Abstract
To define the functional pathways regulating epithelial cell migration, we performed a genome-wide RNAi screen using 55,000 pooled lentiviral shRNAs targeting ∼11,000 genes, selecting for transduced cells with increased motility. A stringent validation protocol generated a set of 31 genes representing diverse pathways whose knockdown dramatically enhances cellular migration. Some of these pathways share features of epithelial-to-mesenchymal transition (EMT), and together they implicate key regulators of transcription, cellular signaling, and metabolism, as well as novel modulators of cellular trafficking, such as DLG5. In delineating downstream pathways mediating these migration phenotypes, we observed universal activation of ERKs and a profound dependence on their RSK effectors. Pharmacological inhibition of RSK dramatically suppresses epithelial cell migration induced by knockdown of all 31 genes, suggesting that convergence of diverse migratory pathways on this kinase may provide a therapeutic opportunity in disorders of cell migration, including cancer metastasis.
Keywords: EMT, MCF10A, RNAi, RSK, cell migration, high throughput
Cell migration is a fundamental process in the formation and maintenance of multicellular organisms. Insufficient cell migration has been linked to a number of congenital birth defects involving craniofacial abnormalities, heart septation defects, and cleft palate, whereas abnormal activation of migratory programs has been connected to cancer metastasis (Yang and Weinberg 2008; Acloque et al. 2009). Mammalian cells may migrate as individual cells, using so-called amoeboid or mesenchymal modes of locomotion, or they can do so as multicellular aggregates known as chain or collective modes (Friedl and Wolf 2010). Cell migration may also reflect baseline cellular motility or may be induced by loss of cell–cell contact within cellular monolayers or the application of secreted chemoattractants, and it may be affected by the composition, density, orientation, and stiffness of the cellular substrate. Signals from such extracellular determinants are integrated through various intracellular pathways, leading to changes in cytoskeletal organization and activation of downstream effectors that regulate cellular motility. Given the critical relevance of cellular migration to the spread of cancer and the complex inputs capable of modulating this process, we undertook a genome-wide screen to identify novel regulators of endogenous locomotion in mammalian epithelial cells.
Genetic screens for genes regulating cell migration have been reported in Drosophila melanogaster and Caenorhabditis elegans (Cram et al. 2006; Wang et al. 2006), but comparable RNAi screens in mammalian cell types have only recently become feasible (Gobeil et al. 2008; Luo et al. 2008; Silva et al. 2008; Hu et al. 2009; Li et al. 2009). Two recent studies analyzed wound scratch filling of cellular monolayers after growth factor stimulation using focused siRNA libraries targeting mainly kinase and phosphatase gene classes (Simpson et al. 2008; Vitorino and Meyer 2008). These studies identified both known and novel “hits,” pointing to a broad set of regulatory pathways, even within these relatively well-annotated gene families.
Beyond interrogating specific gene families, whole-genome RNAi screens offer an unprecedented ability to uncover novel regulators of specific cellular processes. To be successful, such genome-wide screens require a robust cellular endpoint as well as sufficient depth in gene coverage, and extensive post-screen validation to exclude spurious “hits.” While successful screens using the traditional arrayed format whole-genome RNAi have been reported (Hitomi et al. 2008; Hu et al. 2009), they suffer from the high cost and inefficiency of assessing phenotypes one gene knockdown at a time, although miniaturization to 96-well and 384-well plate formats alleviates some of these challenges. The recently developed pooled shRNA format screening offers considerable advantages with respect to ease of assay and cost of analysis. However, pooled shRNA format screening requires an assay in which cells with the desired phenotype can be cleanly enriched from their parental population, thus enabling scoring of relative shRNA abundance using molecular barcodes linked to each shRNA construct.
To apply such a pooled shRNA screening strategy to address cellular migration, we made use of a perforated membrane (Boyden chamber) readily traversed by epithelial cells whose migratory programs have been activated, but not by their poorly motile parental cells. Highly reproducible enrichment of migration-inducing shRNAs was achieved by harvesting cells that had traversed the membrane, identifying genes whose knockdown dramatically enhances baseline migration of epithelial cells. We present a cohort of 31 highly validated genes representing diverse cellular pathways, regulating migration of MCF10A mammary epithelial cells. A remarkable common theme among these otherwise disparate migration gene knockdowns is their shared activation of the ERK signaling pathway and their dependence on the ERK effector kinase RSK. Pharmacological suppression of RSK activity abrogates all shRNA-mediated migratory pathways identified here, without associated cell toxicity, suggesting that it may constitute a therapeutic target for suppressing cellular migration triggered by diverse stimuli.
Results
Screening and candidate gene validation
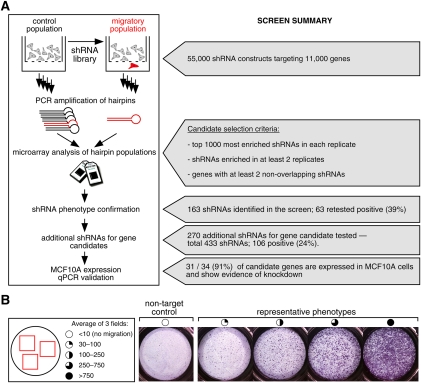
The Boyden chamber assay assesses the ability of cells to traverse across a perforated plastic membrane, providing a physical separation, and thus enrichment, for cells with newly acquired migratory ability (Fig. 1A). To identify novel regulators of cell migration, we targeted 11,000 genes using a lentiviral library containing five hairpins per gene (Luo et al. 2008), comparing in quadruplicate the relative abundance of each shRNA in the enriched migratory versus the unselected cell populations. MCF10A, a nontransformed human breast epithelial cell line with minimal baseline migration in Boyden chamber assays, was used for these experiments. The relative shRNA abundance was measured using microarray hybridization of shRNA barcodes, and the top 1000 shRNAs in each replicate were chosen for further consideration. Genes for which two or more distinct shRNA sequences scored among the top 1000 shRNAs (1.8 percentile) in at least two of the four replicate experiments were selected as candidates for follow-up (Fig. 1A).
Figure 1.
Screen overview. (A) Schematic representation of the screen workflow. Summary of shRNA- and gene-level analysis is presented on the right. (B) Representative images of migratory shRNAs recovered from the screen. The amount of migration was quantitated and shRNAs were classified into four categories, indicated by the extent to which the open circles were filled in.
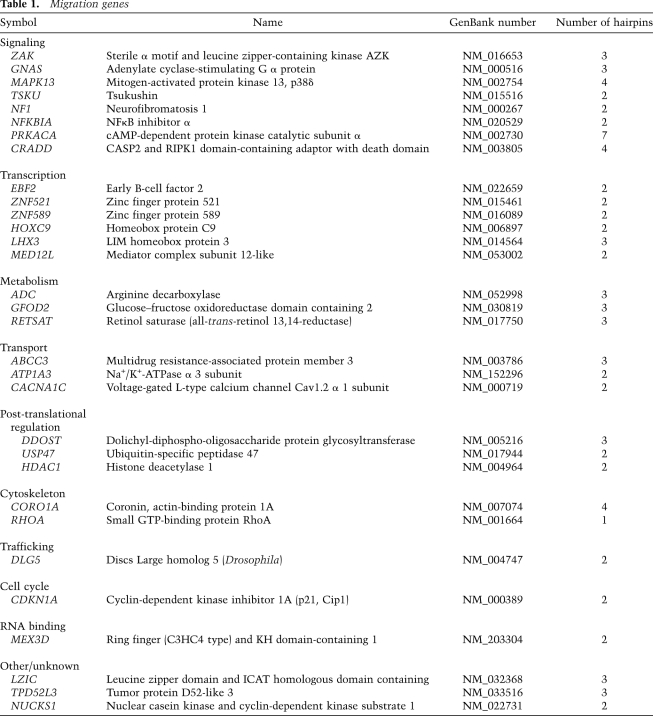
To rigorously validate the candidates identified, we used the following five criteria in deriving the final set of genes: (1) at least two shRNAs per gene had to display a migratory phenotype when retested individually under the original screening conditions (Supplemental Fig. S1). (2) At least two shRNAs per gene had to display a migratory phenotype under different assay conditions (Supplemental Fig. S1; see the Materials and Methods for a detailed description of all assay conditions). Under the conditions used, an shRNA was scored as positive if it induced cell migration more than threefold that of a nontarget control shRNA (Fig. 1B). However, the mean and median fold enrichments for all of the shRNAs shown in Supplemental Figure S1 were 22-fold and 13-fold, respectively. (3) To minimize the chance of off-target effects, hairpins for the same gene had to have nonoverlapping target regions. (4) The candidate genes had to be expressed at baseline in MCF10A cells, as detected by PCR or quantitative RT–PCR (qPCR). (5) Where possible, using qPCR (22 of 31 genes), shRNAs conferring a migratory phenotype had to demonstrate knockdown of their intended target. A list of 31 strong candidate regulators of cell migration is shown in Table 1.
Table 1.
Migration genes
While the above criteria are stringent and aimed at screening out false positives, many genes for which only a single shRNA was identified are presumably valid candidate genes. For example, we noted significant enrichment of a single shRNA targeting RHOA, a small GTPase regulator of the cytoskeleton, previously identified in a number of studies as regulating cell motility (Simpson et al. 2008; Vitorino and Meyer 2008). Thus, the rigorously defined candidate list is likely to underestimate the number of key regulators of cell migration.
As a further test for shRNA knockdown specificity, we validated shRNA-induced phenotypes using chemically synthesized siRNA duplexes targeting sequences distinct from those of the shRNA constructs. Pooled siRNA constructs led to migratory phenotypes in nine of 31 shRNA-defined genes (Supplemental Fig. S2). Of note, the nine genes phenocopied by siRNA were those in which siRNA-mediated knockdown efficiency was comparable with that achieved by shRNAs. For the 22 of 31 genes where expression levels could be monitored by qPCR, we compared the efficiency of knockdown by shRNAs (total of 59 shRNA for 22 genes) to siRNA duplex pools. Target gene knockdown to <10% and to <25% of the transcript remaining was seen with 15 (25%) and 30 (51%), respectively, of the 59 shRNA constructs tested here; in contrast, none of the 22 pooled siRNAs achieved knockdown to the <10% level and only six (27%) reduced target gene expression to 25% of baseline. Thus, we achieved a high confirmation rate in the subset of genes for which siRNA knockdown was successful.
Despite the extensive validation strategy undertaken to confirm that migration phenotypes observed for individual shRNAs within the large pooled screen were specific to the intended target gene, we observed a remarkable case in which multiple shRNAs failed to provide specificity among close gene family members. We identified three positive shRNAs targeting PRKACG, a gene whose endogenous expression could not be documented in MCF10A cells (Supplemental Fig. S3A; data not shown). Instead, PRKACG shRNAs knocked down an abundantly expressed close family member, PRKACA (Supplemental Fig. S3B), despite multiple mismatches in the shRNA target regions (Supplemental Fig. S3C). Knockdown of PRKACA using PRKACA-specific shRNAs and siRNAs yielded the expected migratory phenotypes (Fig. 2; Supplemental Figs. S1, S2, S3D). In summary, within the limits of this genome-wide analysis, as illustrated by these false positive and negative cases, our screening strategy produced a unique set of highly validated regulators of cell migration.
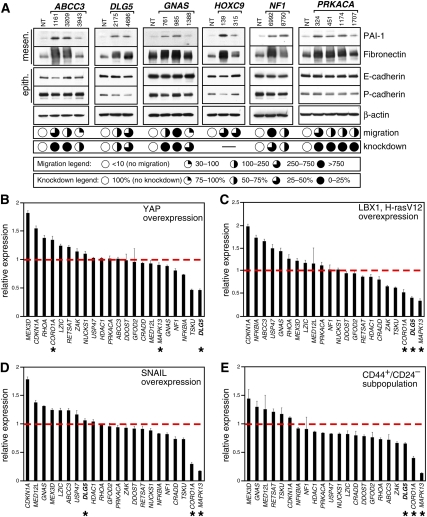
Figure 2.
Migration genes and the EMT program. (A) Immunoblot analysis of EMT markers upon knockdown of migration genes. Selected genes (ABCC3, DLG5, GNAS, HOXC9, NF1, and PRKACA) are shown, and the full analysis is presented in Supplemental Figure S4. Relative strength of the migration phenotype was classified into one of the four categories indicated by the extent to which the open circles were filled in. Similarly, the amount of target gene knockdown is indicated by the extent to which the open circles were filled in. Dashes indicate that the level of expression was below the linear range of a standard qPCR assay. (B) Expression levels of migration genes during EMT induced by retroviral overexpression of YAP in MCF10A cells. (B–D) Relative expression levels of migration genes were derived by comparing cells with EMT treatment to cells transduced with an empty vector. (B–E) Asterisks mark the three most consistently down-regulated genes. (C) Expression levels of migration genes during EMT induced by retroviral overexpression of LBX1 and H-rasV12 in MCF10A cells. (D) Expression levels of migration genes during EMT induced by retroviral overexpression of SNAIL in MCF10A cells. (E) Expression levels of migration genes in the endogenous subpopulation of CD44+/CD24− HMECs. Relative expression levels were derived by comparing the CD44+/CD24− subpopulation (∼5% of the total) to the entire unsorted population.
Genes regulating cell migration
Altogether, we identified a set of 31 validated genes involved in epithelial cell migration (Table 1). Interestingly, only four of these (PRKACA, NUCKS1, MAPK13, and ZAK) belong to the kinase or phosphatase gene families targeted by previous studies, thus highlighting the existence of a rich repertoire of genes that regulate cell migration. Among the entire set of genes identified in earlier RNAi studies, four genes in our cohort (NF1, RHOA, TPD52L3, and PRKACA) were also selected by the study by Simpson et al. (2008) and two genes (RHOA and CDKN1A) were shared with the study by Vitorino and Meyer (2008).
In the analysis of shared cellular pathways implicated by the 31 genes, cAMP signaling emerged as an important regulator of cell migration. GNAS, which encodes a stimulatory G-protein α subunit, links multiple signal transduction pathways to the activation of adenylyl cyclase, and PRKACA, which encodes a catalytic subunit of cAMP-dependent protein kinase (PKA), is a further downstream component of this pathway. Thus, elevated levels of cAMP appear to suppress migration in MCF10A cells. A role of cAMP in cell migration has been proposed based on the effects of pharmacological agents such as forskolin, a direct activator of adenylyl cyclase, or H-89, a specific PKA inhibitor (Chen et al. 2008). Given the postulated redundancy of multiple genes encoding each PKA subunit, the identification of an individual gene, PRKACA, whose knockdown regulates migration, was, perhaps, unexpected. Of note, our screen may have been sensitized to down-regulators of cAMP, given the use of cholera toxin to culture MCF10A cells. A number of unanticipated gene classes were also recovered from this screen. For example, ABCC3 encodes a member of a multidrug resistance-associated protein family, MRP3, that transports a wide variety of organic anions (Borst et al. 2006). DDOST, dolichyl-diphospho-oligosaccharide protein glycosyltransferase, encodes a component of the oligosaccharyltransferase complex, which catalyzes the transfer of high-mannose oligosaccharides to asparagine residues on nascent polypeptides in the lumen of the rough endoplasmic reticulum (Shibatani et al. 2005). The proteins encoded by both of these genes may have specific substrates whose identification may provide pharmacologic opportunities for regulation of cellular migration. Collectively, the list of genes identified in this study (Table 1) identifies a broad range of pathways whose role in cellular migration will warrant detailed functional analyses.
Cell migration genes and the epithelial-to-mesenchymal transition (EMT) program
While epithelial cells can acquire migratory ability through a number of different mechanisms, activation of EMT, a developmental program necessary for proper embryogenesis, has been implicated in an increasing number of diseases and syndromes whose etiology can be traced to aberrations in cell migration (Kurosaka and Kashina 2008). To evaluate the contribution of EMT to the phenotypes observed with the 31 candidate migration genes, we analyzed characteristic epithelial (E-cadherin and P-cadherin) and mesenchymal (Fibronectin and PAI-1) markers implicated in this process (Fig. 2A; Supplemental Fig. S4). More than twofold up-regulation of both mesenchymal markers was observed upon knockdown of eight of 31 (26%) of the migration genes: ABCC3, DLG5, GNAS, HOXC9, NF1, PRKACA, LHX3, and TPD52L3. However, none of these gene knockdowns led to significant reduction in expression of epithelial markers, a coordinated pattern associated with EMT. Thus, EMT does not appear to be a primary mechanism for the induction of migratory properties in our shRNA screen, although it is possible that some identified targets are themselves downstream effectors regulated by the EMT cellular program. However, the fact that our genome-wide loss-of-function screen did not identify master negative regulators of EMT, analogous to known positive transcriptional inducers such as SNAIL, suggests that such regulators are probably rare (Gumireddy et al. 2009) and that, more often, EMT is likely to be a gain-of-function phenomenon.
To test whether some of the identified migration genes were themselves regulated as part of the cellular EMT switch, we measured the levels of individual genes upon induction of EMT by several independent means: overexpression of SNAIL (Cano et al. 2000), YAP (Overholtzer et al. 2006), and LBX1/H-rasV12 (Fig. 2B–E; Yu et al. 2009). Three genes (CORO1A, DLG5, and MAPK13) were identified as being the most significantly down-regulated during EMT (more than twofold reduction in two of three EMT conditions). Of note, EMT in immortalized human mammary epithelial cells (HMECs) has been linked recently to the acquisition of stem cell-like characteristics (Mani et al. 2008) marked by the (CD44+/CD24−) phenotype. All three genes also displayed down-regulation in this purified subset of cells. EMT-associated down-regulation of genes whose knockdown leads to increased cell migration suggests that these could contribute to the overall phenotype associated with EMT.
Regulation of cell migration by DLG5
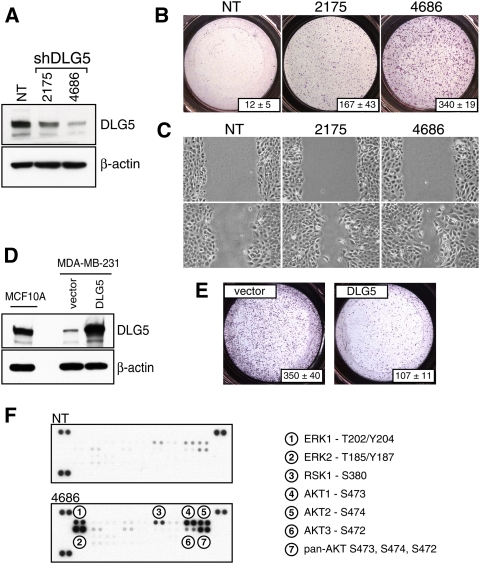
We selected DLG5 for further functional analyses since it highlights a novel pathway not previously implicated in the regulation of cell migration, and DLG5 protein levels are clearly down-regulated during EMT triggered by overexpression of YAP or LBX1/H-rasV12 (Supplemental Fig. S6A). While its name is derived from partial homology with the family of Discs Large (DLG)/Lethal giant larvae/Scribble cell polarity genes, DLG5 has been shown recently to be an adaptor molecule implicated in delivering vesicular cargo to the plasma membrane (Taniuchi et al. 2005; Nechiporuk et al. 2007). In neuroepithelium, N-cadherin appears to be a substrate of DLG5 (Nechiporuk et al. 2007), with other substrates likely to be involved in mammary and other epithelia. In our study, effective DLG5 knockdown using two independent shRNA constructs resulted in significant induction of baseline cell migration in Boyden chambers, as well as wound scratch-induced migration assays (Fig. 3A–C). Similar induction of cell migration upon DLG5 knockdown was observed in two additional cell lines: MCF12A and HCC1954 (Supplemental Fig. S5). While baseline expression of DLG5 is 10-fold lower in the highly migratory mesenchymal breast cancer cell line MDA-MB-231 compared with nonmigratory MCF10A cells, ectopic expression of DLG5 to an equal level resulted in significant inhibition of cell migration without any effect on proliferation rate or the overall levels of epithelial or mesenchymal markers (Fig. 3D,E; data not shown). Thus, suppression of high baseline levels of DLG5 in poorly migratory MCF10A breast epithelial cells increases their migration, whereas overexpression of DLG5 in highly migratory breast cancer cells suppresses migration. To explore a potential role of DLG5 in breast cancer, we examined expression profiles of primary breast tumors in several publicly available data sets (Miller et al. 2005; Chin et al. 2006; Neve et al. 2006; Sotiriou et al. 2006). DLG5 down-regulation was highly correlated with increasing tumor grade in four independent data sets (Supplemental Fig. S6B). Among the various histological subtypes of breast cancer, the lowest levels of DLG5 expression were observed in basal-type (Sarrio et al. 2008) and metaplastic (Hennessy et al. 2009) cancers, which are associated with EMT, stem-like characteristics, and a poor prognosis (Supplemental Fig. S6C).
Figure 3.
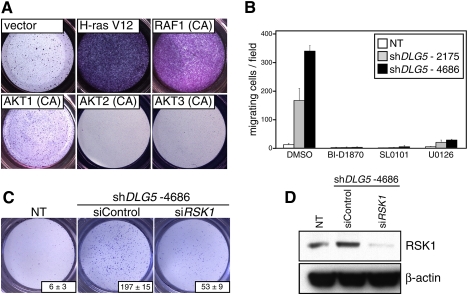
DLG5 regulates cell migration. (A) Immunoblot analysis of DLG5 knockdown in MCF10A cells using two independent shRNAs. (B) DLG5 knockdown in MCF10A cells leads to induction of cell migration as assayed by Boyden chambers. Numbers in the bottom right of each panel represent quantitation of the migration phenotypes and are derived by counting individual cells in three representative fields. (C) DLG5 knockdown in MCF10A cells leads to induction of cell migration, as assessed by wound scratch assays. (D) Immunoblot analysis showing DLG5 overexpression in MDA-MB-231 cells and its relative level in comparison with MCF10A cells. (E) DLG5 overexpression in MDA-MB-231 cells results in inhibition of cell migration. Numbers in the bottom right of each panel represent quantitation of the migration phenotypes and are derived by counting individual cells in three representative fields. (F) The phospho-specific antibody array identifies activation of multiple signaling pathways upon DLG5 knockdown using the best available hairpin.
Since DLG5 has been shown to play a role in the delivery of N-cadherin to the cell surface (Nechiporuk et al. 2007), we first looked at the cell surface localization of candidate molecules such as E-cadherin, P-cadherin, and N-cadherin upon DLG5 knockdown, but observed only relatively minor decreases in their cell surface localization (data not shown). To extend these analyses to an unbiased survey of downstream signaling, we examined the phosphorylation status of multiple pathways using phospho-specific antibody arrays (Fig. 3F). Prominent increases in phosphorylation of ERK1 and ERK2; their downstream kinase, RSK1; and AKT1–3 were observed following knockdown of DLG5. To functionally test the involvement of these pathways, we individually activated these and assessed their effect on cell migration of MCF10A cells. Activation of ERK1/2 by overexpression of H-rasV12 or constitutively active RAF1 resulted in dramatic induction of cell migration (Fig. 4A). In contrast, overexpression of constitutively active AKT1, AKT2, or AKT3 did not result in induction of cell migration. To further dissect the signaling events downstream from H-rasV12 and RAF1, we used common pharmacological inhibitors and tested their ability to suppress cell migration upon DLG5 knockdown. Suppression of ERK1/2 by U0126, an inhibitor of the upstream kinase MEK, abrogated the effect of DLG5 knockdown. A similarly dramatic suppression was observed upon addition of either of two inhibitors of RSK kinases: SL0101 and BI-D1870 (Fig. 4B; Bain et al. 2007). Consistent with the results of pharmacological inhibition, siRNA-mediated knockdown of RSK1 suppressed the migratory phenotype induced by DLG5 knockdown, further suggesting that this kinase appears to be a critical and specific mediator of DLG5 signaling (Fig. 4C,D).
Figure 4.
Activation of the ERK pathway results RSK-dependent cell migration. (A) Activation of multiple signaling cascades points to a critical role of the ERK pathway in the migration of MCF10A cells. (B) Treatment of cells with small molecule inhibitors of the ERK/RSK pathway results in suppression of cell migration phenotypes. Inhibitors were used at previously described concentrations: 10 μM U0126, 10 μM BI-D1870, and 100 μM SL0101. (C) Knockdown of RSK1 can significantly attenuate cell migration due to DLG5 knockdown. (D) Immunoblot analysis showing efficacy of RSK1 knockdown upon siRNA treatment.
RSK is a critical determinant of cell migration
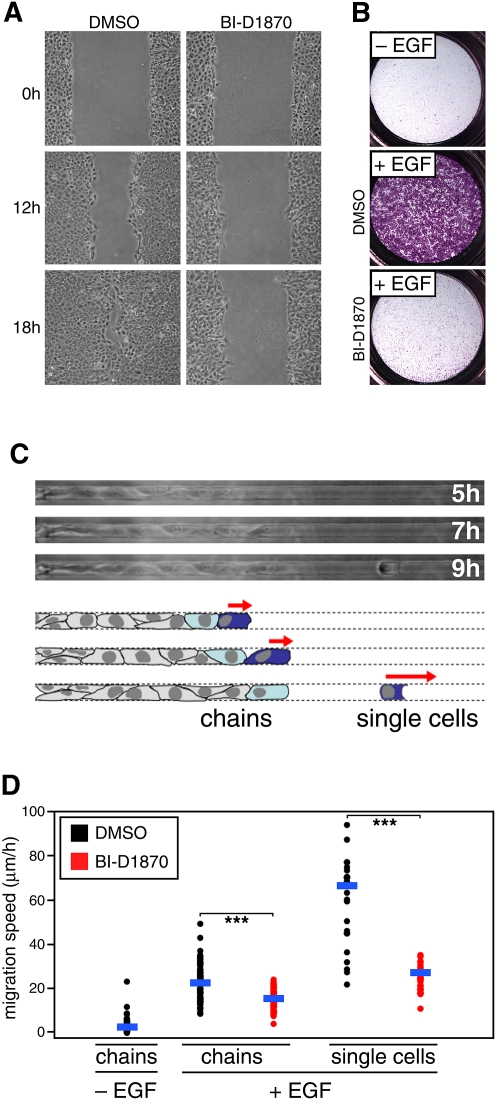
While the functional properties of DLG5 remain to be defined, the dependence of its migratory phenotype on ERK/RSK activation prompted us to investigate the general motility of parental MCF10A cells upon activation of the ERK pathway following the addition of exogenous EGF. Indeed, EGF-induced migration of cells in a Boyden chamber assay and in a wound scratch assay was essentially abrogated by the specific RSK inhibitor BI-D1870 (Fig. 5A,B). To further understand the role of RSK in cell migration, we made use of a microfluidic device in which locomotion is mechanically constrained within an array of linear microcapillaries (Fig. 5C; Irimia and Toner 2009). Consistent with their low migratory baseline, MCF10A cells seeded into the microfluidic device in the absence of EGF did not enter the channels (Fig. 5D; Supplemental Movie 1). In contrast, EGF-treated cells exhibited robust cell migration with two distinct phenotypes: initial migration of epithelial cells as a grouped chain, followed by greatly accelerated locomotion of the frontrunner as a single cell upon its dissociation from the chain (Fig. 5C,D; Supplemental Movies 2–5). Addition of BI-D1870 had a modest effect on migration of cells linked within a chain, but greatly suppressed single-cell migration. Single-cell migration has been implicated in cancer metastasis, although the biological correlates of the effects measured in microfluidic chambers are only starting to emerge (Wolfer et al. 2010). Importantly, MCF10A cells, following treatment with BI-D1870, were visibly viable, confirming RSK suppression of their motility without evidence of cytotoxicity.
Figure 5.
RSK modulates cell migration in multiple cell migration contexts. (A) Wound scratch assay showing decreased cell migration upon addition of RSK inhibitor BI-D1870 (10 μM) in an EGF-stimulated model of cell migration using MCF10A cells. Wound scratch was introduced when MCF10A cells became confluent. Fresh media containing EGF and the inhibitor was added after introducing the wound scratch. Migration status was monitored at the indicated time points. (B) Boyden chamber assay showing decreased cell migration upon addition of RSK inhibitor BI-D1870 (10 μM) in an EGF-stimulated model of cell migration using MCF10A cells. MCF10A cells were resuspended in an EGF-free media and placed in the Boyden chamber. Where indicated, EGF was added to the media in the lower chamber. The assay duration was 24 h. (C) Schematic representation of migration phenotypic categories observed in the microfluidic device. (D) Quantitative assessment of the effects observed on cell migration in the microfluidic device. RSK inhibitor BI-D1870 (10 μM) was used. Horizontal blue bars indicate a mean value for a particular data set. Two-tailed Student's t-test was used to conduct pairwise comparisons of the data. (***) P < 1 × 10−8.
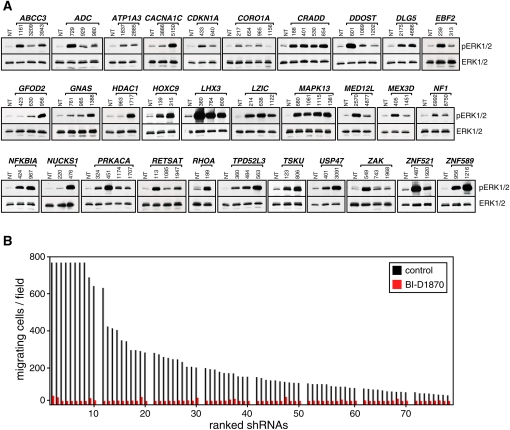
ERK and RSK appear to play a central role in cell migration, raising the possibility that other genes identified in our migration screen, in addition to DLG5, might also share this effector pathway. We first tested all of the migratory candidate gene shRNA knockdowns for activation of the ERK pathway using phospho-specific antibodies. Remarkably, ERK activation was universal upon suppression of all of the genes identified in our migration screen (Fig. 6A). These findings were confirmed using siRNA-mediated knockdown (Supplemental Fig. S7). Given the effect of RSK inhibition of EGF-induced migration of MCF10A parental cells, we tested its effect on all of the shRNA constructs targeting the 31 migration genes identified in our screen. Boyden chamber assays were used for optimal quantitation. Remarkably, BI-D1870 effectively abrogated cell migration induced by all of the hairpins tested (Fig. 6B). Thus, activation of RSK is a critical downstream signal for diverse functional pathways identified in an unbiased global screen for regulators of cell migration.
Figure 6.
Mechanistic convergence of all migration genes identified. (A) Immunoblot analysis showing increased levels of phospho-ERK1/2 upon knockdown of migration genes. (B) Migratory phenotype of all hairpins tested displayed striking dependence on RSK activity. RSK inhibitor BI-D1870 (10 μM) was used.
Discussion
We report a highly validated collection of genes that negatively regulate cell migration, identified through a genome-scale pooled lentiviral shRNA screen. The genes identify rate-limiting steps within diverse cellular pathways whose knockdown has a potent effect in up-regulating cell migration phenotypes. While some of these have well-established links to cellular motility, others are uncharacterized and will benefit from detailed functional studies. The genes identified represent diverse functional classes, yet we observed a striking convergence of their downstream signaling on the ERK pathway, and specifically RSK activation. This finding points to a prominent role for this kinase as a common effector for multiple migratory stimuli, with potential therapeutic implications.
Our genome-wide shRNA screen relied on a high-titer pooled collection of constructs with multiple validated shRNAs per gene, high infectivity of MCF10A cells, and a simple yet rigorous selection criterion; i.e., migration across a membrane by cells with minimal baseline motility. Despite these carefully optimized experimental conditions, we found that extensive validation of hits was essential to distinguish spurious results from bona fide migration regulators. The false positives and false negatives arose from a variety of factors, and their exclusion often required detailed analyses, including assay repetition, confirmation of endogenous gene expression, and matching level of shRNA and siRNA knockdown with degree of phenotype. Given these considerations, the set of 31 genes identified here constitute a highly validated set but are unlikely to represent all rate-limiting hits capable of activating cellular migration. Of note, our gene set shares some candidates with two previous siRNA screens, which targeted kinases and other selected gene families for enhanced activity in wound-healing assays. Differences in cell types used, assay conditions, and established criteria are likely to explain why some hits identified in these assays did not score in our genome-wide screen.
Our specific analysis of cell migration in the context of EMT suggests that epithelial cells are not regulated by a simple loss-of-function switch between epithelial and mesenchymal programs. Loss-of-function hits did lead to pronounced up-regulation of mesenchymal markers, but without the coordinated repression of epithelial markers that is characteristic of EMT. Indeed, a genome-wide shRNA screen may help dissect a complex cell fate phenotype such as EMT, which is likely to comprise a number of coregulated components, including cellular migration and expression of epithelial versus mesenchymal markers. Some shRNA hits such as CORO1A, MAPK13, and DLG5 were identified as potential EMT effectors by virtue of their down-regulation following induction of EMT through ectopic expression of well-known transcriptional master regulators. Interestingly, these migration suppressor genes were differentially repressed by distinct EMT triggers, suggesting a degree of molecular variation between grossly similar EMT phenotypes (Kalluri and Weinberg 2009; Thiery et al. 2009).
The large number of diverse and unexpected gene products identified as negative regulators of epithelial cell migration, which ranged from transcription factors to signaling molecules and cellular transporters, will require in-depth functional analysis. As an example, understanding the mechanism underlying the potent effect of DLG5 knockdown on cellular migration may provide new insight into pathways that are currently poorly defined. While DLG5 has been implicated as an adaptor molecule involved in delivering N-cadherin to the cell surface (Nechiporuk et al. 2007), Dlg5-null mice display abnormalities in E-cadherin localization, suggesting a broader spectrum of substrates. The roles of cadherins in cell migration have been studied extensively, (Onder et al. 2008; Simpson et al. 2008), but the rate-limiting steps in their localization at the cell surface have not been established, nor has the mechanism by which loss of such localization triggers potent ERK activation and cell migration. Future studies of DLG5, along with other novel migration regulators identified here, will help define the full range of pathways with a rate-limiting role in cell motility.
The convergence of diverse cellular migration pathways on the ERK effector RSK is remarkable. This kinase has been identified previously as an important component of RAF1-mediated migration in canine MDCK cells (Doehn et al. 2009), and our genome-wide scan now points to this gene as a common downstream effector for multiple distinct migratory stimuli. RSK kinases are unusual in that they possess two kinase domains sequentially activating one another. ERK1/2 phosphorylates the RSK C-terminal domain, which activates the RSK N-terminal domain and allows it in turn to phosphorylate a large set of target proteins (Anjum and Blenis 2008). While the extent of functional redundancy among the four RSK1–4 family members is not completely understood, all of them are suppressed by the BI-D1870 inhibitor, which targets the common N-terminal kinase (Sapkota et al. 2007). Our siRNA experiments knocking down RSK1 only partially phenocopied the effects of pharmacological inhibitors (Fig. 4C), suggesting that multiple family members may regulate cell migration in MCF10A. In fact, all four members of this family, RSK1–4, are expressed in MCF10A cells (data not shown). While the involvement of individual RSK family members in cancer remains to be elucidated, initial reports strongly suggest roles in tumor growth and metastasis. Overexpression of RSK1 and RSK2 kinases has been reported in a number of human cancers, including breast and prostate (Clark et al. 2005; Smith et al. 2005). Overexpression of constitutively active RSK2 has been shown to elicit a motile phenotype in MDCK cells (Doehn et al. 2009), RSK2 expression enhanced metastastic potential in head and neck squamous cell carcinoma (HNSCC) cells (Kang et al. 2010), and RSK2 knockdown decreased cell invasion in vitro and attenuated lymph node metastases in a xenograft HNSCC model. These reports clearly illustrate specific contexts in which RSKs have been shown to play an important role in cellular migration, and our studies now suggest a broad functional dependence of cell motility phenotypes on RSK activation. Beyond cell migration, RSKs have also been implicated in additional cellular pathways, such as cell survival in some FGFR1 or FGFR3-driven cancer cell lines (Kang et al. 2007; Xian et al. 2009). However, RSK inactivation does not appear to be toxic in most cell types, as demonstrated by treatment with pharmacologic inhibitors and by the viability of Rsk1/Rsk2/Rsk3 triple-knockout mice (Dumont et al. 2005). Should systemic suppression of RSK activity indeed be well tolerated, RSK family members may constitute promising therapeutic targets for the suppression of cancer metastasis.
Materials and methods
Tissue culture
MCF10A cells were cultured as described (Debnath et al. 2003). Immortalized HMECs were a kind gift from Dr. Joan Brugge and were cultured as described previously (Overholtzer et al. 2006). MDA-MB-231, MCF12A, and HCC1954 cells were grown according to American Type Culture Collection (ATCC) recommendations.
Primary screen
Use of the lentiviral pooled library has been described previously (Luo et al. 2008). Briefly, MCF10A were infected at a multiplicity of infection (MOI) of ∼0.3 and selected using 2 μg/mL puromycin. Cells were switched to EGF-free assay media (Debnath et al. 2003) for 24 h before setting up the migration assays. One-hundred-forty-four Boyden chambers were set up in a six-well format with 5 × 105 cells per chamber. Migratory cells were recovered from the bottom of the filters after 48 h, then expanded for 3 d and harvested. Cells from 36 individual wells were combined together, thus providing us with four independent pools of enriched cells. Control cells that did not undergo the enrichment scheme were harvested at the same time to provide a reference population, also in quadruplicate. Isolation of DNA, labeling, microarray hybridization, and data processing were done as described previously (Luo et al. 2008).
Boyden chamber migration conditions
MCF10A condition #1
Cells were switched to an EGF-free assay media for 24 h before the migration assay. Cells were trypsinized, counted, and plated at 5 × 104 cells per chamber in the 24-well plate format. Both the top and bottom of the chamber contained the same assay media. After 48 h, excess cells in the top chamber were removed with cotton swabs, and the cells on the bottom of the filter were fixed and stained with Crystal Violet.
MCF10A condition #2
No pretreatment with EGF-free assay media was done under these conditions. Cells were trypsinized, counted, and plated at 5 × 104 cells per chamber in the 24-well plate format. Both the top and bottom of the chamber contained the same assay media. After 24 h, excess cells in the top chamber were removed with cotton swabs, and the cells on the bottom of the filter were fixed and stained with Crystal Violet. To quantitate the number of cells, three representative nonoverlapping fields, each representing ∼10% of the total membrane surface area, were counted manually. Numbers reported represent the average number of cells per field.
Other cell lines
MDA-MB-231, MCF12A, and HCC1954 cells were set up similarly to MCF10A condition #2 described above, except that 2 × 104, 5 × 104, and 5 × 104 cells, respectively, were used per chamber.
Hairpin level confirmations
MCF10A cells were plated in 24-well dishes at 2 × 104 per well and were spin-infected the next day with individual lentiviruses at an approximate MOI of 1–3. The next day, puromycin selection (at 2 μg/mL) was started to obtain stable cell populations. After 2 d, cells were replated to 6-cm2 dishes and migration assays were set up when confluency of ∼50%–75% was reached. The identity and sequences of positive hairpins for the 31 migration genes are listed in Supplemental Table S1.
siRNA confirmations
ON-TARGETplus SMARTpool duplexes (Dharmacon) were transfected using Lipofectamine2000 reagent (Invitrogen) according to manufacturer's instructions. MCF10A cells were plated in six-well plates and transfected the following day with siRNA duplexes at a final concentration of 25 nM for 48 h. Cells were then expanded into 6-cm dishes and migration assays were set up in the following day using condition #2, outlined above.
Immunoblot analysis
Cells were harvested in 1× RIPA buffer containing 1× protease inhibitor cocktail (Complete, Roche). Cell lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4°C. For immunoblotting analysis, lysates were loaded onto 4%–15% SDS-PAGE gels (ReadyGel, Bio-Rad), and subsequently transferred onto Immobilon PVDF membrane (Millipore). Proteins were visualized with Western Lightning Plus chemiluminescence kit (Perkin Elmer). Antibodies used were CDH1 (610181; BD Biosciences), CDH3 (610227, BD Biosciences), PAI-1 (612024; BD Biosciences), Fibronectin (F3648, Sigma), β-actin (ab6276, Abcam), phospho-ERK1/2 (Thr 202/Tyr 204) (9101, Cell Signaling), total-ERK1/2 (4695, Cell Signaling), and DLG5 (HPA00555, Sigma).
FACS
FACS isolation of the CD44+/CD24− population was done as described previously (Yu et al. 2009).
qPCR
RNA was extracted using RNeasy Minikit (Qiagen), and cDNA synthesis was performed using SuperScript III reverse transcriptase (Invitrogen). Twenty-two of 31 migration genes were expressed at sufficient levels permitting reliable qPCR quantitation using Power SYBR Green PCR Master Mix (Applied Biosystems). The sequences of the PCR primer pairs (all listed from 5′ to 3′) are listed in Supplemental Table S2. All samples were done in triplicate, and the relative abundance was derived by standardizing the input to the control signal, GAPDH.
Microfluidics
To quantify the motility changes of MCF10A cells at single-cell resolution, in the presence of RSK inhibitors, we employed a microfluidic technique developed previously for the study of persistent cancer cell migration (Irimia and Toner 2009). Briefly, microfluidic devices were assembled using polydimethylsiloxane (PDMS) and glass. PDMS was cast on a silicon wafer with photolithographically defined features in the form of parallel arrays of channels (10 × 10 × 600 μm). After casting, devices were cut to 5-mm diameter, and one 1-mm well was punched in the center for cell seeding. PDMS devices were bound to the glass of glass-bottom 24-well plates, two devices per well, using an oxygen plasma protocol. Immediately after bonding, the glass and PDMS surfaces of the channels were coated with collagen IV, deposited by physical absorption from a 2 μg/mL solution in PBS. Before experiments, the wells of the plate were filled with 2 mL of media with and without the drug. A volume of 2 μL from a cell suspension at 1 × 106 cells per milliliter density was loaded into each well of the PDMS devices, and live imaging was started immediately afterward. For this purpose, the plate was placed on the motorized stage of a Nikon Ti microscope, inside the environmentally controlled chamber (37°C, 5% CO2, humidified). Images were acquired automatically every 15 min from the entire plate, under phase-contrast and perfect focusing conditions for 24 h, under the control of Nikon Elements software. At least 400 channels were imaged per condition. The migration of at least 50 cells per condition was tracked manually using ImageJ by tracking the center of the cells moving through the channels. The first cell of a chain and individual cells were tracked and average migration speed was recorded.
Other reagents
All pharmacological inhibitors used were commercially available: BI-D1870 (Stemgent), SL0101 (Tocris Bioscience), U0126 (EMD Biosciences). Expression constructs for constitutively activated RAF1, AKT1, AKT2, and AKT3 were from Addgene. H-rasV12 expression construct was a generous gift from Dr. Julian Downward. Human DLG5 ORF was cloned into the pBABEpuro vector as a BamHI/NotI fragment. A single Flag tag was added to the N terminus during initial PCR. Construct was verified by sequencing.
Acknowledgments
We thank John Doench for critical reading of this manuscript. This work was supported by March of Dimes Foundation (#5-FY09-73) and Charles H. Hood Foundation (to G.A.S); NIH T32 Training Grant CA09361-27 (to J.Z.); Howard Hughes Medical Institute and NIH CA129933 (to D.A.H), and 2P50 CA89393-09 (DF/HCC SPORE in Breast Cancer) and BC05-COE W81XH-06-2-0033 (DOD Center of Excellence in Breast Cancer) (to S.R.); fellowships from Office of Enrichment Programs at Harvard Medical School and AID for Cancer Research (to L.W.N.); and R21CA135601 (to D.I.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1989110.
Supplemental material is available at http://www.genesdev.org.
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA 2009. Epithelial–mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest 119: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum R, Blenis J 2008. The RSK family of kinases: Emerging roles in cellular signalling. Nat Rev Mol Cell Biol 9: 747–758 [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P 2007. The selectivity of protein kinase inhibitors: A further update. Biochem J 408: 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, Zelcer N, van de Wetering K 2006. MRP2 and 3 in health and disease. Cancer Lett 234: 51–61 [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA 2000. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2: 76–83 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang JJ, Huang XY 2008. cAMP inhibits cell migration by interfering with Rac-induced lamellipodium formation. J Biol Chem 283: 13799–13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Clark DE, Errington TM, Smith JA, Frierson HF Jr, Weber MJ, Lannigan DA 2005. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res 65: 3108–3116 [DOI] [PubMed] [Google Scholar]
- Cram EJ, Shang H, Schwarzbauer JE 2006. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci 119: 4811–4818 [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268 [DOI] [PubMed] [Google Scholar]
- Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, Cohen MS, Johansen JV, Winther BR, Lund LR, et al. 2009. RSK is a principal effector of the RAS–ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol Cell 35: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Umbhauer M, Rassinier P, Hanauer A, Verlhac MH 2005. p90Rsk is not involved in cytostatic factor arrest in mouse oocytes. J Cell Biol 169: 227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K 2010. Plasticity of cell migration: A multiscale tuning model. J Cell Biol 188: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S, Zhu X, Doillon CJ, Green MR 2008. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev 22: 2932–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q 2009. KLF17 is a negative regulator of epithelial–mesenchymal transition and metastasis in breast cancer. Nat Cell Biol 11: 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. 2009. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D, Toner M 2009. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol 1: 506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA 2009. The basics of epithelial–mesenchymal transition. J Clin Invest 119: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Dong S, Gu TL, Guo A, Cohen MS, Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, et al. 2007. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell 12: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Elf S, Lythgoe K, Hitosugi T, Taunton J, Zhou W, Xiong L, Wang D, Muller S, Fan S, et al. 2010. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest 120: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka S, Kashina A 2008. Cell biology of embryonic migration. Birth Defects Res C Embryo Today 84: 102–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci 106: 16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. 2005. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci 102: 13550–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk T, Fernandez TE, Vasioukhin V 2007. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5−/− mice. Dev Cell 13: 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA 2008. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 68: 3645–3654 [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci 103: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota GP, Cummings L, Newell FS, Armstrong C, Bain J, Frodin M, Grauert M, Hoffmann M, Schnapp G, Steegmaier M, et al. 2007. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J 401: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J 2008. Epithelial–mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68: 989–997 [DOI] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR 2005. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44: 5982–5992 [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K 2008. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319: 617–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS 2008. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol 10: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Smith JA, Poteet-Smith CE, Xu Y, Errington TM, Hecht SM, Lannigan DA 2005. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res 65: 1027–1034 [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. 2006. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272 [DOI] [PubMed] [Google Scholar]
- Taniuchi K, Nakagawa H, Nakamura T, Eguchi H, Ohigashi H, Ishikawa O, Katagiri T, Nakamura Y 2005. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res 65: 105–112 [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Vitorino P, Meyer T 2008. Modular control of endothelial sheet migration. Genes Dev 22: 3268–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bo J, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ 2006. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell 10: 483–495 [DOI] [PubMed] [Google Scholar]
- Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, et al. 2010. MYC regulation of a ‘poor-prognosis’ metastatic cancer cell state. Proc Natl Acad Sci 107: 3698–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian W, Pappas L, Pandya D, Selfors LM, Derksen PW, de Bruin M, Gray NS, Jonkers J, Rosen JM, Brugge JS 2009. Fibroblast growth factor receptor 1-transformed mammary epithelial cells are dependent on RSK activity for growth and survival. Cancer Res 69: 2244–2251 [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA 2008. Epithelial–mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell 14: 818–829 [DOI] [PubMed] [Google Scholar]
- Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E, Ramaswamy S, Maheswaran S, Haber DA 2009. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev 23: 1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]