Abstract
Isolated, clonal populations of cells are rarely found in nature. The emergent properties of microbial consortia present a challenge for the systems approach to biology, as chances for competition, communication, or collaboration multiply with the number of interacting agents. This review focuses on recent work on intercourse within biofilms, among quorum-sensing populations, and between cross-feeding metabolic cooperators. New tools from synthetic biology allow microbial interactions to be designed and tightly controlled, creating valuable model systems. We address both natural and synthetic partnerships, with an emphasis on how system behaviors derive from the properties of their components. Essential features of microbial biology arose in the context of a very mixed culture and cannot be understood without unscrambling it.
Keywords: Cooperation, coculture, consortium, quorum sensing
Microbes in context
Biology as it is practiced today would not be possible if microbes could not be isolated. Homogenous and clonal microbial cultures are the subject of nearly all microbiological studies. Axenic culture techniques have allowed indispensable gains in technical simplicity, but at the cost of divorcing many microbial strains from their natural context. Laboratory strains are usually exposed only to their clonal sisters, but in evolution most strains spend most of their time embedded in a rich concourse of potential competitors or collaborators.
Adaptations to the concourse are likely to be central to microbial biology. Aggregation, as an evolutionary innovation, occurred near the base of the tree of life. Structured, filamentous microfossils have been found that are >3 billion years old (Rasmussen 2000; Hall-Stoodley et al. 2004). Free-floating plankton are far surpassed by densely packed biofilms, in terms of total global biomass and metabolic activity (Costerton 2007).
Our knowledge of the biology of an individual microbe will remain incomplete until it is connected to a local ecology. More than 99% of microorganisms have not been successfully cultured (Amann et al. 1995; Hugenholtz et al. 1998; Vartoukian et al. 2010). Among them may be strains that require metabolites, signals, or structure provided by the concourse. Approximately one-third of the genes in Escherichai coli remain uncharacterized (Keseler et al. 2009). Most show no obvious fitness defects in rich axenic culture when deleted (Kolisnychenko et al. 2002; Baba et al. 2006). Roughly half of E. coli genes are not expressed in minimal medium (Richmond et al. 1999). The function of certain genes may therefore be revealed only in the context of the concourse.
Microbial interactions are a challenge for the systems approach to biology, which seeks to derive and predict the emergent properties of complex systems from knowledge of their component parts. As individual strains are characterized in detail, it should be possible to extrapolate to higher-order multistrain interactions. Yet the number of possible interactions grows exponentially with the local diversity, and up to hundreds of genotypes may coexist in a community (Kolenbrander 2000). Most single strains cannot be characterized in isolation and occupy structurally complex microenvironments that are difficult to replicate in the laboratory.
One answer to the challenge of this complexity is the development of new technologies that allow natural microbial interactions to be studied in situ. A suite of recently developed tools can inspect the genetic diversity, microstructure, and biochemistry of natural populations without culturing them. These important developments have been reviewed elsewhere and are not discussed here (Raes et al. 2007; Tringe and Hugenholtz 2008; Orphan 2009). Such technologies help to characterize the composition and collective properties of a concourse taken as a whole—a top-down approach.
This review focuses on recently developed model systems for microbial interaction. A bottom-up approach to understanding the concourse begins with the quantitative study of interactions among small numbers of culturable strains. Progress is proceeding toward a set of principles sufficiently general to be applied in natural environments and to interactions that cannot yet be studied directly. This work is a natural extension of the integrative, systems approach to microbiology and draws on the mathematical and conceptual resources of that discipline.
In particular, synthetic biology, in which organisms are rationally engineered to elicit designed behavior, plays an integral role in these studies. Biological interactions can be composed that are otherwise unknown in nature, in species that were previously autonomous. Specific modes of interaction can be produced independently and treated separately from other adaptations that accompany natural relationships. In this way, synthetic biology allows testing of hypotheses that would not otherwise be accessible.
The ability to design novel interactions as well as study existing ones means that microbial experiments can complement and extend classical plant and animal ecology, in which many of the principles of biological interaction were first described. The synthetic relationships we discuss take components and inspiration from natural systems, and in turn can reveal their key functional dynamics.
Natural syntrophy
Cooperative digestion
All metabolites are recycled, with the product of any given class of metabolism becoming the substrate of another. In this sense, the metabolism of each microbe must depend on the nutritive context established by other cells. The term syntrophy is used to describe interactions that are both specific and beneficial, and that open or expand metabolic niches that would not otherwise be available.
Syntrophic metabolism is common in anaerobic marine and freshwater sediments and in environments of extreme pH and temperature (McInerney et al. 2008). The described classes of anaerobic syntrophy generally represent adaptations to the energetic challenge of oxidizing organic molecules in the absence of oxygen. While some syntrophic interactions are revealed only under specific conditions, others appear to be the primary metabolic mode for the participating strains.
The well-described interaction between the Gram-negative bacterium Desulfovibrio vulgaris Hildenborough and the archaeon Methanococcus maripaludis S2 is mutually beneficial and obligate in defined media (Stolyar et al. 2007; Walker et al. 2009). D. vulgaris is an anaerobe that can be cultured axenically when given organic molecules to catabolize and sulfate as an electron sink. M. maripaludis is a methanogen, also strictly anaerobic, that grows in pure culture by fixing CO2 using hydrogen gas as an energy source.
Thus D. vulgaris reduces sulfate, M. maripaludis consumes hydrogen, and neither can grow alone in media lacking both. However, robust growth is observed under the same conditions when the strains are cultured together. With lactate, for example, as the carbon source, methanogenesis drives syntrophic growth
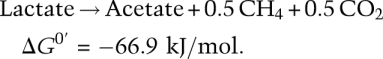 |
D. vulgaris performs the first half of this reaction, fermenting lactate to acetate, CO2, and hydrogen:
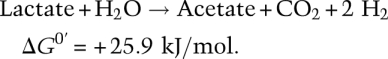 |
M. maripaludis completes the reaction by using hydrogen to reduce CO2 to methane:
M. maripaludis cannot grow alone because H2 is unavailable, and D. vulgaris cannot grow alone because hydrogen production is endergonic under standard conditions. The hydrogen-scavenging activity of the methanogen drives the overall reaction. Thus, both participants benefit from the syntrophy, and together they are able to colonize the new niche.
Even when a syntrophy is not obligate, it can be highly beneficial. Ruminococcus albus will autonomously ferment sugars to alcohol. However, when it is grown in coculture with the hydrogen-scavenging methanogen Wolinella succinogenes, R. albus diverts production from alcohol to secrete hydrogen and acetate (Iannotti et al. 1973). The continuous removal of hydrogen from the system thermodynamically enables the acetate/H2 pathway. This metabolism can, in principle, conserve more energy—four ATP per glucose, up to twice the typical yield.
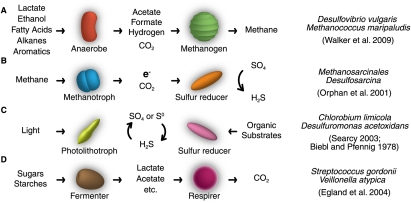
Variations on this theme are common, with a primary digester transferring electrons to a secondary digester via a regenerable electron shuttle (Schink 2006). Intermediates transferred in similar pathways include acetate, hydrogen, or formate (Dong and Stams 1995). The downstream partner in such relationships, invariably a methanogen, also receives a carbon source, in the form of CO2 (Fig. 1A).
Figure 1.
Examples of natural metabolic cooperation. Most natural syntrophic relationships involve the exchange of waste or metabolic by-products. The right column notes an example pair of partners for each class of interaction.
Disposal of excess electrons through syntrophy allows the fermentation of a variety of substrates. Alcohols, formate (Dolfing et al. 2008), fatty acids (Schink and Stieb 1985), alkanes (Zengler et al. 1999), amino acids (Widdel et al. 1988; Plugge et al. 2002), and even aromatic compounds (Kunapuli et al. 2007) can yield energy anaerobically through this mechanism. Globally, the fermentations that break down much of the organic material that falls into anaerobic sediments are in large part carried out syntrophically. This chemistry also contributes a substantial amount of the greenhouse gas methane to the global carbon cycle (Crutzen 1994).
The methanogenesis that drives the above anaerobic fermentation can be reversed when excess sulfate is available as an electron sink (Fig. 1B). In anoxic methane-rich sediments, methane is oxidized by Methanosarcinales archaea in close association with sulfate-reducing Desulfosarcina (Boetius et al. 2000; Orphan et al. 2001). Interestingly, the electron shuttle in this system is yet to be identified, although it does not appear to be any of the typical candidates of hydrogen, formate, or acetate (Nauhaus et al. 2005).
Syntrophic relationships can recycle sulfur as an electron carrier, particularly when sulfur is limiting for growth. Sulfur is a valuable resource anaerobically, and can represent either a source or a sink of electrons, depending on its redox state and the availability of other redox substrates. However, reduced sulfur in the form of sulfide is toxic and will inhibit the growth of even those organisms adapted to produce or consume it (O'Flaherty et al. 1998).
Photolithotrophs such as Prosthecochloris aestuarii or Chlorobium limicola can be cocultured with sulfur-reducing bacteria such as Desulfuromonas and a catalytic amount of sulfur added as a redox shuttle (Fig. 1C). C. limicola use energy from light to oxidize sulfide to sulfate, at light intensities much lower than those needed by typical phototrophs. Sulfur-reducing bacteria such as Desulfuromonas will anaerobically oxidize organic molecules and reduce sulfate back to sulfide, completing the cycle. Either strain growing alone will be inhibited by a toxic excess of sulfide (Pfennig and Biebl 1978).
Under microaerobic, sulfate-rich conditions, Desulfovibrio desulfuricans catabolizes lactate with sulfate to acetate and sulfide. Thiobacillus thioparus exploits residual oxygen to productively reoxidize the sulfide. These bacteria co-occur at oxygen–sulfide interfaces near hydrothermal vents, in marine sediments, or in waste water treatment systems. Because D. deslufiricans is an obligate anaerobe, it benefits from both the recycling of sulfur and the oxygen-scavenging activity of T. thioparus (Ende et al. 1997).
These documented cases probably represent only a fraction of naturally occurring syntrophic associations. Described syntrophs were amenable to discovery and characterization because their interaction is generally facultative and the individual players can be isolated separately given the correct medium. Or, in the case of “Methanobacillus omelianskii,” a two-species syntrophy was isolated jointly and mistaken for a single prototroph (Barker 1939; Wolfe et al. 1967).
The transfer of waste products is a common theme in described metabolic mutualisms. Even when syntrophic association is mutually obligate, the flow of energy and electrons is mostly one-way. In aerobic conditions, the disposal of excess reducing equivalents through respiration is usually unproblematic. The efficient disposal of waste electrons without oxygen is biochemically and energetically challenging, contributing to the emergence of complex syntrophic strategies for anaerobic metabolism.
By-product exchange
A distinct form of waste syntrophy appears to be common in the rich nutritive environment of animal digestive systems. Streptococcus gordonii and Veillonella atypica colonize human dental plaque. S. gordonii ferments starches to lactic acid. V. atypica cannot ferment sugar, but uses lactic acid as a preferred carbon source (Mikx and van der Hoeven 1975). The presence of V. atypica stimulates production of amylase by S. gordonii, breaking down starch to sugar that is fermented to lactic acid (Fig. 1D). This effect appears to be mediated by a small molecule communicated from V. atypica to S. gordonii over short distances within biofilms (Egland et al. 2004). It is not clear if S. gordonii benefits from the increase in amylase expression or the removal of lactic acid by its partner.
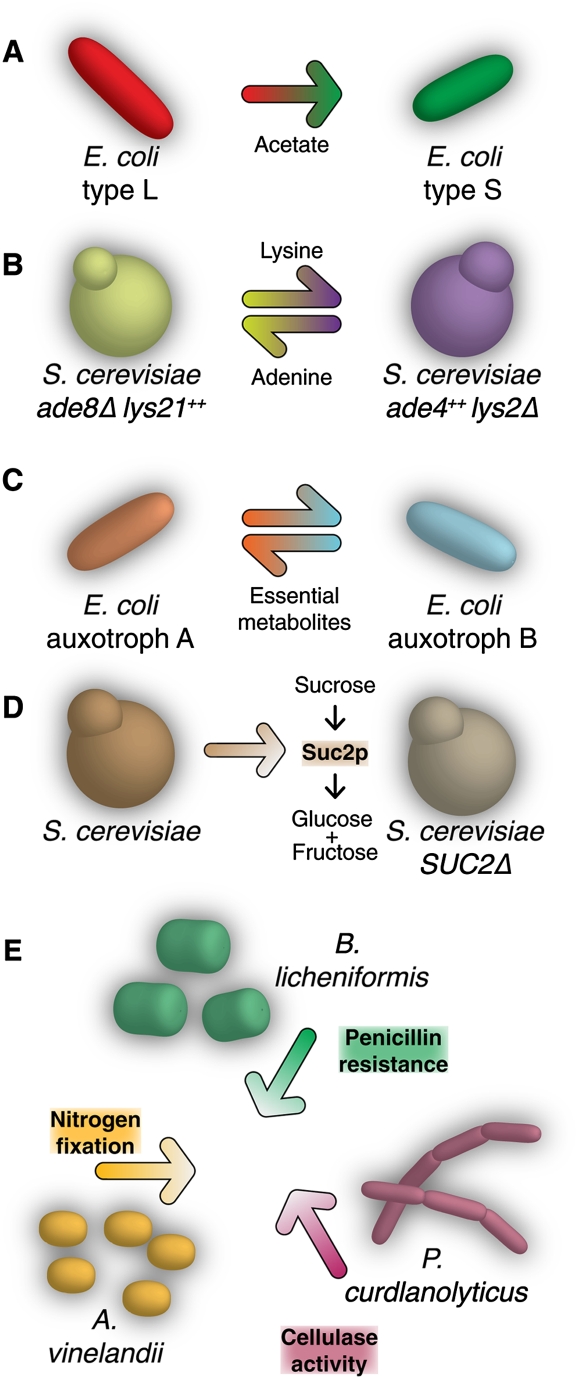
The natural association between V. atypica and S. gordonii, in which the former specializes in consuming the waste products of the latter, resembles a spontaneous polymorphism known to occur in laboratory strains of E. coli. Rozen et al. (2005, 2009) studied the experimental evolution of E. coli serially propagated in defined glucose medium for 18,000 generations. The batch-cultured strains alternately experienced periods of rapid growth, following inoculation in fresh medium, and periods of stasis, when medium was exhausted. After 6000 generations, the ancestral population differentiated into two genotypes, named S and L. The L genotype exhibited a higher maximal growth rate during exponential phase. The S genotype, however, was able to feed on the waste products of L growth, whereas the L type was not (Fig. 2A). Similar spontaneous metabolic polymorphism in E. coli emerges rapidly, stably, and reproducibly in a variety of conditions (Helling et al. 1987; Rosenzweig et al. 1994; Treves et al. 1998; Kinnersley et al. 2009).
Figure 2.
Synthetic metabolic cooperation. Designed microbial partnerships exhibit a range of interactive modes, many not known in nature. Synthetic interactions allow researchers to isolate and test key functional components of interaction dynamics.
The metabolic interaction of these E. coli variants is largely one-directional, with one strain feeding from the waste of the other. It illustrates how easily the metabolic activity of one strain can open a niche for an opportunistic specialist. Interestingly, there is also evidence that the waste specialist S strain accelerated the death and lysis of the L strain in stationary phase (Rozen and Lenski 2000). This would increase the availability of waste products, analogous to the signaling effect of V. atypica on S. gordonii.
Experimental evolution studies of natural syntrophs reveal a contrasting dynamic. The partnership between D. vulgaris and M. maripaludis, discussed above, can be broken and reinitiated under controlled conditions (Hillesland and Stahl 2010). Although these strains derive from syntrophic lineages, they have been isolated from different environments and propagated separately. As a consequence, their interaction is imperfectly adapted to the laboratory environment. Hillesland and Stahl (2010) propagated the partners in well-mixed coculture for 300 generations, with improvements in growth rate and yield generally accumulating as both strains became adapted to syntrophic growth. But the period of adaptation was not without risks. Erratic and stalled growth was sometimes observed as populations adapted, leading to the extinction of the culture in some replicates.
Cooperative metabolism has been hypothesized to originate from the exchange of waste by-products in initially parasitic or commensal relationships (Connor 1995; Sachs et al. 2004). By-product cooperation is appealing as a form of basal cooperation—the initial derivation of a cooperative from a noncooperative state—because it requires no costly investment by one partner in the other. A number of evolutionary scenarios have been proposed that may then favor a transition from waste feeding to bidirectional syntrophy and symbiosis (Searcy 2002).
The different outcomes for the systems of specialized E. coli and D. vulgaris with M. maripaludis illustrate substantial challenges of bidirectional obligate mutualism that do not constrain simple one-way commensalism. While the E. coli interaction emerged spontaneously and robustly, the partnership of D. vulgaris and M. maripaludis was unstable and sensitive to initial conditions. Obligate mutualists must carefully control the parameters of their interaction. Engineered microbes with precisely defined relationships can facilitate the detailed study of metabolite exchange.
Synthetic syntrophy
Efficient exchange
Saccharomyces cerevisiae, a well-characterized prototroph, was redesigned as an obligate mutualist (Shou et al. 2007). Two strains of S. cerevisiae were created. One received the ADE8Δ deletion, conveying a requirement for supplemental adenine. The other a carried the LYS2Δ deletion and required lysine. The two strains were cocultured in defined medium lacking both essential metabolites. Although the capacity existed for metabolic cooperation, no growth was initially observed. Cooperation was synthetically effected by up-regulating production of the shared metabolites. This was accomplished by introducing the LYS21OP and ADE4OP alleles to the ADE8Δ and LYS2Δ strains, respectively, thereby causing each strain to overproduce the metabolite required by its partner (Fig. 2B). With the synthetic cooperators, it was possible to quantitatively investigate the constraints on efficient exchange.
Cooperative metabolism was successful only above a minimum cell density. Cross-fed metabolites must be available at a sufficiently high concentration and cell populations must remain viable while secreted metabolites accumulate. Shou et al. (2007), using parameters measured in their system, were able to derive an expression of this requirement for the lysine auxotroph. The minimum required density (R) was a function of the Michaelis-Menten kinetics of lysine uptake (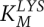 and
and 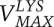 ), the strain's death rate (D), the quantity of lysine required to produce a new cell (LC), and the quantity of lysine produced over the lifetime of a partner cell (LS):
), the strain's death rate (D), the quantity of lysine required to produce a new cell (LC), and the quantity of lysine produced over the lifetime of a partner cell (LS):
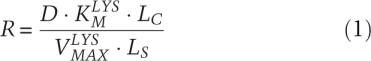 |
In this case, cell populations below 6 × 104 cells per milliliter were inviable. Such a high-density requirement would represent a serious challenge for a natural system. Similar requirements constrain the total initial number of cells and the minimum secretion levels of shared metabolites. Theoretical analysis supports the conclusion that population dynamic constraints severely limit cross-feeding interactions (Bull and Harcombe 2009). This is particularly expected when the metabolites exchanged are valuable resources, such as amino acids, rather than by-products of otherwise selfish metabolisms.
The auxotrophic yeast did not interact spontaneously. Although the potential benefits of metabolite exchange were great, cooperation could be induced only by up-regulating the production of the shared metabolites. This result illustrates another important natural barrier to efficient syntrophy. Microbes are expected to be parsimonious with their internal nutrient stores, and not to share unless specifically adapted to a cooperative lifestyle.
Our recent work examined the capacity of metabolic auxotrophs to spontaneously cross-feed (Wintermute and Silver 2010). A library of 46 conditionally lethal E. coli auxotrophs was cocultured in all pairwise combinations. The selected mutations spanned the metabolic network and included genes required for the biosynthesis of amino acids, nucleotides, and cofactors, as well as genes involved in glycolysis and respiration. Although no strain could grow alone, we observed significant growth in roughly 20% of the mixed pairs (Fig. 2C).
The subset of E. coli that spontaneously cooperates differs from others that do not, in terms of the fitness value of the metabolites that they exchange. In the yeast system, essential determinants of viability included the total amount of metabolites produced and consumed by a cell (LC and LS in Eq. 1). We were able to derive predictions for metabolite production and consumption from purely stoichiometric models of E. coli metabolism. In this way, the behavior of cells in mixed pairs could be derived from the topology of their underlying metabolic networks. Metabolites may be actively secreted, as with waste products, or they may be retained by the cell and released only with lysis following cell death. In general, the metabolites most likely to be spontaneously shared were those with the least value to the secreting strain, in terms of the fitness cost of producing them. This is consistent with a model in which by-product cooperation is the most easily established.
Evolutionary stability
Once metabolic cooperation is established, it must be resistant not only to perturbations in the environment, but to invasion by noncooperative mutants. In a cooperative system, one individual may pay a cost for another to receive a benefit, measured in terms of fitness. Defector strains, which fail to pay the cost of cooperation but receive the benefits, may exhibit higher fitness and quickly dominate the population. A variety of mechanisms exist to circumvent this game-theoretical constraint and allow cooperative phenotypes to thrive (Brännström and Dieckmann 2005; Nowak 2006; MacLean 2008; Chuang et al. 2010).
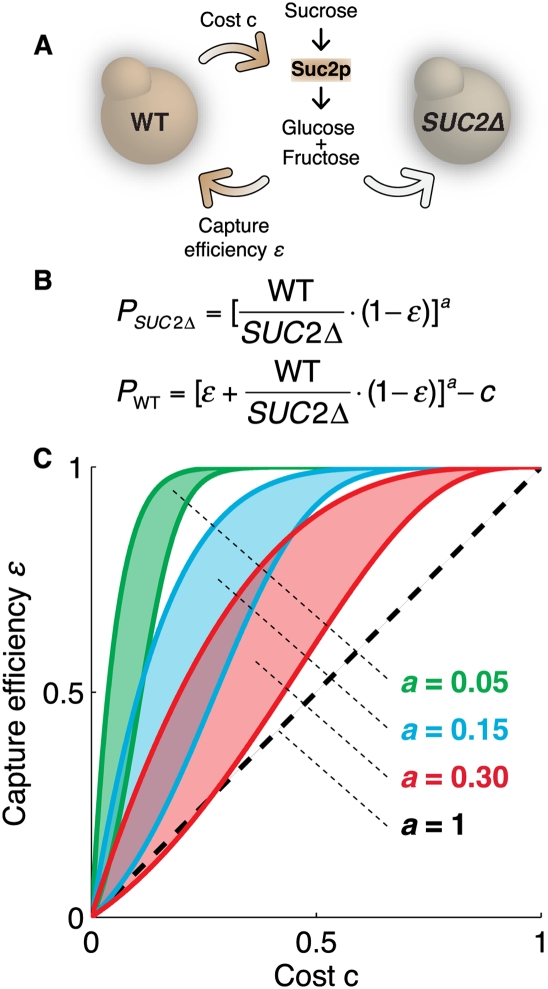
Again, the yeast S. cerevisiae served as the model system (Gore et al. 2009). Yeast carrying the SUC2 gene are able to grow using sucrose as a carbon source through the secretion of the enzyme invertase, which hydrolyzes sucrose to glucose and fructose for uptake. Because invertase is secreted and shared, it can be exploited by SUC2Δ deletion mutants (Fig. 2D). The fitness cost of cooperation is an essential parameter in evolutionary dynamic models that usually cannot be tuned easily by experimenters. To exaggerate and vary this cost, Gore et al. (2009) also deleted the HIS3 gene of the SUC2+ strain. The cost of cooperation could then be controlled by varying the availability of histidine.
Although invertase is secreted, not all of the free glucose and fructose produced by the enzyme is shared uniformly, even in a well-mixed culture. The dynamics of diffusion and sugar import allow a cell to retain roughly 1% of the free sugar yielded by its own efforts. Because of this, an invertase-expressing cooperative strain is able to invade a population of invertase-defective mutants. The 99% of sugars shared by a cooperating strain are a by-product of the selfish pursuit of the 1% of sugars that can be retained (Fig. 3).
Figure 3.
Byproduct cooperation stabilizes a designed yeast population. (A) The synthetic yeast interaction produced by Gore et al. (2009). The cooperator strain secretes Suc2p, which breaks down sucrose into glucose and fructose for uptake. Suc2p production entails a fitness cost c, which is not paid by the SUC2Δ cheater strain. Cooperators are able to capture the sugars produced by their own activity with capture efficiency ɛ before they are lost to diffusion. (B) Cooperator and cheater strains show density-dependant fitness P. Sugars lost to diffusion contribute equally to the fitness of both strains through the term 1 − ɛ. Only the cooperator pays the cost c, and only the cooperator benefits from retained sugars through the term ɛ. Critically, fitness is not linear with sugar availability, but is sublinear with exponent a < 1. The a term was estimated to be 0.15 in this system. (C) Cooperators and cheaters can coexist for certain values of a. Regions indicate parameter values for which the frequency of cooperators is between 1% and 99% of the population in the steady state. For a = 1, either cooperators or cheaters must eventually dominate the population and the mixed region shrinks to 0. For a < 1, frequency-dependent selection stabilizes population heterogeneity.
Spatial structure: natural and synthetic
The above example demonstrates an essential role for the structural and diffusive properties of the local environment in cooperative interactions. In well-mixed systems, shared metabolites are easily lost. More precisely structured environments can facilitate cooperative exchange. A natural microbial concourse rarely takes the form of the well-mixed suspensions that are commonly studied in the laboratory. Structured associations in the form of flocs, consortia, and biofilms predominate.
Syntrophic hydrogen-producing fermenters and methanogens, in anoxic lake sediments or sewage sludge, produce large aggregative flocs. By forming close associations in macroscopic structures, flocculating syntrophs are able to retain most of the shared metabolites that would otherwise be lost to diffusion. Roughly 95% of the hydrogen produced within such flocs is consumed by them (Conrad et al. 1985). The close associations not only help to conserve energy, but expedite the scavenging of hydrogen from the proximity of the primary fermenter, making hydrogen production more energetically favorable.
Other conjugate syntrophs form elegant microstructures referred to as consortia. Methanotrophic archaea and sulfate-reducing bacteria often co-occur as an outer layer of bacteria encapsulating a core of archaea. Acidithiobacillus ferrooxidans and Acidiphilium subspecies form pods of regular sizes and shapes on the scale of a few microns, held together by an extracellular matrix. These strains syntrophically metabolize organic molecules, exchanging sulfur as a redox shuttle. Pod formation would allow the recycling of the sulfur shuttle, while keeping low the concentration of that potentially toxic metabolite (Orphan et al. 2001; Norlund et al. 2009). Physical aggregation of syntrophs is associated with changes of metabolism and gene expression (Walker et al. 2009). The mechanisms that coordinate the association, dissolution, and shape of consortia are beginning to be described (Xavier and Foster 2007; Parsek and Tolker-Nielsen 2008; Neu et al. 2010).
Spatial structure was able to stabilize an artificial symbiosis between three strains (Kim et al. 2008). Azotobacter vinelandii fixes nitrogen, Bacillus licheniformis degrades penicillin, and Paenibacillus curdlanolyticus breaks down cellulose into a usable carbon source (Fig. 2E). In defined medium, all three capacities are required for any strain to be viable. Kim et al. (2008) found that cultures were more productive when spatially segregated through microfluidic channels than when well mixed. Separated strains, under nutrient and growth limitations, tended to produce more of the essential nutrient than they themselves consumed, allowing an excess to sustain their syntrophic partners. Efficient cooperative relationships may therefore require a delicate balance of metabolite production, consumption and exchange.
Natural communication
Quorum sensing
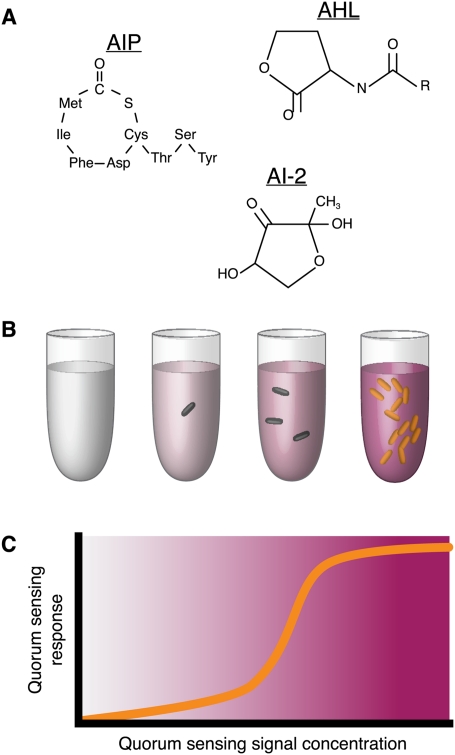
All microbes alter the chemistry of their local environment and sense chemical changes that may be produced by neighbors. Microbial communication is the exchange of signals specialized for that purpose. The chemical signals involved have a negligible role as nutrients or antibiotics, or exhibit a signaling function that can be distinguished from their other effects (Fig. 4A).
Figure 4.
Quorum sensing. (A) Example of communication molecules of three classes. AHLs are found in Gram-negative bacteria such as V. fischeri. AIPs are produced by Gram-positive strains like Staphylococcus. AI-2 is an intermediate of methionine metabolism, found among both Gram-positive and Gram-negative bacteria. (B) QS bacteria produce signal molecules at a basal level that accumulate locally. Signal levels reflect the local population density and trigger a collective response at high densities. Responding cells are indicated in orange. (C) The QS response is often nonlinear with signal concentration. Typically, response levels increase sharply above a critical signal threshold.
Many microbial communication mechanisms exhibit a characteristic threshold-dependent response. Cells do not respond proportionally to the chemical input, but instead defer a response until a critical high level of signal is detected. Systems of this kind can allow the synchronization of behaviors in a dense population that would be ineffective if performed by isolated individuals. Although some communication systems may perform other functions (Redfield 2002), they are often referred to generally as quorum-sensing (QS) mechanisms (Fig. 4B,C).
The best known QS system was first described in the bioluminescent Vibrio fischeri (Nealson 1977; Meighen 1999). N-acyl-homoserine lactones (AHLs) serve as the signal molecule. The core function is provided by two proteins that, respectively, synthesize and recognize the AHL. LuxI catalyzes the formation of N-(3-oxohexanoyl)-homoserine lactone from S-adenosylmethionine and an acyl group from the fatty acid biosynthesis pathway. The AHL diffuses freely through the cell membrane, around the local environment, and into neighboring cells. It binds and activates the LuxR protein, a transcription factor that induces the light-producing lux operon. The concentration of the AHL signal increases with the density of signaling bacteria in the local environment. Thus, light is produced by V. fischeri only when it can be done at high levels from a dense population.
QS systems of the AHL class are functionally diverse. The opportunistic human pathogen Pseudomonas aeruginosa operates two such systems in series. The LasI/LasR and RhlI/RhlR pairs play roles analogous to LuxI/LuxR, transmitting and receiving two distinct AHL compounds. A wide range of virulence factors are induced when a quorum is detected, including proteases, exotoxins, and biofilm-formation pathways (Willcox et al. 2008).
An orthologous AHL system in Agrobacterium tumefaciens is integral to the onset of crown gall disease in plants. Virulent A. tumefaciens transfer a segment of the tumor-inducing (Ti) plasmid to the nuclear genome of their plant host. Transformed plant cells produce opines, which stimulate the synthesis of QS AHL molecules among the infecting Agrobacterium. The bacterial quorum responds to the signal by increasing the copy number of the Ti plasmid and spreading it through the population by conjugation (Zhang et al. 2002; Haudecoeur and Faure 2010). Proliferation of the Ti plasmid expands the capacity of the bacterial population to transform plant cells with the gall-forming plasmid-borne DNA cassette.
AHL-type signaling systems are also known to control collective swarming motility in Serratia (Daniels et al. 2004) and Yersinia (Atkinson et al. 2008). They effect biofilm polysaccharide production in Pantoea (Koutsoudis et al. 2006). The broad distribution and myriad functions of AHL communication in Gram-negative bacteria are matched by structural diversity in the signaling molecules. A core homoserine lactone ring is shared generally, but the composition of the attached acyl chain varies with species and system. AHL signaling molecules are often produced as mixtures of similar compounds, with a species-specific principle component. Similarly, response to AHL signals typically shows preference for the main product of a given species, with some sensitivity to related compounds (Dickschat 2010). Thus, there is potential for cross-talk, even between species with divergent systems.
Oligopeptide signaling in Gram-positive bacteria plays a role similar to AHL signaling in the Gram-negative bacteria. Among pathogenic Staphylococcus, the best-studied examples, the preprotein AgrD is cleaved, processed, and secreted as an autoinducing peptide (AIP). The AIP is detected by a two-component-like signaling system comprised of the receptor AgrC and the transcription factor AgrA. AgrA in turn activates a variety of virulence factors and enterotoxins (Lyon and Novick 2004). Peptide-based QS is employed by Bacillus subtilis and Streptococcus pneumoniae in the decision between competence and sporulation (Schultz et al. 2009; Yang et al. 2010). It has been postulated that signaling in these cases allows for coordinated population-level strategic bet hedging, with only a fraction of the population pursuing a potentially risky strategy like genetic competence (Dubnau and Losick 2006).
A third distinct class of signaling pathway employs the LuxS enzyme to produce autoinducer-2 (AI-2). AI-2 is a general term for 4,5-dihydroxy-2,3-petanedione and its spontaneous cyclic derivatives (Federle 2009). AI-2 production elements are found in both Gram-positive and Gram-negative bacteria, including Vibrio harveyi, Salmonella enterica, Helicobacter pylori, and Clostridium perfringens. Systems for detecting and reacting to the AI-2 signal diverge broadly among taxa (Vendeville et al. 2005). Interestingly, the AI-2 molecule itself does not appear to show the diversity and specialization that are found in the AIP and AHL-type signaling systems. Rather, AI-2 appears to be chemically identical in all systems of this class, even when production and detection mechanisms differ. This has led to the hypothesis that AI-2 represents a kind of universal signal for interspecies communication (Xavier and Bassler 2003).
AI-2 signaling occupies a middle ground between metabolic interaction and pure communication. Because AI-2 is a stoichiometric by-product of methionine metabolism, it can report directly on the metabolic activity or the growth rates of cells that secrete it (DeLisa et al. 2001). It is derived from the toxic metabolic pathway intermediate S-adenosylhomocysteine, suggesting it may have a role simply as a waste product in some contexts (Schauder et al. 2001). If secreted AI-2 or its degradation products were later metabolized as an energy source, AI-2 exchange would match the above description of metabolic by-product cooperation. Such considerations have led to the hypothesis that AI-2 exchange represents metabolic interaction and not communication in many of the species that produce AI-2 (Winzer et al. 2002). Other signaling molecules also have nonsignaling roles, suggesting that, in some cases, communication may have originated as the exchange of waste or other by-products (Vendeville et al. 2005; Schertzer et al. 2009).
Specificity and cross-talk
The density and diversity of signals that can coexist in the concourse challenges the ability of cells to coordinate precise responses with specific partners. A given communicating strain may find itself mixed with unrelated cells using the same or similar signals for their own purposes. The composition of many QS networks reveals adaptations that appear to either mitigate or exploit the inherent difficulties of chemical signaling in a crowded, noisy concourse.
AI-2 signaling, for example, appears to be completely nonspecific. Not only is the same signal produced by each QS lineage, AI-2 is produced by a wide range of strains for which it is probably only a metabolite and not a signal (Rezzonico and Duffy 2008).
Peptide signal–receptor interactions, in contrast, are highly specific. Single amino acid substitutions of the seven to nine that compose an AIP are often sufficient to disrupt interaction (Geisinger et al. 2008). AIPs coevolve with their conjugate receptors and diverge among communicating strains. Four specificity groups are discernible among isolates of the model Staphylococcus aureus, defined functionally as sets of receptor/ligand pairs that can cross-activate. Signal molecules from other groups not only fail to activate the response, but often actively inhibit the receptor (Ji et al. 1997; Geisinger et al. 2009). The result is the competitive inhibition of biofilm formation in one strain by another, a kind of evolutionary arms race. As they establish their own quorum, Staphylococcus biotypes suppress competing quora within the same niche.
AHL-type signaling systems show intermediate and varying levels of cross-talk. Reporter strains in which LuxR-binding sites are cloned upstream of a detectable marker like GFP indicate ranges of sensitivity for individual LuxR homologs. Specificity varies with the length and functional groups of the AHL side chain. For example, while the A. tumefaciens system responds to short acyl groups (C6–C12), the Ensifer meliloti homolog prefers longer chains (C12–C16), with some overlap in sensitivity to intermediate chain lengths (Dickschat 2010). P. aeruginosa and Burkholderia cepacia employ AHL systems with partially overlapping sensitivity, and are known to co-occur as mixed biofilms in cystic fibrosis patients (Riedel et al. 2001).
Synthetic communication
Natural QS systems are invariably connected to complex transcriptional and regulatory networks that complicate attempts to quantitatively characterize their function. P. aeruginosa, for example, employ two AHL-based communication systems wired in series. More than 300 genes are controlled by one or both systems, with responses varying with the level and duration of signal (Schuster et al. 2003). QS populations may effect sophisticated and subtle regulation of communication by integrating it with other signaling pathways. V. harveyi integrate three separate QS pathways in parallel (Waters and Bassler 2005). A. tumefaciens initiate communication behavior only in the presence of opine signals from their plant hosts (Haudecoeur and Faure 2010).
Synthetic biologists can reverse-engineer complex natural networks by breaking them down into functional submodules. Submodules are heterologously expressed in model strains, usually E. coli, without the complex suite of specific adaptations that accompanies natural communication systems. The core components of QS systems, particularly the AHL-based systems, have proven remarkably amenable to this approach.
Signaling modules
QS systems can be redesigned to control behaviors not found in nature. In E. coli, heterologous LuxI is sufficient to broadcast the AHL signal and LuxR is sufficient to receive it. In principle, any gene of interest can be introduced with a LuxR-responsive promoter and controlled by QS. You et al. (2004) coupled expression of the suicide gene ccdB to a synthetic QS system. At a sufficiently high cell density, induction of ccdB killed QS E. coli by poisoning DNA gyrase. In this way, the population stabilized at a density lower than that imposed by nutrient restrictions of the medium.
Other well-known synthetic modules can be interfaced with communication circuits, creating complex behaviors that emerge rationally from their constituent parts. Kobayashi et al. (2004) linked QS to cellular differentiation. The classic synthetic toggle switch employs positive feedback to sustain one of two possible transcriptional steady states (Gardner et al. 2000). When the toggle switch was induced by the AHL signal, E. coli epigenetically retained memory of a past quorum.
LuxI and LuxR from the Vibrio QS system can be also adapted to form macroscopic spatial patterns (Basu et al. 2005). In this case, the signal production and response element were assigned to different cell populations. The AHL signal was produced by LuxI in a patch of sender E. coli on an agar plate. Surrounding that patch were receiver cells wired with LuxR to sense the signal and produce GFP in response. A transcription network was rationally designed to repress GFP expression at both high and low levels of signal, but release repression at intermediate levels. The result was a ring of fluorescence in the receiver cells where the diffusive signal gradient had reached an optimal concentration.
Pattern formation, controlled cell division, differentiation, and memory are functions often perceived to be associated only with sophisticated multicellular eukaryotes. Yet basic versions of each were introduced de novo into bacteria as remarkably simple transcriptional networks. The dynamics in natural communicating populations, with their associated large and complex regulatory networks, may be much more sophisticated still. The soil bacterium Myxocuccus xanthus recapitulates features analogous to multicellular eukaryotes (Zusman et al. 2007). Microbial biofilms exhibit remarkable internal organization (Costerton 2007; Parsek and Tolker-Nielsen 2008). Synthetic studies present clear examples of how such organization can be achieved.
Positive feedback and cross-talk
Natural systems can inspire design refinements for synthetic systems. Important functional features of natural communication systems become evident when the proteins that send and receive signals are seen in the context of larger functional networks. For example, many natural systems incorporate feedback loops in their core QS circuits.
In V. fischeri, the luxICDABE operon is transcribed in response to an AHL signal. The gene products LuxCDABE chemically produce the bioluminescence that appears to be the primary functional output of the QS system in this species. The sixth gene product, LuxI, is the AHL synthase that first initiated signaling. Thus, high levels of AHL positively feed back to further increase AHL production.
Positive feedback is also a ubiquitous feature of peptide-based signaling systems (Novick and Geisinger 2008). In Staphylococcus, the AIP signal activates a variety of virulence factors, and also the four-gene agrBDCA operon. The agrD product is processed by the agrB product to produce more AIP, while agrC and agrA encode the receptor and the transcription factor that relay the AIP signal. Thus, AIP positively feeds back on its own production and that of its conjugate receptor.
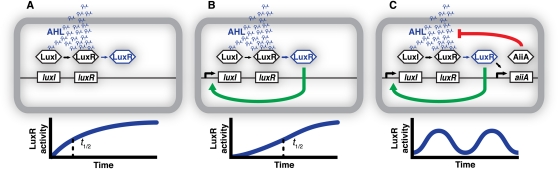
The recurrence of positive feedback in signaling systems suggests an important functional role that qualitative descriptions alone cannot adequately capture (Alon 2007). Feedback was not incorporated into the synthetic designs for population control or pattern formation, described above, yet both systems achieved basic signaling functions. Quantitative models reveal an important role for positive feedback in slowing the activation and improving the fidelity of such systems. Response to the QS signal is initially slow, then accelerates as signal production increases with autoactivation. The result is a concave signal activation curve that responds less to transient exposure to QS signal and reaches half-maximal activation more slowly than a similar network with no feedback (Fig. 5A,B).
Figure 5.
Natural and synthetic feedback loops in QS. (A) A simple topology for a functional communication network. The AHL signal is produced constitutively by LuxI and is received by LuxR, which may activate downstream genes. LuxR activity shows a simple saturating increase with time, as cells proliferate and AHL accumulates. This design was used successfully in early synthetic QS networks (You et al. 2004; Basu et al. 2005). (B) Positive feedback slows response time and improves response fidelity. With LuxI expression under the control of LuxR, the increase in LuxR activity with time becomes slower and more concave. Note the relatively flat response for early time points with low AHL levels, reflecting insensitivity to transient induction. The dotted line indicates the time required for half-maximal LuxR activity, which is extended in this case. Natural QS systems widely employ this kind of positive feedback, and it has been useful in synthetic systems as well (Brenner et al. 2007). (C) Negative feedback and temporal control. By introducing AiiA, which degrades AHL, under the control of LuxR, Danino et al. (2010) produced cells that respond to a quorum by destroying the QS signal. The result was synchronized, population-level oscillations in QS activity. Negative feedback is also a feature in many natural QS systems, where it may provide fine temporal response control.
Positive feedback proved a valuable feature when two synthetic signaling pathways were designed to operate in parallel. Brenner et al. (2007) adapted two QS systems of P. aeruginosa to operate in parallel in E. coli. Cells with one synthetic circuit produced 3-oxododecanoyl-AHL (3OC12-AHL) and responded to butanoyl-AHL (C4-AHL). For cells with the other circuit, the signal and response compounds were reversed. Rewired in this way, the cells each respond to a quorum of the other, with the response of both occurring only when both are at sufficiently high density.
With two signaling compounds present simultaneously, the minimization of cross-talk became an essential design consideration. Specifically, a cell was intended not to respond to its own signal, only that of its partner. Dynamic modeling results led to the adoption of positive feedback, a feature of the natural system, as a mechanism to suppress cross-talk. Transcription of the AHL-producing LasI and RhlI proteins was therefore set to be activated by the presence of the conjugate signal, rather than simply constitutively expressed.
When the artificial partners were cultivated in mixed biofilms, the QS response became activated in clusters of intimate heterologous contact. Mixed QS could therefore be a way for syntrophic or otherwise cooperating strains to identify their collaborators within a biofilm.
Negative feedback and temporal dynamics
Negative feedback, also a powerful and ubiquitous feature of many natural and artificial systems, can further enhance the dynamic potential of communication systems. Negative feedback was integrated with a basic QS system with the help of another new component from nature. The AiiA enzyme from Bacillus thurigiensis degrades AHL compounds by hydrolyzing the homoserine ring (Liu et al. 2008). The degradation product is no longer able to activate a QS response.
By driving the expression of AiiA lactonase from an AHL-responsive promoter, Danino et al. (2010) were able to produce temporally and spatially synchronized gene expression in a population of E. coli. This system incorporated the now familiar V. fischeri signaling system, including the natural positive feedback feature linking LuxI production to AHL levels. As AHL accumulated in the quorum, AiiA and GFP were expressed. Eventually, AHL degradation by AiiA exceeded production by LuxI, and the system returned to the initial prequorum state. Feedback loops of this kind are known to be capable of producing biological oscillations (Elowitz and Leibler 2000; Balagaddé et al. 2005; Swinburne et al. 2008). By linking intracellular signaling to the negative feedback loop, the oscillations became synchronized across the population (Fig. 5C). Population-level oscillations were also more stable and robust than can be attained in single cells.
Coupled oscillations may be a case of synthetic biology preceding what is known in natural systems. The concept and theoretical design for a system of synthetic coupled oscillators long anticipated its living implementation (McMillen et al. 2002). Temporally synchronized oscillations are not known in populations that use QS of the AHL type, nor is it clear what biological function such behavior might serve. Although AHL-degrading enzymes can be found in a variety of bacteria, relatively little is known of their functional role (Czajkowski and Jafra 2009). Recent work has uncovered other kinds of negative feedback in AHL signaling systems (Tu et al. 2010). These results suggest that such feedback may allow fine temporal and spatial control of communication.
Conclusion
Assigning a function to a network feature like a feedback loop is difficult in its complex natural context. By reconstituting model circuits in naive hosts, it is possible to establish when specific genetic components are sufficient for an observed behavior. This, in turn, can imply a functional role for analogous natural features. Similarly, engineering novel biological interactions allows the synthetic biologist to isolate their essential attributes from the rich suite of adaptations that accompanies natural partnerships. The ambitious research program of systems biology seeks to aggregate knowledge of these features into a comprehensive model of living microbial communities.
Dynamic behavior in the mixed concourse represents a new layer of complexity, emerging from the behavior of many distinct individuals. The prospects for a complete theory of microbial communities may appear daunting, given the inherent complexity of such systems and the sheer number of moving parts. An understanding of microbial interaction will have clear implications for medicine and biotechnology (Bjarnsholt and Givskov 2008; Brenner et al. 2008). More importantly, adaptations to mixed culture are central to basic microbial biology. Integrative and systems approaches raise the possibility that laboratory microbes, so long orphaned, can be returned to their natural home in the concourse.
Acknowledgments
We thank Patrick Boyle for reviewing the manuscript, and the UotH group for input on content. This work was supported by funding from the NSF Graduate Research Fellowship Program to E.W.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1985210.
References
- Alon U 2007. An introduction to systems biology: Design principles of biological circuits. CRC Press, Boca Raton, FL [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Chang CY, Patrick HL, Buckley CM, Wang Y, Sockett RE, Cámara M, Williams P 2008. Functional interplay between the Yersinia pseudotuberculosis YpsRI and YtbRI quorum sensing systems modulates swimming motility by controlling expression of flhDC and fliA. Mol Microbiol 69: 137–151 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: 2006.0008 doi: 10.1038.msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagaddé FK, You L, Hansen CL, Arnold FH, Quake SR 2005. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 309: 137–140 [DOI] [PubMed] [Google Scholar]
- Barker HA 1939. Studies upon the methane fermentation IV. The isolation and culture of Methanobacterium omelianskii. Antonie van Leeuwenhoek 6: 201–220 [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R 2005. A synthetic multicellular system for programmed pattern formation. Nature 434: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Givskov M 2008. Quorum sensing inhibitory drugs as next generation antimicrobials: Worth the effort? Curr Infect Dis Rep 10: 22–28 [DOI] [PubMed] [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407: 623–626 [DOI] [PubMed] [Google Scholar]
- Brännström A, Dieckmann U 2005. Evolutionary dynamics of altruism and cheating among social amoebas. Proc Biol Sci 272: 1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, Karig DK, Weiss R, Arnold FH 2007. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci 104: 17300–17304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, You L, Arnold FH 2008. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol 26: 483–489 [DOI] [PubMed] [Google Scholar]
- Bull JJ, Harcombe WR 2009. Population dynamics constrain the cooperative evolution of cross-feeding. PLoS ONE 4: e4115 doi: 10.1371/journal.pone.0004115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S 2010. Cooperation and Hamilton's rule in a simple synthetic microbial system. Mol Syst Biol 6: 398 doi: 10.1038/msb.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RC 1995. The benefits of mutualism: A conceptual framework. Biol Rev Camb Philos Soc 70: 427–457 [Google Scholar]
- Conrad R, Phelps TJ, Zeikus JG 1985. Gas metabolism evidence in support of the juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl Environ Microbiol 50: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW 2007. The biofilm primer. Springer, Berlin [Google Scholar]
- Crutzen PJ 1994. Global budgets for non-CO2 greenhouse gases. Environ Monit Assess 31: 1–15 [DOI] [PubMed] [Google Scholar]
- Czajkowski R, Jafra S 2009. Quenching of acyl-homoserine lactone-dependent quorum sensing by enzymatic disruption of signal molecules. Acta Biochim Pol 56: 1–16 [PubMed] [Google Scholar]
- Daniels R, Vanderleyden J, Michiels J 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev 28: 261–289 [DOI] [PubMed] [Google Scholar]
- Danino T, Mondragón-Palomino O, Tsimring L, Hasty J 2010. A synchronized quorum of genetic clocks. Nature 463: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisa MP, Valdes JJ, Bentley WE 2001. Quorum signaling via AI-2 communicates the ‘metabolic burden’ associated with heterologous protein production in Escherichia coli. Biotechnol Bioeng 75: 439–450 [DOI] [PubMed] [Google Scholar]
- Dickschat JS 2010. Quorum sensing and bacterial biofilms. Nat Prod Rep 27: 343–369 [DOI] [PubMed] [Google Scholar]
- Dolfing J, Jiang B, Henstra AM, Stams AJ, Plugge CM 2008. Syntrophic growth on formate: A new microbial niche in anoxic environments. Appl Environ Microbiol 74: 6126–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Stams AJ 1995. Evidence for H2 and formate formation during syntrophic butyrate and propionate degradation. Anaerobe 1: 35–39 [DOI] [PubMed] [Google Scholar]
- Dubnau D, Losick R 2006. Bistability in bacteria. Mol Microbiol 61: 564–572 [DOI] [PubMed] [Google Scholar]
- Egland PG, Palmer RJ, Kolenbrander PE 2004. Interspecies communication in Streptococcus gordonii–Veillonella atypica biofilms: Signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci 101: 16917–16922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Ende FP, Meier J, Gemerden H 1997. Syntrophic growth of sulfate-reducing bacteria and colorless sulfur bacteria during oxygen limitation. FEMS Microbiol Ecol 23: 65–80 [Google Scholar]
- Federle MJ 2009. Autoinducer-2-based chemical communication in bacteria: Complexities of interspecies signaling. Contrib Microbiol 16: 18–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342 [DOI] [PubMed] [Google Scholar]
- Geisinger E, George EA, Muir TW, Novick RP 2008. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem 283: 8930–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP 2009. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci 106: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Youk H, van Oudenaarden A 2009. Snowdrift game dynamics and facultative cheating in yeast. Nature 459: 253–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P 2004. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108 [DOI] [PubMed] [Google Scholar]
- Haudecoeur E, Faure D 2010. A fine control of quorum-sensing communication in Agrobacterium tumefaciens. Commun Integr Biol 3: 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling RB, Vargas CN, Adams J 1987. Evolution of Escherichia coli during growth in a constant environment. Genetics 116: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA 2010. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci 107: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti EL, Kafkewitz D, Wolin MJ, Bryant MP 1973. Glucose fermentation products in Ruminococcus albus grown in continuous culture with Vibrio succinogenes: Changes caused by interspecies transfer of H 2. J Bacteriol 114: 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G, Beavis R, Novick RP 1997. Bacterial interference caused by autoinducing peptide variants. Science 276: 2027–2030 [DOI] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. 2009. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res 37: D464–D470 doi: 10.1093/nar/gkn751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF 2008. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci 105: 18188–18193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley MA, Holben WE, Rosenzweig F 2009. E Unibus Plurum: Genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet 5: e1000713 doi: 10.1371/journal.pgen.000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ 2004. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci 101: 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE 2000. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu Rev Microbiol 54: 413–437 [DOI] [PubMed] [Google Scholar]
- Kolisnychenko V, Plunkett G, Herring CD, Fehér T, Pósfai J, Blattner FR, Pósfai G 2002. Engineering a reduced Escherichia coli genome. Genome Res 12: 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc Natl Acad Sci 103: 5983–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunapuli U, Lueders T, Meckenstock RU 2007. The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J 1: 643–653 [DOI] [PubMed] [Google Scholar]
- Liu D, Momb J, Thomas PW, Moulin A, Petsko GA, Fast W, Ringe D 2008. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47: 7706–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25: 1389–1403 [DOI] [PubMed] [Google Scholar]
- MacLean RC 2008. The tragedy of the commons in microbial populations: Insights from theoretical, comparative and experimental studies. Heredity 100: 471–477 [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci 1125: 58–72 [DOI] [PubMed] [Google Scholar]
- McMillen D, Kopell N, Hasty J, Collins JJ 2002. Synchronizing genetic relaxation oscillators by intercell signaling. Proc Natl Acad Sci 99: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighen EA 1999. Autoinduction of light emission in different species of bioluminescent bacteria. Luminescence 14: 3–9 [DOI] [PubMed] [Google Scholar]
- Mikx FHM, van der Hoeven JS 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch Oral Biol 20: 407–410 [DOI] [PubMed] [Google Scholar]
- Nauhaus K, Treude T, Boetius A, Krüger M 2005. Environmental regulation of the anaerobic oxidation of methane: A comparison of ANME-I and ANME-II communities. Environ Microbiol 7: 98–106 [DOI] [PubMed] [Google Scholar]
- Nealson KH 1977. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch Microbiol 112: 73–79 [DOI] [PubMed] [Google Scholar]
- Neu TR, Manz B, Volke F, Dynes JJ, Hitchcock AP, Lawrence JR 2010. Advanced imaging techniques for assessment of structure, composition and function in biofilm systems. FEMS Microbiol Ecol 72: 1–21 [DOI] [PubMed] [Google Scholar]
- Norlund KL, Southam G, Tyliszczak T, Hu Y, Karunakaran C, Obst M, Hitchcock AP, Warren LA 2009. Microbial architecture of environmental sulfur processes: A novel syntrophic sulfur-metabolizing consortia. Environ Sci Technol 43: 8781–8786 [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E 2008. Quorum sensing in staphylococci. Annu Rev Genet 42: 541–564 [DOI] [PubMed] [Google Scholar]
- Nowak MA 2006. Five rules for the evolution of cooperation. Science 314: 1560–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Flaherty V, Mahony T, O'Kennedy R, Colleran E 1998. Effect of pH on growth kinetics and sulphide toxicity thresholds of a range of methanogenic, syntrophic and sulphate-reducing bacteria. Process Biochem 33: 555–569 [Google Scholar]
- Orphan VJ 2009. Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol 12: 231–237 [DOI] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293: 484–487 [DOI] [PubMed] [Google Scholar]
- Parsek MR, Tolker-Nielsen T 2008. Pattern formation in Pseudomonas aeruginosa biofilms. Curr Opin Microbiol 11: 560–566 [DOI] [PubMed] [Google Scholar]
- Pfennig N, Biebl H 1978. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch Microbiol 117: 9–16 [Google Scholar]
- Plugge CM, Balk M, Zoetendal EG, Stams AJ 2002. Gelria glutamica gen. nov., sp. nov., a thermophilic, obligately syntrophic, glutamate-degrading anaerobe. Int J Syst Evol Microbiol 52: 401–407 [DOI] [PubMed] [Google Scholar]
- Raes J, Foerstner KU, Bork P 2007. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr Opin Microbiol 10: 490–498 [DOI] [PubMed] [Google Scholar]
- Rasmussen B 2000. Filamentous microfossils in a 3,235-million-year-old volcanogenic massive sulphide deposit. Nature 405: 676–679 [DOI] [PubMed] [Google Scholar]
- Redfield RJ 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol 10: 365–370 [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Duffy B 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol 8: 154 doi: 10.1186/1471-2180-8-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res 27: 3821–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Høiby N, Givskov M, Molin S, Eberl L 2001. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147: 3249–3262 [DOI] [PubMed] [Google Scholar]
- Rosenzweig RF, Sharp RR, Treves DS, Adams J 1994. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics 137: 903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Lenski RE 2000. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat 155: 24–35 [DOI] [PubMed] [Google Scholar]
- Rozen DE, Schneider D, Lenski RE 2005. Long-term experimental evolution in Escherichia coli. XIII. Phylogenetic history of a balanced polymorphism. J Mol Evol 61: 171–180 [DOI] [PubMed] [Google Scholar]
- Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D 2009. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett 12: 34–44 [DOI] [PubMed] [Google Scholar]
- Sachs JL, Mueller UG, Wilcox TP, Bull JJ 2004. The evolution of cooperation. Q Rev Biol 79: 135–160 [DOI] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL 2001. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol 41: 463–476 [DOI] [PubMed] [Google Scholar]
- Schertzer JW, Boulette ML, Whiteley M 2009. More than a signal: Non-signaling properties of quorum sensing molecules. Trends Microbiol 17: 189–195 [DOI] [PubMed] [Google Scholar]
- Schink B 2006. Syntrophism among prokaryotes. In The prokaryotes (ed. Dworkin M), pp. 309–335 Springer, New York [Google Scholar]
- Schink B, Stieb M 1985. Anaerobic oxidation of fatty acids by Clostridium bryantii sp. nov., a sporeforming, obligately syntrophic bacterium. Arch Microbiol 140: 387–390 [Google Scholar]
- Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN 2009. Deciding fate in adverse times: Sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci 106: 21027–21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol 185: 2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searcy G 2002. Nutritional syntrophies and consortia as models for the origin of mitochondria. In Symbiosis: Mechanisms and model systems (ed. Seckbach J), p. 796 Springer, Dordrecht [Google Scholar]
- Shou W, Ram S, Vilar JM 2007. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci 104: 1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA 2007. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol 3: 92 doi: 10.1038/msb4100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne IA, Miguez DG, Landgraf D, Silver PA 2008. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev 22: 2342–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J 1998. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol 15: 789–797 [DOI] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P 2008. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11: 442–446 [DOI] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL 2010. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG 2010. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 309: 1–7 [DOI] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR 2005. Making ‘sense’ of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3: 383–396 [DOI] [PubMed] [Google Scholar]
- Walker CB, He Z, Yang ZK, Ringbauer JA, He Q, Zhou J, Voordouw G, Wall JD, Arkin AP, Hazen TC, et al. 2009. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol 191: 5793–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL 2005. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346 [DOI] [PubMed] [Google Scholar]
- Widdel F, Zindel U, Freudenberg W, Rieth M, Andreesen JR, Schnell J 1988. Eubacterium acidaminophilum sp. nov., a versatile amino acid-degrading anaerobe producing or utilizing H2 or formate. Arch Microbiol 150: 254–266 [Google Scholar]
- Willcox MD, Zhu H, Conibear TC, Hume EB, Givskov M, Kjelleberg S, Rice SA 2008. Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology 154: 2184–2194 [DOI] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA 2010. Emergent cooperation in microbial metabolism. Mol Syst Biol 6: 407 doi: 10.1038/msb.2010.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P 2002. Bacterial cell-to-cell communication: Sorry, can't talk now—Gone to lunch! Curr Opin Microbiol 5: 216–222 [DOI] [PubMed] [Google Scholar]
- Wolfe RS, Bryant MP, Wolin EA, Wolin MJ 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Microbiol 59: 20–31 [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL 2003. LuxS quorum sensing: More than just a numbers game. Curr Opin Microbiol 6: 191–197 [DOI] [PubMed] [Google Scholar]
- Xavier JB, Foster KR 2007. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci 104: 876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Evans BA, Rozen DE 2010. Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc Biol Sci 277: 2991–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Cox RS, Weiss R, Arnold FH 2004. Programmed population control by cell–cell communication and regulated killing. Nature 428: 868–871 [DOI] [PubMed] [Google Scholar]
- Zengler K, Richnow HH, Rosselló-Mora R, Michaelis W, Widdel F 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401: 266–269 [DOI] [PubMed] [Google Scholar]
- Zhang HB, Wang LH, Zhang LH 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci 99: 4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5: 862–872 [DOI] [PubMed] [Google Scholar]