrestrial environments compared with fungal lignin degradation (Peng et al., 2002). This is supported by the fact that no laccase activity was found on non-mycorrhizal root tips. Among bacteria, Collimonas sp. display chitinolytic activities (de Boer et al., 2004). However, these bacteria are present under specific conditions, completely different from greenhouse experiments with artificial substrate. Thus, despite the fact that this experiment was performed in non-axenic conditions, it can be assumed that the secreted enzymes that were measured in mycorrhizas were mainly due to fungal activity.
Host genetic control of ECM enzyme secretion
The enzymatic activities expressed in mycorrhizal roots differed significantly between the two parents, while it did not differ in non-mycorrhizal roots. Significant differences were found between poplar genotypes for all enzymatic activities measured on ECMs except for laccase activity. In contrast, no significant differences were found between poplar genotypes for enzymatic activities of non-mycorrhizal root tips, except for acid phosphatase activity.
Heritability values of enzymatic activities were similar for ectomycorrhizal and non-mycorrhizal root tips, except for β-glucuronidase in both types of roots and for laccase that was not detected on non-mycorrhizal root tips. It is remarkable to find a high heritability value among the poplar genotypes for the enzymatic secretions of mycorrhizal roots, which are mainly due to fungal activity.
Several previous studies have demonstrated significant genetic variability within plants and/or fungal species for symbiotic capability in mycorrhizal interactions. Rosado et al. (1994) reported a high value of heritability for colonization of Pinus ellilotii by the ECMf Pisolithus tinctorius, and moderate heritability for the development of P. tinctorius extramatrical mycelium. Eucalyptus grandis, E. globulus, E. marginata, and Pinus muricata varied greatly in their growth response to different Pisolithus and Rhizopogon genotypes, respectively (Tonkin et al., 1989; Burgess et al., 1994; Piculell et al., 2008). Tagu et al. (2005) have already shown that the ability of poplar to form ECMs is under its genetic control. Other studies with contrasting results have found that the plant genotype can play a dominant role in controlling the associated soil microbial communities (Mari et al., 2003; Korkama et al., 2007). Short-term experiment have either shown variations in mycorrhizal colonization, in microbial and in mycorrhizal communities (Gehring and Whitham, 2002; Gehring et al., 2006; Barbour et al., 2009; Lojewski et al., 2009), or few differences in arbuscular fungal and bacterial communities (Bever et al., 1996; Madritch and Hunter, 2002). In the present study, the degree of fungal enzymatic secretion is modulated according to the poplar genotype. An explanation could be that the host genotype controls the amount of fungal tissue in the mantle and that enzyme activity is determined by the amount of fungal tissue present on the root. However, no significant differences were found in the projected area of mycorrhized or non-mycorrhized root tips between genotypes. Therefore, this means that the amount of mycelium in the root tip is similar whatever the genotype. These results suggest the potential for the poplar genome to drive the microbial–plant interaction, to create environments to which ECMf can respond and that could be explained by the ‘extended phenotype’ phenomenom (Schweitzer et al., 2008; Whitham et al., 2008). As defined by Whitham et al. (2003), the heritable genetic variation within individual species (poplar in this study) has community and ecosystem consequences. In addition, a high positive heterosis was found for the capacity of poplar to form mycorrhizas (h2 a= +4 %). Positive heterosis was found for characters such as LMA, dry weight of roots, and for five of the seven enzymatic activities of mycorrhizal roots. Heterosis for poplar hybrids is a well-known phenomenon (Li and Wu, 1997; Marron et al., 2006). Heterosis is determined by non-mutually exclusive mechanisms, including genome-wide dominance complementation, locus-specific overdominance effects, and epistasis, although the relative contribution of each of these mechanisms is still unclear (Lippman and Zamir, 2007). However, it is also the first time that it is has been shown that plant heterosis could be expressed through the physiological activity of the fungal partner.
Conclusion
The genetic diversity in tree species can influence fluxes of nutrients as well as interactions with soil microorganisms. Assessing tree genotype×environment interactions is a major challenge in functional ecology. In this study, the data linked and quantified the general relationships between poplar plant genetics, ECM fungal infection, and physiological parameters. In the association L. bicolor/poplar, variations in plant and fungal responses in these controlled conditions illustrate the broad plasticity of the interaction. In this study, the role of poplar genetics in determining both poplar growth characteristics and fungal activities has been highlighted.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Heterosis calculated as the ratio between the average of the hybrids and the best parent or between the average of the hybrids and the average of the two parents.
Supplementary Material
Acknowledgments
PEC was supported by a grant from the French Ministry of Ecology and Sustainable Development, and JL by a scholarship from the Région Lorraine/INRA; part of this research has been supported by the Biological Invasions program of the same Ministry. This project was also supported by the European Network of Excellence EVOLTREE. PEC gratefully acknowledges the Swiss National Science Foundation for current support. We thank Joseph Armento and Krista Plett for language corrections and helpful comments on the manuscript, and Francis Martin for valuable discussion.
References
- Barbour RC, Baker SC, O'Reilly-Wapstra JM, Harvest TM, Potts BM. A footprint of tree-genetics on the biota of the forest floor. Oikos. 2009;118:1917–1923. [Google Scholar]
- Barker SJ, Duplessis S, Tagu D. The application of genetic approaches for investigations of mycorrhizal symbioses. Plant and Soil. 2002;244:85–95. [Google Scholar]
- Bever JD, Morton J, Antonovics J, Schultz P. Host specificity and diversity of glomalean fungi: an experimental approach in an old-field community. Journal of Ecology. 1996;84:71–82. [Google Scholar]
- de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiology Review. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- de Boer W, Leveau JHJ, Kowalchuk GA, Gunnewiek PJAK, Abeln ECA, Figge MJ, Sjollema K, Janse JD, van Veen JA. Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. International Journal of Systematic and Evolutionary Microbiology. 2004;54:857–864. doi: 10.1099/ijs.0.02920-0. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Busov VB, Strauss SH. Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends in Plant Science. 2004;9:49–56. doi: 10.1016/j.tplants.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Buée M, Courty PE, Mignot D, Garbaye J. Soil niche effect on species diversity and catabolic activities in an ectomycorrhizal fungal community. Soil Biology and Biochemistry. 2007;39:1947–1955. [Google Scholar]
- Burgess T, Dell B, Malajczuk Thompson N. Variation in mycorrhizal development and growth stimulation by 20 Pisolithus isolates inoculated on to Eucalyptus grandis W. Hill ex Maiden. New Phytologist. 1994;127:731–739. doi: 10.1111/j.1469-8137.1994.tb02977.x. [DOI] [PubMed] [Google Scholar]
- Cervera MT, Storme V, Ivens B, Gusmao J, Liu BH, Hostyn V, Hostyn Van Slycken J, Van Montagu M, Boerjan W. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. doi: 10.1093/genetics/158.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert JV, van Laere A, van Tichelen KK, van Assche JA. The use of inositol hexaphosphate as a phosphorus source by mycorrhizal and non-mycorrhizal Scots pine (Pinus sylvestris) Functional Ecology. 1997;11:407–415. [Google Scholar]
- Conn C, Dighton J. Litter quality influences on decomposition, ectomycorrhizal community structure and mycorrhizal root surface acid phosphatase activity. Soil Biology and Biochemistry. 2000;32:489–496. [Google Scholar]
- Cornelius J. Heritabilities and additive genetic coefficients of variation in forest trees. Canadian Journal of Forest Research. 1994;24:372–379. [Google Scholar]
- Courty PE, Buée M, Diedhiou AG, Frey-Klett P, Le Tacon F, Rineau F, Turpault MP, Uroz S, Garbaye J. The role of ectomycorrhizal communities in forest ecosystem processes: new perspectives and emerging concepts. Soil Biology and Biochemistry. 2010b;42:679–698. [Google Scholar]
- Courty PE, Franc A, Garbaye J. Temporal and functional pattern of secreted enzyme activities in an ectomycorrhizal community. Soil Biology and Biochemistry. 2010a;42:2022–2025. [Google Scholar]
- Courty PE, Hoegger P, Kilaru S, Kohler A, Buée M, Garbaye J, Martin F, Kües U. Phylogenetic analysis, genomic organization and expression analysis of multicopper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytologist. 2009;182:736–750. doi: 10.1111/j.1469-8137.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- Courty PE, Pouysegur R, Buée M, Garbaye J. Laccase and phosphatase activities of the dominant ectomycorrhizal types in a lowland oak forest. Soil Biology and Biochemistry. 2006;38:1219–1222. [Google Scholar]
- Courty PE, Pritsch K, Schloter M, Hartmann A, Garbaye J. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytologist. 2005;167:309–319. doi: 10.1111/j.1469-8137.2005.01401.x. [DOI] [PubMed] [Google Scholar]
- Cullings K, Courty PE. Saprotrophic capabilities as functional traits to study functional diversity and resilience of ectomycorrhizal communities. Oecologia. 2009;161:661–664. doi: 10.1007/s00442-009-1434-6. [DOI] [PubMed] [Google Scholar]
- Cumming JR. Phosphate-limitation physiology in ectomycorrhizal pitch pine (Pinus rigida) seedlings. Tree Physiology. 1996;16:977–983. doi: 10.1093/treephys/16.11-12.977. [DOI] [PubMed] [Google Scholar]
- Di Battista C, Selosse MA, Bouchard D, Stenström E, Le Tacon F. Variations in symbiotic efficiency, phenotypic characters and ploidy level among different isolates of the ectomycorrhizal basidiomycete Laccaria bicolor strain S238. Mycological Research. 1996;100:1315–1324. [Google Scholar]
- Dighton J. Phosphatase production by mycorrhizal fungi. Plant and Soil. 1983;71:455–462. [Google Scholar]
- Dillen SY, Marron N, Bastien C, Ricciotti L, Salani F, Sabatti M, Pinel MPC, Rae AM, Taylor G, Ceulemans R. Effects of environment and progeny on biomass estimations of five hybrid poplar families grown at three contrasting sites across Europe. Forest Ecology and Management. 2007;252:12–23. [Google Scholar]
- Ezawa T, Hayatsu M, Saito M. A new hypothesis on the strategy for acquisition of phosphorus in arbuscular mycorrhiza: up-regulation of secreted acid phosphatase gene in the host plant. Molecular Plant-Microbe Interactions. 2005;18:1046–1053. doi: 10.1094/MPMI-18-1046. [DOI] [PubMed] [Google Scholar]
- Frey-Klett P, Pierrat JC, Garbaye J. Location and survival of mycorrhiza helper Pseudomonas fluorescens during establishment of ectomycorrhizal symbiosis between Laccaria bicolor and Douglas fir. Applied and Environmental Microbiology. 1997;63:139–144. doi: 10.1128/aem.63.1.139-144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring CA, Mueller R, Whitham TG. Environmental and genetic effects on the formation of ectomycorrhizal and arbuscular mycorrhizal associations in cottonwoods. Oecologia. 2006;149:158–164. doi: 10.1007/s00442-006-0437-9. [DOI] [PubMed] [Google Scholar]
- Gehring CA, Whitham TG. Mycorrhiza–herbivore interactions: population and community consequences. In: van der Heijden MGA, Sanders IR, editors. Mycorrhizal ecology. Berlin: Springer; 2002. pp. 295–320. [Google Scholar]
- Jorge V, Dowkiw A, Faivre-Rampant P, Bastien C. Genetic architecture of qualitative and quantitative Melampsora larici-populina leaf rust resistance in hybrid poplar: genetic mapping and QTL detection. New Phytologist. 2005;167:113–127. doi: 10.1111/j.1469-8137.2005.01424.x. [DOI] [PubMed] [Google Scholar]
- Karst J, Jones MD, Turkington R. Ectomycorrhizal colonization and intraspecific variation in growth responses of lodgepole pine. Plant Ecology. 2009;200:161–165. [Google Scholar]
- Kellner H, Luis P, Zimdars D, Kiesel B, Buscot F. Diversity of laccase-like multicopper-oxidase genes in forest and grassland Cambisol soil samples. Soil Biology and Biochemistry. 2008;40:638–648. [Google Scholar]
- Khasa DP, Hambling B, Kernaghan G, Fung M, Ngimbi E. Genetic variability in salt tolerance of selected boreal woody seedlings. Forest Ecology and Management. 2002;165:257–269. [Google Scholar]
- Koide RT, Sharda JN, Herr JR, Malcolm GM. Ectomycorrhizal fungi and the biotrophy–saprotrophy continuum. New Phytologist. 2008;178:230–233. doi: 10.1111/j.1469-8137.2008.02401.x. [DOI] [PubMed] [Google Scholar]
- Korkama T, Fritze H, Pakkanen A, Pennanen T. Interactions between extraradical ectomycorrhizal mycelia, microbes associated with the mycelia and growth rate of Norway spruce (Picea abies) clones. New Phytologist. 2007;173:798–807. doi: 10.1111/j.1469-8137.2006.01957.x. [DOI] [PubMed] [Google Scholar]
- Le Tacon F, Bouchard D. Effects of different ectomycorrhizal fungi on growth of larch, Douglas fir, Scots pine and Norway spruce seedlings in fumigated nursery soil. Oecologia Applicata. 1986;7:389–402. [Google Scholar]
- Li B, Wu R. Heterosis and genotype×environment interactions of juvenile aspens in two contrasting sites. Canadian Journal of Forest Research. 1997;27:1525–1537. [Google Scholar]
- Lindahl BD, Finlay RD, Cairney JWG. Enzymatic activities of mycelia in mycorrhizal fungal communities. In: Dighton J, White JF, Oudemans P, editors. The fungal community: its organization and role in the ecosystem. 3rd edn. Boca Raton, FL: CRC Press; 2005. pp. 331–348. [Google Scholar]
- Linderman RG, Davis EA. Varied response of marigold (Tagetes spp.) genotypes to inoculation with different arbuscular mycorrhizal fungi. Scientia Horticulturae. 2004;99:67–78. [Google Scholar]
- Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends in Genetics. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Lojewski NR, Fischer DG, Bailey JK, Schweitzer JA, Whitham TG, Hart SC. Genetic basis of aboveground productivity in two native Populus species and their hybrids. Tree Physiology. 2009;29:1133–1142. doi: 10.1093/treephys/tpp046. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiology Molecular Biology Review. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madritch MD, Hunter MD. Phenotypic diversity influences ecosystem functioning in an oak sandhills community. Ecology. 2002;83:2084–2090. [Google Scholar]
- Mari S, Jonsson A, Finlay R, Ericsson T, Kähr M, Eriksson G. Genetic variation in nitrogen uptake and growth in mycorrhizal and nonmycorrhizal Picea abies (L.) Karst. seedlings. Forest Science. 2003;49:258–267. [Google Scholar]
- Marron N, Bastien C, Sabatti M, Taylor G, Ceulemans R. Plasticity of growth and sylleptic branchiness in two poplar families grown at three sites across Europe. Tree Physiology. 2006;26:935–946. doi: 10.1093/treephys/26.7.935. [DOI] [PubMed] [Google Scholar]
- Marron N, Villar M, Dreyer E, Delay D, Boudouresque E, Petit JM, Delmotte FM, Guehl JM, Brignolas F. Diversity of leaf traits related to productivity in 31 Populus deltoides– Populus nigra clones. Tree Physiology. 2005;25:425–435. doi: 10.1093/treephys/25.4.425. [DOI] [PubMed] [Google Scholar]
- Martin F, Nehls U. Harnessing ectomycorrhizal genomics for ecological insights. Current Opinion in Plant Biology. 2009;12:508–515. doi: 10.1016/j.pbi.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahrén D, et al. The genome sequence of the basidiomycete fungus Laccaria bicolor provides insights into the mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Monje OA, Bugbee B. Inherent limitations of nondestructive chlorophyll meters: a comparison of two types of meters. Hortscience. 1992;27:69–71. [PubMed] [Google Scholar]
- McCaig B, Meagher RB, Dean JFD. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta. 2005;221:619–636. doi: 10.1007/s00425-004-1472-6. [DOI] [PubMed] [Google Scholar]
- Nygren CMR, Rosling A. Localisation of phosphomonoesterase activity in ectomycorrhizal fungi grown on different phosphorus sources. Mycorrhiza. 2009;19:197–204. doi: 10.1007/s00572-008-0223-0. [DOI] [PubMed] [Google Scholar]
- Peng X, Masai E, Kitayama H, Harada K, Katayama Y, Fukuda M. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Applied and Environmental Microbiology. 2002;68:4407–4415. doi: 10.1128/AEM.68.9.4407-4415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piculell BJ, Hoeksema JD, Thompson JN. Interactions of biotic and abiotic environmental factors in an ectomycorrhizal symbiosis, and the potential for selection mosaics. BMC Biology. 2008;6:23. doi: 10.1186/1741-7007-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot O, Viterbo A, Friesem D, Oppenheim A, Chet I. Regulation of two homodimer hexosaminidases in the mycoparasitic fungus Trichoderma asperellum by glucosamine. Current Genetics. 2004;45:205–213. doi: 10.1007/s00294-003-0478-0. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- Richardson AD, Duigan SP, Berlyn GP. An estimation of non-invasive methods to estimate foliar chlorophyll content. New Phytologist. 2002;153:185–194. [Google Scholar]
- Rosado SCS, Kropp BR, Piche Y. Genetics of ectomycorrhizal symbiosis. I. Host plant variability and heritability of ectomycorrhizal and root traits. New Phytologist. 1994;126:105–110. [Google Scholar]
- Schweitzer JA, Bailey JK, Fischer DG, LeRoy CJ, Lonsdorf EV, Whitham TG, Hart SC. Plant–soil–microorganism interactions: heritable relationship between plant genotype and associated soil microorganisms. Ecology. 2008;89:773–781. doi: 10.1890/07-0337.1. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal symbiosis. 3rd edn. London: Academic Press; 2008. [Google Scholar]
- Tagu D, Bastien C, Faivre-Rampant P, Garbaye J, Vion P, Villar M, Martin F. Genetic analysis of phenotypic variation for ectomycorrhiza formation in an interspecific F1 poplar full-sib family. Mycorrhiza. 2005;15:87–91. doi: 10.1007/s00572-004-0302-9. [DOI] [PubMed] [Google Scholar]
- Tagu D, Faivre Rampant P, Lapeyrie F, Frey-Klett P, Vion P, Villar M. Variation in the ability to form ectomycorrhizas in the F1 progeny of an interspecific poplar (Populus spp.) cross. Mycorrhiza. 2001;10:237–240. [Google Scholar]
- Tonkin CM, Malajczuk N, McComb JA. Ectomycorrhizal formation by micropropagated clones of Eucalyptus marginata inoculated with isolates of Pisolithus tinctorius. New Phytologist. 1989;111:209–214. doi: 10.1111/j.1469-8137.1989.tb00684.x. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa. Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- van der Heijden EW, Kuyper TW. Laboratory experiments imply the conditionality of mycorrhizal benefits for Salix repens: role of pH and nitrogen to phosphorus ratios. Plant and Soil. 2001;228:275–290. [Google Scholar]
- van der Heijden M, Sanders I. Mycorrhizal ecological studies. Berlin: Springer Verlag; 2002. [Google Scholar]
- Whitham TG, Young WP, Martinsen GD, et al. Community and ecosystem genetics: a consequence of the extended phenotype. Ecology. 2003;84:559–573. [Google Scholar]
- Whitham TG, DiFazio SP, Schweitzer JA, Shuster SM, Allan GJ, Bailey JK, Woolbright SA. Extending genomics to natural communities and ecosystems. Science. 2008;320:492–495. doi: 10.1126/science.1153918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.