Abstract
Upon intracellular bacterial exposure, the Crohn's disease and sarcoidosis susceptibility protein NOD2 (nucleotide oligomerization domain protein 2) binds to the protein kinase RIP2 (receptor-interacting protein 2) to coordinate NF-κB (nuclear factor κ B)-mediated cytokine responses. While RIP2 clearly has kinase activity, the function of its kinase domain has been enigmatic. Although originally classified as a serine–threonine kinase based on homology scans, we find that RIP2 also has tyrosine kinase activity. RIP2 undergoes autophosphorylation on Tyr 474 (Y474). This phosphorylation event is necessary for effective NOD2 signaling and does not occur in the presence of the most common Crohn's disease-associated NOD2 allele. Given this tyrosine kinase activity, a small-molecule inhibitor screen designed to identify pharmacologic agents that inhibit RIP2's tyrosine kinase activity was performed. At nanomolar concentrations, the EGFR (epidermal growth factor receptor) tyrosine kinase inhibitors gefitinib (Iressa) and erlotinib (Tarceva) were found to inhibit both RIP2 tyrosine phosphorylation and MDP (muramyl dipeptide)-induced cytokine release in a variety of NOD2 hyperactivation states. This effect is specific for RIP2 and does not depend on EGFR. The finding that RIP2 has tyrosine kinase activity and the finding that gefitinib and erlotinib, two agents already used clinically for cancer chemotherapy, can inhibit this activity suggest that RIP2's tyrosine kinase activity could be targeted specifically in the treatment of inflammatory diseases.
Keywords: NOD2, RIP2, ITCH, innate immunity, tyrosine kinase
NOD2 (nucleotide oligomerization domain protein 2) is an essential regulator of immunologic mucosal homeostasis (Chen et al. 2009; Rosenstiel and Schreiber 2009). In patients harboring loss-of-function NOD2 alleles, this decreased function causes genetic Crohn's disease, an autoinflammatory disorder of the gastrointestinal tract (Hugot et al. 2001; Ogura et al. 2001a; Girardin et al. 2003; Inohara et al. 2003). Conversely, patients who harbor gain-of-function NOD2 alleles develop Blau Syndrome and sarcoidosis, two autoinflammatory granulomatous diseases (Miceli-Richard et al. 2001; Chamaillard et al. 2003; Schurmann et al. 2003; Tanabe et al. 2004; Kanazawa et al. 2005). In addition to diseases of genetic NOD2 alteration, NOD2 has also been implicated in the pathogenesis of Crohn's disease in patients who do not harbor loss-of-function NOD2 alleles (Stronati et al. 2008; Negroni et al. 2009). In these patients, wild-type NOD2 expression and activity are increased, and it has been argued that this increase may help drive the inflammation present in pediatric Crohn's disease patients (Stronati et al. 2008; Negroni et al. 2009). Thus, wild-type NOD2 alleles, gain-of-function NOD2 mutant alleles, and loss-of-function NOD2 polymorphic alleles all influence the development of autoinflammatory diseases, and these clinical genotype–phenotype correlations strongly suggest that NOD2 is essential to maintaining inflammatory and immunologic homeostasis.
Structurally, NOD2 contains two N-terminal CARDs (caspase activation recruitment domains) for interaction with other CARD-containing proteins, an intermediary NOD for oligomerization, and 11 leucine-rich repeats (LRRs) responsible for sensing the intracellular presence of MDP (muramyl dipeptide). It has been proposed that, in the absence of agonist, NOD2 (like other NLRP proteins) (Duncan et al. 2007; Faustin et al. 2007) is in an inactive state, with the LRRs folded over the NOD region. Upon sensing MDP, NOD2 undergoes a conformational change that exposes the NOD, promoting NOD2 oligomerization and exposure of its CARDs (Tanabe et al. 2004; Strober et al. 2006). Once this occurs, NOD2 then binds the protein kinase RIP2 (receptor-interacting protein 2) (RICK and CARDIAK) via homotypic CARD–CARD interactions (for review, see Strober et al. 2006). Recruitment of RIP2 to activated NOD2 is a critical step in allowing the activation of downstream signal transduction pathways, as both genetic loss of RIP2 and loss of RIP2 expression cause loss of NOD2-induced signaling responses (Kobayashi et al. 2002; Park et al. 2007). Current models suggest that, upon activation, RIP2 is K63-polyubiquitinated by the E3 ubiquitin ligases cIAP1, cIAP2, and xIAP (Bertrand et al. 2009; Krieg et al. 2009). This activated RIP2 induces the K63-linked polyubiquitination of NEMO, and the K63-linked polyubiquitination of both RIP2 and NEMO allows the TAK1–TAB1/2/3 complex to bind and phosphorylate IKKβ to stimulate NF-κB (nuclear factor κ B) and drive cytokine responses (Abbott et al. 2007; Windheim et al. 2007; Yang et al. 2007; Hasegawa et al. 2008). Once NF-κB is activated, the E3 ubiquitin ligase ITCH specifically ubiquitinates activated RIP2 to down-regulate NOD2-induced signal transduction events and NOD2-induced cytokine responses (Tao et al. 2009). As such, cells genetically deficient in Itch show increased MDP-induced NF-κB activity, increased MDP-induced cytokine responses, and an inability to appropriately down-regulate the NOD2 signaling pathway (Tao et al. 2009).
One of the most enigmatic features of the NOD2 signaling pathway is the role of the kinase activity of the NOD2-binding protein RIP2. RIP2 was originally classified as a serine–threonine kinase based on homology scans performed in the 1990s (Inohara et al. 1998; McCarthy et al. 1998; Thome et al. 1998). While the kinase activity of RIP2 was clearly demonstrated, it was unclear whether RIP2 had specific kinase activity toward only serines or threonines. In addition, it was initially thought that the kinase activity of RIP2 was dispensable altogether for NOD2-mediated signal transduction, as kinase-inactive mutants of RIP2 (K47A and D146N) displayed equal or greater capacity to induce NF-κB activation compared with wild-type RIP2 in overexpression systems (Inohara et al. 1998; McCarthy et al. 1998; Thome et al. 1998; Eickhoff et al. 2004). However, as the kinase domain makes up ∼50%–60% of the RIP2 protein, and as RIP2 is essential for NOD2 signaling, it seems unlikely that the kinase domain is completely inert. More recent studies have more clearly highlighted the role that the kinase domain plays in NOD2 signaling. A knock-in mouse containing kinase-dead (K47A) RIP2 showed that RIP2 kinase activity was necessary for optimal RIP2 expression (Nembrini et al. 2009). Separate work using a pharmacologic inhibitor of both RIP2 and p38 (SB203580) showed that RIP2 kinase activity was necessary for both RIP2's expression and NOD2's activity, and that this inhibitor could dampen NOD2-dependent signaling responses (Windheim et al. 2007). Collectively, these more recent findings have helped clarify RIP2's kinase activity in NOD2 signaling, and have suggested that pharmacologically targeting RIP2 may have efficacy in the autoinflammatory disorders in which NOD2:RIP2 activity is aberrantly high.
These somewhat contradictory findings concerning the kinase domain of RIP2 prompted us to more formally evaluate RIP2's kinase activity. In this study, we find that, upon NOD2 activation, RIP2 is inducibly tyrosine-phosphorylated. We map the phosphorylation site to Tyr 474 (Y474) and show that phosphorylation of this tyrosine is necessary for maximal RIP2-induced NF-κB activation and maximal signaling synergy with NOD2. We further show that RIP2 possesses tyrosine kinase activity and is capable of tyrosine autophosphorylation in response to NOD2 activation. Lastly, we show that RIP2's tyrosine kinase activity can be inhibited by the epidermal growth factor receptor (EGFR) inhibitors gefitinib and erlotinib, and that this inhibition can dampen NOD2-induced cytokine release and NOD2-induced NF-κB activation in a variety of NOD2 hyperactive states. These findings suggest that inhibition of NOD2 signaling may have efficacy in inflammatory diseases in which NOD2 is active. Taken together, this study highlights an unexpected and novel finding regarding the tyrosine kinase activity of RIP2. This work establishes the tyrosine phosphorylation site Y474 as a key activator and marker of activated RIP2, and it identifies two pharmacologic agents already in clinical use as potential chemotherapeutics in clinical autoinflammatory disorders.
Results
RIP2 is tyrosine-phosphorylated in response to NOD2 activation
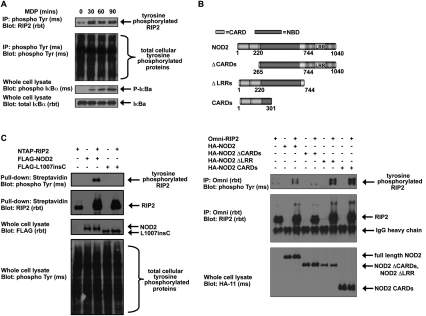
RIP2 is heavily post-translationally modified, and our own mass spectrometry (MS) data have found ∼20–25 phosphorylation sites on RIP2 (DW Abbott and JM Asara, unpubl.). While most of these occur on serine or threonine, we also found through proteomic mapping by liquid chromatography and tandem MS (LC/MS/MS) that RIP2 was also phosphorylated on tyrosine. Since only a small proportion (∼0.5%–1.8%) (Hunter and Sefton 1980; Olsen et al. 2006) of cellular phosphorylation occurs on tyrosine, we attempted to determine whether RIP2 could be tyrosine-phosphorylated upon activation of NOD2. To determine this, we first treated human intestinal epithelial cells (HT-29) that stably express NOD2 with the NOD2 agonist MDP (a breakdown product of bacterial peptidoglycan). We found that RIP2 could be tyrosine-phosphorylated in response to MDP, and the kinetics of this MDP-induced tyrosine phosphorylation coincided with activation of the NF-κB pathway (as shown by the appearance of phospho-IκBα) (Fig. 1A). Transient transfection experiments also showed that overexpression of NOD2 can itself cause RIP2 tyrosine phosphorylation (Fig. 1B). This RIP2 modification is dependent on the NOD2 CARDs. These CARDs are important for mediating the NOD2:RIP2 interaction (Ogura et al. 2001b), and use of a NOD2 mutant lacking this region (ΔCARDs) abolishes the NOD2-induced RIP2 tyrosine phosphorylation (Fig. 1B). Consistent with these results, transient transfections of a NOD2 Crohn's disease-associated polymorphism, L1007insC NOD2, failed to induce RIP2 tyrosine phosphorylation (Fig. 1C). L1007insC NOD2 has been found by multiple groups to bind RIP2 less efficiently than its wild-type counterpart (Abbott et al. 2004; Barnich et al. 2005; Lecine et al. 2007), and this finding again suggests that NOD2 binding is required for RIP2 tyrosine phosphorylation. Collectively, these findings suggest that activation of NOD2 (through exposure to MDP or overexpression of NOD2) and a direct interaction between NOD2 and RIP2 are required for tyrosine phosphorylation of RIP2.
Figure 1.
RIP2 is tyrosine-phosphorylated in response to NOD2 activation. (A) HT-29 cells were stimulated with 10 μg/mL MDP, and lysates were harvested at the indicated time points. Tyrosine-phosphorylated proteins were immunoprecipitated, and Western blotting was performed using the indicated antibodies. NOD2 activation stimulated RIP2 tyrosine phosphorylation. (B) HEK293 cells were transfected with wild-type NOD2 or with NOD2 deletion mutants as shown in the top schematic. RIP2 was immunoprecipitated, and Western blotting was performed using the indicated antibodies. NOD2-induced RIP2 tyrosine phosphorylation required the presence of the CARDs of NOD2, indicating that NOD2:RIP2 binding was essential. (C) HEK293 cells were transfected with the indicated constructs. RIP2 was immunoprecipitated and Western blotting was performed. RIP2 tyrosine phosphorylation is lost with the loss-of-function Crohn's disease-associated NOD2 polymorphism L1007insC.
RIP2 is phosphorylated on Tyr 474
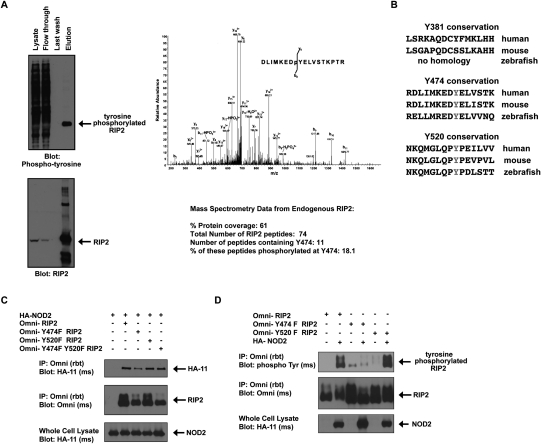
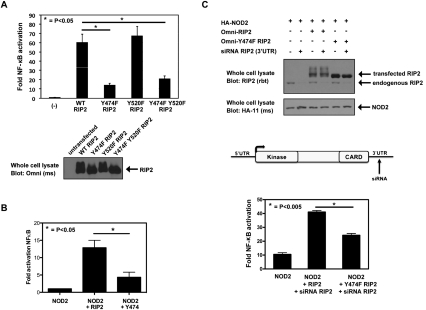
To determine the site at which the MDP/NOD2-induced RIP2 tyrosine phosphorylation occurs, HT-29 cells that express endogenous RIP2 were exposed to MDP for 45 min. Lysates were harvested, and endogenous RIP2 was immunoprecipitated under stringent conditions (1% SDS, 1 M NaCl). While the whole lysate contained a multitude of tyrosine-phosphorylated proteins, a single tyrosine-phosphorylated protein was identified in the RIP2 immunoprecipitate (Fig. 2A). LC/MS/MS analyses showed that this band contained almost exclusively peptides from RIP2 (74 total RIP2 peptides) and contained phosphorylation of endogenous RIP2 at Tyr 474 (Y474) (Fig. 2A). Additional experiments using transfected 293s were also performed and resulted in the identification of two additional tyrosine phosphorylation sites (Y381 and Y520). These sets of experiments, coupled with the findings from published post-translational modification databases (Daub et al. 2008; Oppermann et al. 2009), ultimately resulted in the identification of three tyrosine phosphorylation sites (Y381, Y474, and Y527) on RIP2. As only Y474 and Y527 are conserved in zebrafish (the earliest organism in which RIP2 is expressed) (Fig. 2B), we focused on these two sites. In order to confirm the identity of the MDP/NOD2-induced RIP2 tyrosine phosphorylation site, we performed site-directed mutagenesis on Y474 and Y520 and mutated each of the tyrosines to phenylalanine (Y474F and Y520F). Y474F RIP2, Y520F RIP2, and the double mutant Y474FY520F RIP2 all bind to NOD2; however, we consistently find slightly decreased binding of Y474F to NOD2 (Fig. 2C), and transient transfection assays show that mutation on Y474 but not Y520 abolished NOD2-induced RIP2 tyrosine phosphorylation (Fig. 2D). To then determine whether downstream NF-κB activation is affected by phosphorylation of RIP2 at Y474, we transfected HEK293 cells with NF-κB-driven luciferase, CMV-driven renilla, and each of the following: wild-type RIP2, Y474F RIP2, Y520F RIP2, or Y474FY520F RIP2. Prevention of phosphorylation at Y474 reduced (but did not abolish) NF-κB activation significantly compared with the activation induced by wild-type RIP2 or the activation induced by Y520F RIP2 (Fig. 3A). In addition, RIP2−/− mouse embryonic fibroblasts (MEFs) showed a severely decreased NF-κB response when NOD2 was cotransfected into the cell in the presence of Y474F RIP2, but not when cotransfected into the cell in the presence of wild-type RIP2. Lastly, in a third set of experiments, endogenous RIP2 expression was inhibited by using an siRNA that targets RIP2's 3′ untranslated region (UTR). While this siRNA was ∼80%–90% effective at inhibiting endogenous RIP2 expression, the wild-type RIP2 and Y474F RIP2 constructs that did not have this 3′UTR were unaffected by this siRNA (Fig. 3B). Use of the endogenously targeting RIP2 siRNA showed that the synergy between NOD2 and RIP2 was significantly decreased when Y474F RIP2 was transfected into the cell (Fig. 3B). Collectively, these data suggest that Y474 is a functional tyrosine phosphorylation site, as Y474 phosphorylation is induced by NOD2 activation and is necessary for maximal NOD2-driven NF-κB activation.
Figure 2.
RIP2 is phosphorylated on Tyr 474. (A) HT-29 cells were treated with 10 μg/mL MDP for 45 min. Lysates were generated and endogenous RIP2 was immunoprecipitated. Western blotting showed that RIP2 was purified from the lysate (bottom panel), and that this contained extensive tyrosine phosphorylation (top panel). This tyrosine-phosphorylated RIP2 was excised from the gel and subjected to MS analysis. Overall, in this experiment, 74 peptides corresponding to RIP2 were identified and 61% protein coverage was obtained. The MS/MS spectrum of the RIP2 peptide DLIMKEDpYELVSTKPTR showing a single phosphorylation site at Tyr 8. Fragment ions are consistent with the phosphorylation site localized to Tyr 8 in the above peptide. The phosphate loss at b8 proves that phosphorylation is at Tyr 8, as does the lack of phosphate losses from y1 through y9. However, phosphate losses at y12 and y15 also add credibility to phosphorylation at Tyr 8. These findings show that phosphorylation can occur at Y474 in endogenous RIP2. (B) Additional MS analysis by our laboratory and others (Daub et al. 2008; Oppermann et al. 2009) (in total covering collectively 90% of the RIP2 protein) found a total of three tyrosine phosphorylation sites on RIP2 (Y381, Y474, and Y527). Conservation of these sites is shown. Y474 and Y527 are conserved in zebrafish, while Y381 is not conserved. (C) HA-tagged NOD2 was coexpressed with RIP2, Y474F RIP2, Y520F RIP2, or Y474FY520F RIP2. Immunoprecipitations showed that all four RIP2 proteins could coprecipitate with NOD2. (D) HEK293 cells were transfected with or without NOD2 in the presence of wild-type (WT), Y474F, or Y520F RIP2. RIP2 was immunoprecipitated, and Western blotting was performed using the indicated antibodies. Mutation of Tyr 474 but not Tyr 520 on RIP2 inhibits NOD2-induced RIP2 tyrosine phosphorylation.
Figure 3.
Mutation of Y474 causes a decrease in RIP2's ability to induce NF-κB. (A) NF-κB luciferase assays were performed to measure RIP2 or the RIP2 mutants' ability to activate NF-κB. Mutation of Tyr 474 but not Tyr 520 strongly inhibited RIP2's ability to activate NF-κB (P < 0.03, N = 4). (B) A similar experiment was performed in RIP2−/− MEFs. NF-κB luciferase assays showed that, in these cells, NOD2 could not activate NF-κB without the presence of wild-type RIP2. Activation with NOD2 and Y474F RIP2 was significantly lower (P < 0.015, N = 5). (C) To determine the effect of loss of wild-type, endogenous RIP2 expression on NOD2-induced NF-κB activity, endogenous RIP2 expression was inhibited through the use of an siRNA that targets the 3′UTR of RIP2. (Top panels) Western blotting showed that, while endogenous RIP2 could be silenced using this siRNA, the cDNA constructs expressing either wild-type RIP2 or Y474F RIP2 (both lacking the 3′UTR) could rescue the expression. NF-κB-driven luciferase assays were then performed to determine the synergy between NOD2 and either RIP2 or Y474F RIP2. NOD2 cDNA transfection expression was titrated (75 ng) to a dose in which we found synergy with RIP2 (350 ng) to be maximal, and NF-κB luciferase assays were performed in the presence of the siRNA targeting endogenous RIP2. Synergy with Y474F RIP2 was significantly lower when compared with wild-type RIP2 (P < 0.005, N = 4).
RIP2 autophosphorylates on tyrosines in response to NOD2 activation
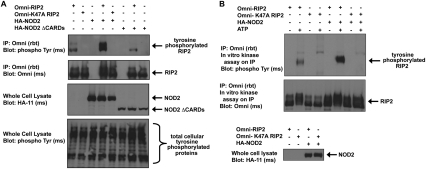
RIP2 was originally classified as a serine/threonine kinase based on homology scans (Inohara et al. 1998; McCarthy et al. 1998; Thome et al. 1998); however, searches using updated pfam and BLAST software now classify RIP2 as a tyrosine-like kinase. While the kinase activity of RIP2 has been shown previously, specific kinase activity toward only serines or threonines has never been formally demonstrated. Since we found that RIP2 is tyrosine-phosphorylated in response to MDP/NOD2 activation, and since RIP2 is a tyrosine-like kinase, we tested the hypothesis that RIP2 might be capable of tyrosine autophosphorylation. To address this, we used a kinase-inactive form of RIP2 (K47A). Transient transfection assays showed that wild-type but not K47A RIP2 is tyrosine-phosphorylated in response to overexpression of NOD2 (Fig. 4A). As a control, this inducible tyrosine phosphorylation is also lost if we cotransfect NOD2 lacking the CARD regions (and which is therefore unable to recruit and activate RIP2) with either the wild-type or kinase-inactive RIP2 (Fig. 3A). To demonstrate in vitro tyrosine kinase autophosphorylation activity, we performed in vitro kinase assays using RIP2 as the substrate. After transient transfection with NOD2 and either wild-type or kinase-inactive RIP2, RIP2 was stringently immunoprecipitated, and this immunoprecipitate was incubated in the presence or absence of ATP in a standard in vitro kinase assay. While wild-type RIP2 was capable of autophosphorylation on tyrosine, kinase-inactive (K47A) RIP2 was unable to induce autophosphorylation on tyrosine either basally or upon cotransfection with NOD2 (Fig. 4B). Collectively, these data imply that it is, in fact, the intrinsic tyrosine kinase activity of RIP2 that is responsible for MDP/NOD2-induced RIP2 tyrosine phosphorylation.
Figure 4.
RIP2 autophosphorylates on tyrosine. (A) HEK293 cells were transfected with NOD2 in the presence of wild-type or K47A (kinase-inactive) RIP2. RIP2 was immunoprecipitated and Western blotting was performed. RIP2 kinase activity is required for NOD2-induced tyrosine phosphorylation of RIP2. (B) HEK293 cells were transfected with the indicated constructs, and RIP2 was immunoprecipitated under stringent washing conditions. Purified RIP2 or kinase-dead RIP2 (K47A) was then incubated with or without ATP in an in vitro kinase assay. Addition of SDS-PAGE sample buffer followed by boiling stopped the reaction. Western blotting was then performed using the indicated antibodies. RIP2 kinase activity is required for in vitro tyrosine autophosphorylation.
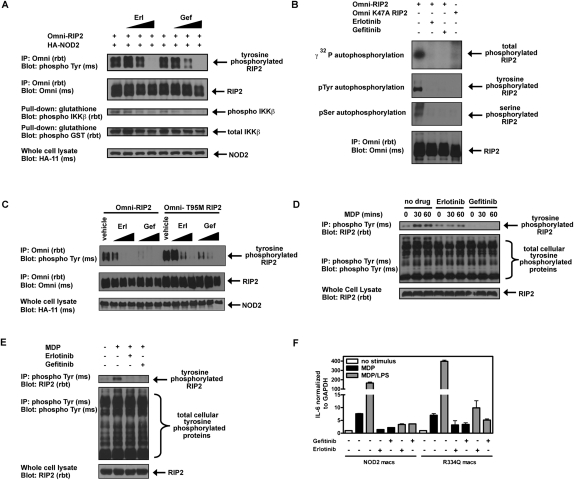
Erlotinib and gefitinib inhibit RIP2 tyrosine phosphorylation
Our findings show that RIP2 autophosphorylates on tyrosine in response to NOD2 activation and that such tyrosine phosphorylation correlates with NF-κB activity. Given that NOD2:RIP2 inhibition might be efficacious in hyperactive NOD2 signaling state (e.g., in individuals with Blau Syndrome and Early Onset Sarcoidosis who harbor an activating NOD2 mutation, in wild-type NOD2 pediatric Crohn's disease patients who are reported to have increased RIP2 kinase activity, and in patients harboring ITCH mutations who exhibit increased mucosal inflammatory pathology) (Lohr et al. 2010), we performed a limited small-molecule inhibitor screen to identify RIP2 tyrosine kinase inhibitors. Three compounds were found to strongly inhibit the tyrosine kinase activity of RIP2. These compounds included two that had been demonstrated previously to inhibit RIP2's kinase activity (SB203580 [a p38 inhibitor] and gefitinib [an EGFR inhibitor]) (Argast et al. 2005; Brehmer et al. 2005; Windheim et al. 2007). We additionally identified an additional EGFR inhibitor (erlotinib) that has not been found previously to inhibit the kinase activity of RIP2. Erlotinib and gefitinib were found to inhibit tyrosine phosphorylation of RIP2 and RIP2-induced IKKβ activation with similar efficacy (Fig. 5A). In vitro kinase assays indicated that these drugs could inhibit both tyrosine and serine–threonine kinase activity of RIP2, as shown by both Western blot and loss of total 32P incorporation in radioactive autophosphorylation assays (Fig. 5B). While these drugs act as competitive inhibitors for ATP by binding to the ATP-binding pocket of their target kinase, neither gefitinib (Iressa) nor erlotinib (Tarceva) has been shown previously to have an effect on NOD2:RIP2 signaling or MDP-induced cytokine release. For this reason, we next tested the specificity of these agents for RIP2. As shown in Figure 5C, the inhibition of RIP2 tyrosine phosphorylation by both erlotinib and gefitinib occurs at nanomolar concentrations. These doses are similar to the doses shown to have clinical efficacy when targeting the EGFR. Patients who develop resistance to erlotinib and gefitinib harbor a mutation in the ATP-binding pocket of EGFR (T790M) that prevents binding of the drug and subsequently causes insensitivity to these compounds (Kobayashi et al. 2005). In order to determine whether these EGFR inhibitors act on RIP2 directly, we generated RIP2 containing the homologous desensitizing mutation in the ATP-binding pocket (T95M). This mutation partially desensitized RIP2 to the inhibitory effects of erlotinib and gefitinib (Fig. 5C), suggesting that the effect of erlotinib and gefitinib on RIP2 was direct. In addition, we also show that these drugs can inhibit endogenous MDP-induced RIP2 tyrosine phosphorylation in both an intestinal epithelial cell line and a macrophage cell line, both of which stably express NOD2 (Fig. 5D,E). While these drugs have no effect on the NOD2:RIP2 interaction (Supplemental Fig. 1A), they can inhibit endogenous RIP2 tyrosine phosphorylation in a dose-dependent manner (Supplemental Fig. 1B). Lastly, to mimic a hyperactive NOD2 state, we studied the effect of erlotinib and gefitinib on cells expressing activating Blau Syndrome-causing NOD2 mutations. RAW264.7 cells were stably transduced with either wild-type NOD2 or NOD2 harboring the gain-of-function Blau Syndrome-causing mutation R334Q. Cells were then treated with MDP alone or LPS (lipopolysaccharide) + MDP to study the well-described NOD2/TLR (Toll-like receptor) synergy. If RIP2 tyrosine phosphorylation is a necessary precursor to NF-κB activation, then inhibition of RIP2 tyrosine phosphorylation using erlotinib and gefitinib may serve to correct the excessive activation seen with the R334Q polymorphism. Under stimulatory conditions, both erlotinib and gefitinib reduced the levels of IL-6 (interleukin-6) in response to MDP or synergistic NOD2–TLR stimulation, suggesting that the use of such drugs is efficacious in vitro (Fig. 5F).
Figure 5.
Erlotinib (Tarceva) and gefitinib (Iressa) inhibit RIP2 tyrosine phosphorylation. (A) HEK293 cells were transfected with RIP2, IKKβ, and NOD2. Lysates were generated and immunoprecipitations were performed. Western blotting was then performed using the indicated antibodies. Increasing doses of erlotinib and gefitinib (10 nM, 100 nM, and 1 μM) inhibited RIP2 tyrosine phosphorylation, and this inhibition matched the inhibition of RIP2-induced IKKβ activation. (B) To determine whether gefitinib and erlotinib could inhibit RIP2's kinase activity in vitro and whether these compounds inhibited both serine–threonine and tyrosine kinase activity of RIP2, in vitro kinase assays were performed using either immunoprecipitated RIP2 or kinase-dead (K47A) RIP2. One-hundred nanomolar gefitinib and 100 nM erlotinib could inhibit total autophosphorylation activity of RIP2 (32P incorporation, top autoradiograph) and autophosphorylation on both tyrosines and serines, as shown by Western blotting with anti-phosphoserine and anti-phosphotyrosine antibodies. (C) HEK293 cells were transfected with NOD2 and wild-type or T95M RIP2 for 6 h before treatment with erlotinib and gefitinib (100 nM, 500 nM, and 2 μM). The T95M mutation is homologous to the EGFR erlotinib and gefitinib-resistant mutation (T790M) that occurs in resistant cancer patients. After 16 h, lysates were harvested for immunoprecipitation and Western blotting. Erlotinib and gefitinib inhibited RIP2 tyrosine phosphorylation in a dose-dependent manner and are potent at nanomolar concentrations. The sensitivity decreased at least fivefold when T95M RIP2 was used. (D,E) HT-29 cells overexpressing NOD2 (D) or RAW264.7 macrophages overexpressing NOD2 (E) either were not pretreated or were pretreated with erlotinib and gefitinib (500 nM). One hour later, the cells were stimulated with 10 μg/mL MDP for the time points indicated before harvesting lysates for immunoprecipitation and Western blotting. While there was basal tyrosine phosphorylation of RIP2 in untreated cells, both erlotinib and gefitinib inhibit MDP-induced RIP2 tyrosine phosphorylation in both these divergent cell lines. (F) RAW264.7 macrophages overexpressing wild-type NOD2 or the activating R334Q NOD2 mutation associated with Blau Syndrome were pretreated with erlotinib and gefitinib for 1 h before stimulation with MDP (10 μg/mL) or MDP (10 μg/mL) + LPS (5 ng/mL) . Sixteen hours later, cells were harvested to obtain RNA for RT–PCR analysis. Erlotinib and gefitinib inhibit the NOD2–TLR synergy seen in either wild-type NOD2 cells or R334Q Blau Syndrome NOD2 cells. This finding was also true for TNF-α (not shown).
While the above results suggest a specificity of erlotinib and gefitinib for RIP2, we had two additional concerns regarding the specificity of erlotinib and gefitinib for the NOD2:RIP2 signaling pathway. First, erlotinib and gefitinib could be affecting RIP2-independent as well as RIP2-dependent inflammatory signaling pathways. To rule this out, we used well-known agonists for two separate NF-κB-activating signaling pathways. LPS activates TLR4 on macrophages, and TNF-α (tumor necrosis factor α) activates the TNF receptor (TNFR) on a variety of cells. Both macrophages and HT-29 cells were pretreated with erlotinib and gefitinib prior to exposure to LPS or TNF. Analysis of NF-κB signaling revealed that neither TLR4 nor TNF signaling was affected by gefitinib or erlotinib (Supplemental Fig. 2). A second concern centered on the fact that erlotinib and gefitinib may also be inhibiting the EGFR in these cells and indirectly affecting the tyrosine phosphorylation state of RIP2. Given this, we tested these cell lines for EGFR expression. We were unable to detect EGFR expression on bone marrow-derived macrophages (BMDM) from wild-type or ITCH−/− mice, and we detected expression of only a limited amount of EGFR on the NOD2-expressing HT-29 intestinal epithelial cell line when compared with our positive control A549 (lung adenocarcinoma cell line) (Supplemental Fig. 3A,B, represented graphically as change in mean fluorescence intensity [MFI]). To further rule out any possible contribution of EGF signaling in induction of RIP2 tyrosine phosphorylation, we treated serum-starved NOD2-expressing HT-29 cells with recombinant EGF in the presence or absence of a neutralizing antibody to the EGFR (LA1). Despite the fact that EGF did induce weak signaling in this cell line (evidenced by phosphorylated EGFR), we demonstrate no increase in RIP2 tyrosine phosphorylation over basal levels and no decrease when using an EGFR inhibitor with a completely different mode of inhibition relative to erlotinib or gefitinib (cf. Supplemental Fig. 3C and Fig. 1A). The fact that genetic manipulation of RIP2 directly changed the inhibitory capacity of erlotinib and gefitinib (Fig. 5C), the cell lines under study show minimal EGFR expression (Supplemental Fig. 3A), and addition of exogenous EGF or blocking such signaling using a EGFR-neutralizing antibody do not affect RIP2 tyrosine phosphorylation levels (Supplemental Fig. 3C) suggests that the effect of erlotinib and gefitinib on NOD2:RIP2 signaling is due to direct RIP2 inhibition.
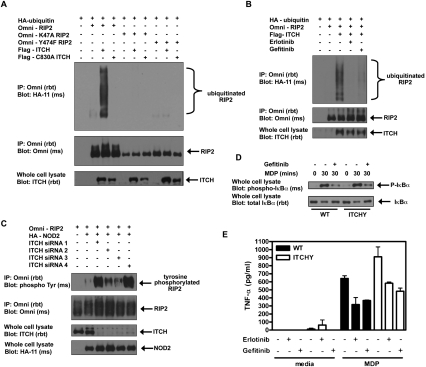
Tyrosine phosphorylation of RIP2 promotes its ubiquitination by the E3 ligase ITCH, and inhibition of this tyrosine phosphorylation inhibits the exaggerated MDP responses seen in ITCH−/− cells
ITCH is a HECT-type E3 ligase that we described recently as being an important negative regulator of NOD2-induced NF-κB activation (Tao et al. 2009). It accomplishes such regulation by ubiquitinating previously activated RIP2 and targeting it for inactivation. Depending on the genetic background, ITCH-deficient mice can exhibit inflammation of a number of organs, including the skin, gastrointestinal tract, and lungs (Perry et al. 1998). BMDMs from ITCH-deficient mice show increased NF-κB activation and increased cytokine release upon MDP treatment (Tao et al. 2009). Here we demonstrate that ITCH-induced ubiquitination of RIP2 is lost if RIP2 cannot be tyrosine-phosphorylated at 474 (Fig. 6A), consistent with phosphorylation of Y474 being necessary for RIP2 activation and subsequent deactivation by ITCH. Likewise, loss of its kinase activity via K47A RIP2 (and, therefore, loss of its ability to autophosphorylate at Y474), also results in abrogation of the ITCH-induced RIP2 ubiquitination (Fig. 6A). The requirement for tyrosine phosphorylation of RIP2 prior to its ubiquitination by ITCH was also confirmed with the use of gefitinib and erlotinib, as both gefitinib and erlotinib were able to inhibit ITCH-induced RIP2 ubiquitination at nanomolar concentrations (Fig. 6B). Lastly, as our data suggest that ITCH targets phosphorylated Y474 to ubiquitinate and down-regulate RIP2, we hypothesized that inhibition of ITCH expression might increase RIP2 tyrosine phosphorylation. To this end, we inhibited ITCH expression through the use of siRNA. These results showed that, when lower amounts of NOD2 were transfected into the cell (to increase the sensitivity of ITCH loss), knockdown of ITCH showed increased RIP2 tyrosine phosphorylation (Fig. 6C).
Figure 6.
Tyrosine phosphorylation of RIP2 promotes its ubiquitination by the E3 ligase ITCH, and inhibition of this tyrosine phosphorylation inhibits the exaggerated MDP responses seen in ITCH−/− cells. (A) HEK293 cells were transfected with the indicated constructs. RIP2 was immunoprecipitated, and immunoblotting with the indicated antibodies was performed. Both the kinase activity of RIP2 and phosphorylation at Tyr 474 are required for ITCH-induced ubiquitination of RIP2. (B) HEK293 cells were transfected with the indicated constructs. Six hours later, cells were treated with erlotinib and gefitinib overnight before harvesting cell lysates for immunoprecipitation. Western blotting showed that both erlotinib and gefitinib inhibit ITCH-induced ubiquitination of RIP2. (C) HEK293 cells were transfected with RIP2 and NOD2 in the absence or presence of four different siRNAs against ITCH. Cell lysates were harvested 24 h after transfection for immunoprecipitation and Western blotting. siRNA knockdown of ITCH enhances NOD2-induced RIP2 tyrosine phosphorylation, suggesting that ITCH ubiquitinates and down-regulates the tyrosine-phosphorylated form of RIP2. (D) BMDMs from wild-type or ITCH−/− mice were treated with gefitinib overnight before stimulation with 10 μg/mL MDP. Cell lysates were collected at the time points indicated for immunoprecipitation and Western blotting. Gefitinib inhibits MDP-induced NF-κB activation in both wild-type and ITCH−/− mice. (E) BMDMs from wild-type or ITCH−/− mice were treated with erlotinib and gefitinib for 1 h before stimulation with 10 μg/mL MDP. After 24 h, supernatants were harvested for TNF-α ELISA. Erlotinib and gefitinib can reduce the exacerbated MDP-induced cytokine responses in the ITCH−/− mice to wild-type levels.
Given these results, we formulated the following hypothesis. MDP induces activation of NOD2. Activated NOD2 binds RIP2 and causes RIP2 activation and RIP2 tyrosine autophosphorylation. This activated tyrosine-phosphorylated RIP2 allows for subsequent NF-κB activation. To terminate NOD2-induced NF-κB activation, ITCH targets only the activated tyrosine-phosphorylated form of RIP2 for ubiquitination and subsequent down-regulation. In the absence of ITCH, RIP2 inhibition is down-regulated, and increased levels of tyrosine-phosphorylated RIP2 allow for increased NOD2-induced NF-κB activation and increased NOD2-induced cytokine responses. If this model is true, then inhibiting RIP2 tyrosine phosphorylation may work to reduce both NF-κB activation and NF-κB-associated inflammation in the MDP-hypersensitive macrophages from ITCH−/− mice. To test this hypothesis, we pretreated BMDMs from wild-type or ITCH−/− mice with erlotinib or gefitinib before stimulation with MDP. We show a reduction in the resulting NOD2-induced NF-κB activation in the ITCH−/− mice (Fig. 6D). Likewise, use of either erlotinib or gefitinib was able to lower the exacerbated MDP-induced TNF-α response seen in the ITCH−/− mice to wild-type levels (Fig. 6E). Because we are using a stimulus (MDP) specific for NOD2 activation, and because the cells used show low levels of EGFR expression (Supplemental Fig. 3A), these results suggest that both erlotinib and gefitinib could be used to inhibit RIP2 tyrosine phosphorylation directly to dampen NOD2-dependent signaling responses under conditions in which they are abnormally up-regulated.
Discussion
In humans, the majority of cellular phosphorylation occurs on serine and threonine (86.4% and 11.8%, respectively), while only 0.5%–1.8% of phosphorylation occurs on tyrosine (Hunter and Sefton 1980; Olsen et al. 2006). A general problem with MS proteomic phosphorylation mapping is identifying phosphorylated serines and threonines that are ultimately not necessarily functional. Due to the low overall abundance of tyrosine phosphorylation within the cell (relative to the total number of tyrosines present), however, tyrosine phosphorylation is somewhat more likely to be biologically relevant. RIP2 has been traditionally classified as a serine/threonine kinase, and our own unpublished work has shown that RIP2 can phosphorylate myelin basic protein on serines and threonines in in vitro kinase assays. Despite this, BLAST and pfam domain searches using more updated software show that RIP2 has the most homology with tyrosine-like kinases rather than serine–threonine kinases. In addition, there is precedence in the literature that certain serine–threonine kinases (such as the DYRK kinase family) can autophosphorylate on tyrosine while performing substrate phosphorylation on serines and threonines (Kentrup et al. 1996; Lochhead et al. 2005). In this study, we show that RIP2 is tyrosine-phosphorylated after exposure to the NOD2-activating agonist MDP (Fig. 1). NOD2 overexpression induces RIP2 tyrosine phosphorylation, and the most common NOD2 Crohn's disease-associated susceptibility allele, L1007insC, does not induce RIP2 tyrosine phosphorylation (Fig. 1). We further show that RIP2 can autophosphorylate on tyrosine (Fig. 4). We map sites of RIP2 tyrosine phosphorylation and find that one, Y474, is responsible for transmitting downstream NF-κB signaling (Figs. 2, 3). Lastly, we screened tyrosine kinase inhibitors for their ability to inhibit the tyrosine kinase activity of RIP2, and found that both erlotinib (Tarceva) and gefitinib (Iressa), two agents used clinically, could inhibit the tyrosine kinase ability of RIP2 and inhibit NOD2-dependent signaling in a number of NOD2 hyperactivation states (Figs. 5, 6). These findings help classify RIP2's kinase activity in NOD2 signaling. While overexpression of kinase-defective RIP2 does lead to NF-κB activation, this feature could be due to high stoichiometry that amplifies RIP2's scaffolding role in coordinating this pathway. However, our results suggest that full NOD2 activity requires active RIP2 kinase activity, and our data highlight the unexpected finding of RIP2 tyrosine kinase activity. The data also show that this activity might be targeted for pharmacologic effect.
Integrating these findings with our own previously published work and those of others (Tanabe et al. 2004; Windheim et al. 2007; Yang et al. 2007; Hasegawa et al. 2008; Bertrand et al. 2009; for review, see Strober et al. 2006; Chen et al. 2009) suggests the following model for NOD2:RIP2 signaling and deactivation. Upon intracellular exposure to MDP, NOD2 oligomerizes and binds to RIP2 via homotypic CARD interactions. RIP2 is then ubiquitinated by the IAP proteins and autophosphorylates on Y474. This active NOD2:RIP2 complex then helps coordinate cytokine release through NF-κB activation. Because too much NOD2:RIP2 activation could be deleterious to the cell, deactivation must occur. Our data suggest that, like other signaling pathways, RIP2 activation occurs at low stoichiometry and, for this reason, the cell needs a mechanism to target only activated RIP2. This could occur through the recognition of Y474 by a ubiquitin-editing complex containing the E3 ubiquitin ligase ITCH. Upon recognizing phosphorylated RIP2, ITCH further ubiquitinates RIP2 to target activated RIP2 for deactivation through an as-yet-undefined mechanism. This model is consistent with our data, in that NOD2 requires the CARDs to both bind RIP2 and induce RIP2 tyrosine phosphorylation (Fig. 1). Inhibition of Y474 phosphorylation very strongly inhibits NF-κB signaling (Figs. 3–5), and loss of Y474 phosphorylation does not allow ITCH to target RIP2 for deactivation (Fig. 6).
The finding that phosphorylation of Y474 correlates with NOD2:RIP2-induced activation of NF-κB also highlights the importance of the study of basic signal transduction pathways for potential clinical benefit. For example, recent reports have demonstrated increased expression of both NOD2 and RIP2 and increased activation of RIP2 in the inflamed intestinal mucosa of a pediatric population with Crohn's disease (Stronati et al. 2008; Negroni et al. 2009). Likewise, the identified Blau Syndrome and Early Onset Sarcoidosis NOD2 polymorphisms have been described as gain-of-function mutations showing increased NF-κB activation basally and in response to NOD2 activation, presumably occurring through increased activation of RIP2 (Kanazawa et al. 2005; for review, see Strober et al. 2006). In these clinical conditions, inhibition of the NOD2:RIP2 signal transduction pathway might be efficacious, and in this study we describe off-target effects of two EGFR inhibitors—gefitinib and erlotinib—on the tyrosine kinase activity of RIP2. We believe that this inhibition is direct, as evidenced by (1) the decreased sensitivity of the T95M RIP2 to the EGFR inhibitors, (2) the low expression of EGFR on the cells used in this study, (3) the lack of inducible RIP2 tyrosine phosphorylation in cell lines stimulated with EGF, and (4) the lack of an effect of erlotinib or gefitinib on LPS or TNF signaling pathways. These findings could broaden the use of already FDA-approved, clinically available drugs, and could potentially lead to alternative therapies for NOD2 hyperactivation states.
Materials and methods
Cell culture, transfections, immunoprecipitations, and Western blotting
HEK293 cells were grown in DMEM (Mediatech) supplemented with 5% FBS (Hyclone) and 1% antibiotic–antimycotic (Invitrogen). RAW264.7 macrophages stably expressing NOD2 were grown in DMEM (Mediatech) supplemented with 10% FBS (Hyclone), 1% antibiotic–antimycotic (Invitrogen), and 150 μg/mL G418 (InvivoGen). HT-29 cells stably expressing NOD2 were grown in RPMI (Mediatech) supplemented with 10% FBS (Hyclone), 1% antibiotic–antimycotic (Invitrogen), and 150 μg/mL G418 (InvivoGen). BMDMs were generated by culturing bone marrow from wild-type or ITCH−/− mice in Ladmac-conditioned medium for 7 d before passaging cells in DMEM with 10% FBS for downstream assays. All cell lines were obtained from American Type Tissue Collection (ATCC). Stably expressing cell lines were generated through infection with virus made by transfection of a retrovirus packaging cell line, Amphopak-293, with pMXneo-based constructs. Transient transfections were performed by calcium phosphate precipitation or by Lipofectamine LTX (RIP2−/− MEFs). Lysates were obtained using Cell Signaling lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1 mM β-glycerophosphate, 1 mM PMSF, 1 mM NaVO4, 10 nM Calyculin A, protease inhibitor cocktail [Sigma]). After adding protein G sepharose beads, immunoprecipitations were washed at least five times before Western blotting. Western blotting was performed on nitrocellulose membranes (Bio-Rad) or on PVDF membranes (Whatmann) as described previously (Abbott et al. 2004).
Antibodies, plasmids, and reagents
Anti-phosphotyrosine (P-Tyr-100), anti-phospho IκBα (5A5), anti-total IκBα (44D4), anti-phospho EGFR (53A5), and anti-EGFR Alexa 488 (D38B1) were obtained from Cell Signaling Technology. Anti-Omni rabbit (M-21), anti-Omni mouse (D-8), anti-RICK (H-300), and anti-ITCH (H-100) were obtained from Santa Cruz Biotechnology. Anti-HA (16B12) was obtained from Covance. Streptavidin agarose was obtained from Sigma, while protein G agarose was obtained from Invitrogen. The K47A, T95M, Y474F, Y520, and Y520F/Y474F RIP2 constructs were generated by QuickChange site-directed mutagenesis of the Omni-RIP2 construct (Stratagene) and were all sequence-verified. Likewise, the R334Q NOD2 construct was also generated by site-directed mutagenesis of pMXneo-Flag-Nod2 (McDonald et al. 2005). HA-Ubiquitin, Omni-RIP2, NTAP-RIP2, Omni-NOD2, Flag-NOD2, Flag-L1007insC NOD2, Flag-ITCH, and Flag-C830A ITCH were used as described previously (Inohara et al. 1999; Abbott et al. 2004, 2007; Hutti et al. 2007; Clark et al. 2008; Tao et al. 2009). The Omni-tagged NOD2 construct was originally subcloned from the mouse EST as described previously (Abbott et al. 2004). Unlike the human, the insertion of cytosine at L1007 in the mouse allele causes a frameshift, leading to a nonsense transcript encoding 41 additional base pairs. HA-NOD2 and HA-NOD2 deletion mutants (ΔCARDs, ΔLRRs, and CARDs) were a kind gift from Christine McDonald (Cleveland Clinic Foundation). MDP was obtained from Bachem, and LPS was obtained in a highly purified form from InvivoGen. Gefitinib and erlotinib were obtained from LC Laboratories.
Mass spectrometry
HT-29 cells (1 × 109) were treated with 10 μg/mL MDP prior to lysis in Cell Signaling lysis buffer (recipe above) containing pervanadate. Endogenous RIP2 was immunoprecipitated using an anti-RIP2 antibody (H300; Santa Cruz Biotechnology) with stringent washing (four washes with lysis buffer, four washes with lysis buffer containing 1 M NaCl and 1% SDS, four washes with PBS, and two washes with lysis buffer). SDS-PAGE was performed, and the predominant band (as stained by Coomassie G) was excised from the gel reduced with DTT, alkylated with iodoacetamide, and digested overnight with trypsin. Tryptic peptides were analyzed by data-dependent reversed-phase microcapillary tandem MS (LC/MS/MS) via CID using a hybrid linear ion trap-LTQ Orbitrap XL mass spectrometer (Thermo Scientific) operated in positive ion mode at a flow rate of 300 nL/min using a 75-μm (ID) × 15-μm (ID tip) × 15-cm (length) C18 microcapillary column. The column was equilibrated and peptides were loaded using buffer A (0.1% formic acid, 0.9% acetonitrile, 99% water) then eluted with a gradient from 3% buffer B (acetonitrile) to 38% B. One MS survey scan was followed by six MS/MS scans. The Sequest algorithm (Thermo Scientific) was used for database searching of all MS/MS spectra against the reversed SWISS-PROT protein database and single entry RIP2 database with the differential modifications: oxidation (+15.99 Da) of Met and phosphorylation (+79.97 Da) of Ser/Thr/Tyr. Peptide sequences were accepted if they matched the forward database and passed the following scoring thresholds: 2+ ions, Xcorr ≥ 2.0, Sf ≥ 0.4, P ≥ 0; 3+ ions, Xcorr ≥ 2.65, Sf ≥ 0.5, P ≥ 5. Peptides with gas phase charges of 1+ and 4+ were generally not accepted as valid due to difficulty of interpretation. After passing the scoring thresholds, all MS/MS were then manually inspected rigorously to be sure that all b− (fragment ions resulting from the peptide's N terminus) and y− ions (fragment ions resulting from the peptide's C terminus) aligned with the assigned protein database sequence. Peptide false discovery rates (FDR) were calculated to be ∼1.5% based on reversed database hits.
In vitro kinase assay
Wild-type RIP2 or K47A RIP2 was immunoprecipitated via its Omni-tag as described above. The kinase buffer contained 50 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM β-glycerophosphate, 1 mM DTT, 1 mM NaVO4, and 100 μM ATP/reaction mix. Reaction mixes were incubated for 30 min at 30°C. 32γATP was added to the reaction as indicated in the text.
siRNA transfections
Four separate siRNAs against ITCH were purchased from Qiagen. The sequences of these were as follows: siRNA1, CACGGGCGAGTTTACTATGTA; siRNA2, CAAGAGCTATGAGCAACTGAA; siRNA3, ATGGGTAGCCTCACCATGAAA; siRNA4, TGCCGCCGACAAATACAAATA. The siRNA targeting the 3′UTR or endogenous RIP2 had the following sequence: AAGAAGAAATGTGTTTCATAA. Six-centimeter plates of HEK293 cells were transfected with 2 nM each siRNA using calcium phosphate.
NF-κB dual luciferase assay
The dual luciferase reporter assay system was purchased from Promega and used according to the manufacturer's instructions. HEK293 cells were transfected with 100 ng of NF-κB-firefly luciferase and 50 ng of CMV-renilla luciferase along with 3 μg of the construct of interest. For RIP2−/− MEFs, 2 μg of total DNA was transfected using Lipofectamine LTX (Invitrogen). Lysates were harvested 24 h after transfection using 1× passive lysis buffer (Promega). Twenty microliters of lysate was used for each assay. A Victor Plate Reader (Perkin Elmer) was used for detection of luminescence.
Quantitative RT–PCR
RNA was harvested using a Qiagen Total RNA Isolation kit. First-strand DNA synthesis was performed using Qiagen's Quantitect Reverse Transcription kit according to the manufacturer's instructions. Real-time PCR was carried out using primers against human IL-6 (forward, 5′-TCCACAAGCGCCTTCGGTCC-3′; reverse, 5′-GTGGCTGTCTGTGTGGGGCG-3′) and human GAPDH (forward, 5′-GACCTGACCTGCCGTCTA-3′; reverse, 5′-GTTGCTGTAGCCAAATTCGTT-3′) along with the iQ SYBR Green Supermix (Bio-Rad), and was detected using a Bio-Rad iCycler. The data shown are normalized to GAPDH.
ELISA
A mouse TNF-α ELISA kit was purchased from eBioscience and used according to the manufacturer's instructions. Supernatants from stimulated BMDM were harvested 24 h after agonist stimulation and plated overnight on ELISA plates coated with TNF-α capture antibody. Development was performed as suggested by the manufacturer. A Victor Plate Reader (Perkin Elmer) was used for detection of fluorescence.
Flow cytometry
One-million cells from each cell line were fixed and permeabilized using Fix/Perm and permabilization buffer reagents (eBioscience). One microgram of anti-EGFR Alexa 488 antibody (Cell Signaling) was used to stain for intracellular EGFR. After 1 h of staining, cells were thoroughly washed with permeabilization buffer and resuspended in flow cytometry buffer before acquisition of data on a BD FACscan. Data were analyzed using FlowJo software.
Graphs and statistical analysis
All graphs and statistical analysis were generated and performed using GraphPad Prism software. Significance was determined using a Student's t-test with P < 0.05 selected as the cutoff for significance.
Acknowledgments
We thank MingFang Tao and Xuemei Yang for technical assistance, and Jill Marinis for her critical discussions. We thank Dr. Michelle Kelliher (University of Massachusetts, Worcester, MA) for the gift of immortalized B6 macrophages. We also thank Dr. Christine McDonald (Cleveland Clinic Foundation, Cleveland, OH) for her gift of various NOD2 constructs, and Dr. Christine McDonald and Dr. Gabriel Nunez (University of Michigan, Ann Arbor, MI) for providing RIP2−/− MEFs. This work was supported by NIH grants R01 GM086550 (to D.W.A.) and R21 AI076886 (to D.W.A.), and the Burroughs Wellcome Career Award to Biomedical Scientists (to D.W.A.). This work was additionally supported by an ARRA supplement grant, R01GM086550S1 (to D.W.A.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1964410.
Supplemental material is available at http://www.genesdev.org.
References
- Abbott DW, Wilkins A, Asara JM, Cantley LC 2004. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol 14: 2217–2227 [DOI] [PubMed] [Google Scholar]
- Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC 2007. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol 27: 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argast GM, Fausto N, Campbell JS 2005. Inhibition of RIP2/RIck/CARDIAK activity by pyridinyl imidazole inhibitors of p38 MAPK. Mol Cell Biochem 268: 129–140 [DOI] [PubMed] [Google Scholar]
- Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK 2005. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for NF-κB activation in muramyl dipeptide recognition. J Cell Biol 170: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M 2009. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30: 789–801 [DOI] [PubMed] [Google Scholar]
- Brehmer D, Greff Z, Godl K, Blencke S, Kurtenbach A, Weber M, Müller S, Klebl B, Cotton M, Kéri G, et al. 2005. Cellular targets of gefitinib. Cancer Res 65: 379–382 [PubMed] [Google Scholar]
- Chamaillard M, Philpott D, Girardin SE, Zouali H, Lesage S, Chareyre F, Bui TH, Giovannini M, Zaehringer U, Penard-Lacronique V, et al. 2003. Gene–environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc Natl Acad Sci 100: 3455–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Shaw MH, Kim YG, Nunez G 2009. NOD-like receptors: Role in innate immunity and inflammatory disease. Annu Rev Pathol 4: 365–398 [DOI] [PubMed] [Google Scholar]
- Clark NM, Marinis JM, Cobb BA, Abbott DW 2008. MEKK4 sequesters RIP2 to dictate NOD2 signal specificity. Curr Biol 18: 1402–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Körner R, Greff Z, Kéri G, Stemmann O, Mann M 2008. Kinase-selective enrichment enables qualtitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 31: 438–448 [DOI] [PubMed] [Google Scholar]
- Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, Ting JP 2007. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci 104: 8041–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff J, Hanke M, Stein-Gerlach M, Kiang TP, Herzberger K, Habenberger P, Muller S, Klebl B, Marschall M, Stamminger T, et al. 2004. RICK activates a NF-κB-dependent anti-human cytomegalovirus response. J Biol Chem 279: 9642–9652 [DOI] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC 2007. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 25: 713–724 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278: 8869–8872 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Núñez G, Inohara N 2008. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J 27: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599–603 [DOI] [PubMed] [Google Scholar]
- Hunter T, Sefton BM 1980. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci 77: 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti JE, Turk BE, Asara JM, Ma A, Cantley LC, Abbott DW 2007. IκB kinase β phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol Cell Biol 27: 7451–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, del Peso L, Koseki T, Chen S, Núñez G 1998. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem 273: 12296–12300 [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, et al. 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J Biol Chem 274: 14560–14567 [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, et al. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem 278: 5509–5512 [DOI] [PubMed] [Google Scholar]
- Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, et al. 2005. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-κB activation: Common genetic etiology with Blau syndrome. Blood 105: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Kentrup H, Becker W, Heukelbach J, Wilmes A, Schürmann A, Huppertz C, Kainulainen H, Joost HG 1996. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J Biol Chem 271: 3488–3495 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416: 194–199 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B 2005. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352: 786–792 [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC 2009. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci 106: 14524–14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécine P, Esmiol S, Métais JY, Nicoletti C, Nourry C, McDonald C, Nunez G, Hugot JP, Borg JP, Ollendorff V 2007. The NOD2–RICK complex signals from the plasma membrane. J Biol Chem 282: 15197–15207 [DOI] [PubMed] [Google Scholar]
- Lochhead PA, Sibbet G, Morrice N, Cleghon V 2005. Activation-loop autophosphorylation is mediated by novel transitional intermediate form of DYRKs. Cell 121: 925–936 [DOI] [PubMed] [Google Scholar]
- Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider NL, Chikwava KR, Cummings OW, Morton DH, et al. 2010. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet 86: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JV, Ni J, Dixit VM 1998. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J Biol Chem 273: 16968–16975 [DOI] [PubMed] [Google Scholar]
- McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, Lecine P, Borg JP, Nunez G 2005. A role for Erbin in the regulation of Nod2-dependent NF-κB signaling. J Biol Chem 280: 40301–40309 [DOI] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP 2001. CARD15 mutations in Blau syndrome. Nat Genet 29: 19–20 [DOI] [PubMed] [Google Scholar]
- Negroni A, Stronati L, Pierdomenico M, Tirindelli D, Di Nardo G, Mancini V, Maiella G, Cucchiara S 2009. Activation of NOD2-mediated intestinal pathway in a pediatric population with Crohn's disease. Inflamm Bowel Dis 15: 1145–1154 [DOI] [PubMed] [Google Scholar]
- Nembrini C, Kisielow J, Shamshiev AT, Tortola L, Coyle AJ, Kopf M, Marsland BJ 2009. The kinase activity of Rip2 determines its stability and consequently Nod1- and Nod2-mediated immune responses. J Biol Chem 284: 19183–19188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. 2001a. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603–606 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G 2001b. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J Biol Chem 276: 4812–4818 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Kéri G, Mann M, Daub H 2009. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 8: 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Nunez G 2007. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol 178: 2380–2386 [DOI] [PubMed] [Google Scholar]
- Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG 1998. The ITCH−/− locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet 18: 143–146 [DOI] [PubMed] [Google Scholar]
- Rosenstiel P, Schreiber S 2009. NOD-like receptors—Pivotal guardians of the immunological integrity of barrier organs. Adv Exp Med Biol 653: 35–47 [DOI] [PubMed] [Google Scholar]
- Schurmann M, Valentonyte R, Hampe J, Muller-Quernheim J, Schwinger E, Schreiber S 2003. CARD15 gene mutations in sarcoidosis. Eur Respir J 22: 748–754 [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani A, Watanabe T 2006. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol 6: 9–21 [DOI] [PubMed] [Google Scholar]
- Stronati L, Negroni A, Merola P, Pannone V, Borrelli O, Cirulli M, Annese V, Cucchiara S 2008. Mucosal NOD2 expression and NF-κB activation in pediatric Crohn's disease. Inflamm Bowel Dis 14: 295–302 [DOI] [PubMed] [Google Scholar]
- Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. 2004. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J 23: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW 2009. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol 19: 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C, Tschopp J 1998. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol 8: 885–888 [DOI] [PubMed] [Google Scholar]
- Windheim M, Lang C, Peggie M, Plater LA, Cohen P 2007. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J 404: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA 2007. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem 282: 36223–36229 [DOI] [PubMed] [Google Scholar]