Abstract
Arabidopsis embryos lacking DICER-LIKE1 (DCL1), which is required for microRNA (miRNA) biogenesis, arrest early in development. To assess the functions of embryonic miRNAs, we determined the developmental and molecular consequences of DCL1 loss. We found that DCL1 is required for cell differentiation events as early as the eight-cell stage and soon thereafter for proper division of the hypophysis and subprotoderm cells. By the early globular (∼32-cell) stage, dcl1-null mutant embryos overexpress ∼50 miRNA targets. In dcl1 eight-cell embryos, the two most up-regulated targets are those of miR156 and encode SPL10 and SPL11 transcription factors. SPL10 and SPL11 are derepressed >150-fold in dcl1 embryos and are redundantly required for the dcl1 early patterning defects. Moreover, as early as the eight-cell stage, miR156-mediated repression of zygotic SPL transcripts prevents premature accumulation of transcripts from genes normally induced during the embryonic maturation phase. Thus, the first perceptible molecular function of plant embryonic miRNAs is the opposite of that in vertebrates; in vertebrates, miRNAs sharpen the first developmental transition, whereas in plants, they forestall developmental transitions by repressing mRNAs that act later. We propose that, by preventing precocious expression of differentiation-promoting transcription factors, miRNAs enable proper embryonic patterning.
Keywords: MicroRNA, embryogenesis, high-throughput sequencing, plant development, post-transcriptional gene regulation, Arabidopsis thaliana
MicroRNAs (miRNAs) are ∼21-nucleotide (nt) RNAs that guide the post-transcriptional regulation of target genes during plant and animal development (Bartel 2004). Plant miRNAs recognize nearly perfect complementary binding sites in target mRNAs and mediate RNA cleavage (Llave et al. 2002; Rhoades et al. 2002; Tang et al. 2003). The high degree of complementarity between plant miRNAs and their binding sites in target mRNAs has allowed confident miRNA target predictions (Rhoades et al. 2002; Jones-Rhoades and Bartel 2004; Fahlgren and Carrington 2010). Plant miRNA targets tend to encode key developmental regulators, including many transcription factors (Rhoades et al. 2002; Jones-Rhoades et al. 2006). Accordingly, several studies have demonstrated that the miRNA-mediated repression of target transcripts is essential for correct cell differentiation and developmental timing during post-embryonic development (Jones-Rhoades et al. 2006; Chen 2009). However, relatively little is known regarding the roles of miRNAs in embryonic cell differentiation, and miRNA functions during embryo developmental timing have not been characterized.
DICER-LIKE1 (DCL1) encodes an RNaseIII domain-containing protein that is nuclear-localized and required for processing primary miRNA transcripts into mature miRNAs (Park et al. 2002; Reinhart et al. 2002; Papp et al. 2003; Fang and Spector 2007). With this realization that DCL1 is necessary for miRNA biogenesis, the recovery and analysis of dcl1 mutant alleles from previous forward genetic screens performed using Arabidopsis thaliana (Schauer et al. 2002) provide insight into the roles of miRNAs during plant development. Null dcl1 alleles (originally named emb76, then sus1) were recovered by Meinke's group (Errampalli et al. 1991; Castle et al. 1993) more than 15 years ago in screens for embryos with defective development. dcl1 embryos are developmentally arrested at the globular stage of embryogenesis and exhibit abnormal divisions throughout the extraembryonic suspensor (Schwartz et al. 1994). These results suggest that DCL1 and, by implication, miRNAs are required for embryo development and viability. Null mutations in other genes required for normal miRNA biogenesis or function also produce defects during embryogenesis (Lynn et al. 1999; Lobbes et al. 2006; Grigg et al. 2009). In addition, specific miRNA/target interactions are required for proper cotyledon formation during embryo development (Palatnik et al. 2003; Laufs et al. 2004; Mallory et al. 2004, 2005). Moreover, several plant miRNA targets are required for proper embryogenesis (Aida et al. 1997; Emery et al. 2003; Dharmasiri et al. 2005; Prigge et al. 2005). Collectively, these results strongly indicate that miRNAs have important functions during embryo development.
Morphogenesis is the phase of embryo development when the basic plant body plan is established. Embryonic cell types are specified in particular locations at precise developmental time points. Shoot meristem precursors and root meristem precursors are specified at the apical and basal poles of the early embryo, respectively (Mayer et al. 1991). The post-embryonic activity of these meristems generates the adult plant body. Patterning along the radial axis during early embryogenesis generates the outermost protoderm layer, the innermost vascular primordium, and a middle layer of ground tissue precursors (Laux et al. 2004). After morphogenesis, embryos transition to a maturation phase, where they accumulate storage proteins, undergo desiccation tolerance, and prepare to enter into a state of dormancy prior to germination (Gutierrez et al. 2007; Holdsworth et al. 2008). Although progress has been made (Weber et al. 2005; Braybrook and Harada 2008; Park and Harada 2008), the molecular basis of early embryonic patterning and the morphogenesis-to-maturation-phase transition are not completely understood.
To assess the regulatory functions of miRNAs during embryogenesis, we revisited the phenotypic characterization of dcl1 embryos, with a focus on early morphogenesis and cell specification defects. DCL1 was required for multiple embryonic cell differentiation events as early as the eight-cell stage. Genome-wide transcript profiling revealed that DCL1 was required for the early embryonic repression of nearly 50 miRNA targets. Several of the miRNA targets up-regulated in eight-cell dcl1 embryos encode transcription factors that promote differentiation during later stages of embryogenesis. Moreover, dcl1 embryos prematurely accumulated transcripts from hundreds of genes typically expressed during the maturation phase of embryo development. The disruption of miR156-mediated regulation of two redundant SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors was both responsible for the premature gene expression observed in dcl1 embryos and partially required for the dcl1 embryo patterning defects. Therefore, some of the earliest roles of miRNAs are to prevent the precocious expression of differentiation-promoting transcription factors during early embryogenesis and to enable proper pattern formation.
Results
DCL1 is required for early embryonic pattern formation
dcl1-null mutant embryos exhibit morphological defects and arrest at the globular stage of development (Schwartz et al. 1994). To identify the earliest dcl1 morphological defects, we systematically analyzed dcl1 embryos throughout early embryogenesis. Because dcl1 embryos are not viable, embryos from self-pollinated plants heterozygous for dcl1-5 and dcl1-10, two presumed null alleles (McElver et al. 2001; Schauer et al. 2002), were examined.
Embryos developing within a single silique are approximately at the same developmental stage, which enabled an estimate of dcl1 embryonic stages based on the morphology of their wild-type siblings. It was reported previously that sus1/dcl1 embryos exhibit abnormal cell divisions at the base of the embryo proper beginning at the globular stage and in the extraembryonic suspensor beginning at the heart stage (Schwartz et al. 1994). Our morphological analysis of dcl1 embryos confirmed these findings and revealed previously unreported phenotypes (Fig. 1A). We identified initial morphological defects in the presumptive hypophysis cells (i.e., the suspensor cell most proximal to the embryo proper) of 19% (29 of 154) of dermatogen stage embryos from self-pollinated dcl1-5/+ plants (Fig. 1A). By the early globular stage, morphological defects were observed in ∼25% of embryos from selfed dcl1-5/+ plants (Fig. 1A). These embryos with defects were presumably homozygous for dcl1-5-null alleles, and their defects included abnormal hypophysis cell divisions as well as the previously unreported loss of periclinal subprotoderm cell divisions in the embryo proper (Fig. 1A). In subsequent stages, these abnormal embryos did not produce cotyledons and were developmentally arrested and nonviable (Fig. 1A; Schwartz et al. 1994). Furthermore, dcl1-5 homozygous seedlings could not be recovered from selfed dcl1-5/+ plants, which suggested that the developmentally arrested embryos derived from selfed dcl1-5/+ plants were indeed homozygous for dcl1-5 (data not shown). Because embryos from selfed dcl1-5/+ and selfed dcl1-10/+ plants exhibited the same morphological defects at indistinguishable frequencies, subsequent analyses focused on embryos from selfed dcl1-5/+ plants (Table 1; data not shown). DCL1 transcripts were detected by quantitative RT–PCT (qRT–PCR) in both early globular embryos from wild-type plants and early globular embryos from selfed dcl1-5/+ plants that developed normally, but were not detected in abnormal early globular embryos from selfed dcl1-5/+ plants (Supplemental Fig. 1). These expression analyses further confirmed that abnormal embryos from selfed dcl1/+ plants were homozygous for dcl1-null alleles.
Figure 1.
DCL1 is required for pattern formation during early embryogenesis. (A) Representative Nomarski images of embryos from wild-type and dcl1-5/+ self-pollinated plants. Phenotypic classes are denoted directly above each column, and developmental stages are indicated at the left of each row. The percent of embryos in each phenotypic class is noted below each image, and total numbers of embryos examined are in parentheses. Asterisks indicate products of abnormal divisions, and carets designate positions where cell divisions did not take place in the mutants. Bars, 20 μm. (B) Representative micrographs of RNA in situ hybridizations performed with WOX2, WOX8, and ATML1 antisense (AS) probes, and CSLM images with WOX5p∷NLS-YFP and DR5rev∷GFP reporters. Presumptive embryo genotypes are indicated above each column, and developmental stages are indicated to the left of each row. Probes/reporters are indicated at the bottom of each image. For the CSLM images, GFP and FM4-64 fluorescence are shown in green and red, respectively. See also Supplemental Figure 2.
Table 1.
Frequencies of embryos with Dcl1 phenotype
aEmbryo phenotypes were determined when wild-type-like embryos were at the late globular/transition stages.
bBased on χ2 tests.
cP-value for frequency ≠ 25%.
dP-value for frequency ≠ frequency of dcl1-5/+ spl11-1 (16.8%).
We next tested whether DCL1 was required for embryonic cell differentiation by examining the expression of cell-specific markers in dcl1-5 embryos. WUSCHEL-related HOMEOBOX2 (WOX2) transcripts are localized in the apical cell lineage of wild-type preglobular embryos (Haecker et al. 2004). After the two/four-cell stage, WOX2 transcripts were not detected in about a quarter of eight-cell and dermatogen stage embryos (seven of 35 and 14 of 52, respectively) from self-pollinated dcl1-5/+ plants (Fig. 1B), which implied that dcl1-5 embryos had undetectable WOX2 expression. Similar to WOX2, PINHEAD/ZWILLE/ARGONAUTE10 transcripts, hereafter referred to as PNH, are localized in the apical cell lineage of preglobular embryos (Lynn et al. 1999). Approximately 89% (72 of 81) of eight-cell and dermatogen stage embryos from selfed dcl1-5/+ plants had detectable levels of PNH transcripts, suggesting that apical cell fate is not completely abolished in all preglobular dcl1 embryos (Supplemental Fig. 2).
In contrast to WOX2 and PNH, WOX8 transcripts are localized to the basal cell lineage of preglobular wild-type embryos (Haecker et al. 2004). WOX8 transcripts were detected in 90% (62 of 69) of eight-cell and dermatogen stage embryos from selfed dcl1-5/+ plants, suggesting that basal cell lineage differentiation is not perturbed in all preglobular dcl1 embryos (Fig. 1B). Thus, dcl1 embryos had detectable defects in the differentiation of apical cell but not basal cell descendents as early as the eight-cell stage.
To test whether DCL1 is required for radial patterning, we examined a series of markers that are differentially expressed along the radial axis. A. thaliana MERISTEM LAYER1 (ATML1) and PROTODERMAL FACTOR1 (PDF1) transcripts are detectable in the protoderm, but not the subprotoderm, of globular stage embryos (Lu et al. 1996; Abe et al. 1999). ATML1 and PDF1 transcripts were appropriately localized to the protoderm of globular stage dcl1-5 embryos, but were ectopically localized in the suspensors of dcl1-5 embryos whose wild-type siblings were at the early heart stage (Fig. 1B; Supplemental Fig. 2). This observation was consistent with the previously reported ectopic localization of embryo proper-specific starch grains and protein bodies in the suspensors of late stage sus1/dcl1 embryos (Schwartz et al. 1994). Consistent with the reduction/loss of subprotodermal cell divisions in dcl1 late globular/early heart stage embryos, we detected reduced levels of RPS5A transcripts in these cell types (Supplemental Fig. 2). Moreover, vascular primordium and ground tissue initial markers were absent from these cell types (Supplemental Fig. 2).
In addition to the ectopic localization of protoderm markers in the suspensor, globular dcl1-5 suspensors had reduced levels of WOX8 transcripts (Fig. 1B). Furthermore, hypophysis markers were not detectable in dcl1 globular embryos (Fig. 1B; Supplemental Fig. 2). Considered together with the defective hypophysis cell divisions observed in dcl1 embryos, the lack of hypophysis markers indicates that the hypophysis is misspecified in dcl1 embryos.
The plant hormone auxin is transported from the embryo proper to the presumptive hypophysis and contributes to hypophysis specification (Friml et al. 2003; Aida et al. 2004; Weijers et al. 2006). Because the auxin signaling-responsive DR5rev promoter fused to GFP (DR5rev∷GFP) is specifically active in the hypophysis cells of wild-type globular embryos (Friml et al. 2003), and hypophysis specification is perturbed in dcl1 embryos, we examined the activity of DR5rev∷GFP in dcl1-5 embryos to test whether DCL1 was required for appropriate auxin signaling. Although presumptive dcl1-5 hypophysis cells often expressed DR5rev∷GFP, cells located above the presumptive hypophysis ectopically expressed DR5rev∷GFP (Fig. 1B). Therefore, DCL1 is required for the hypophysis-specific increase in auxin signaling. Given the severe subprotodermal defects observed in dcl1 embryos, the inappropriate auxin signaling observed in dcl1 embryos was likely due to the incorrect expression of auxin transport and/or auxin signaling factors. Therefore, the auxin response defects observed in dcl1 embryos were probably secondary consequences of subprotoderm cell differentiation defects, which in turn were probably due to the apical cell lineage defects observed at the preglobular stages.
Many miRNA targets have increased transcript levels in dcl1 embryos
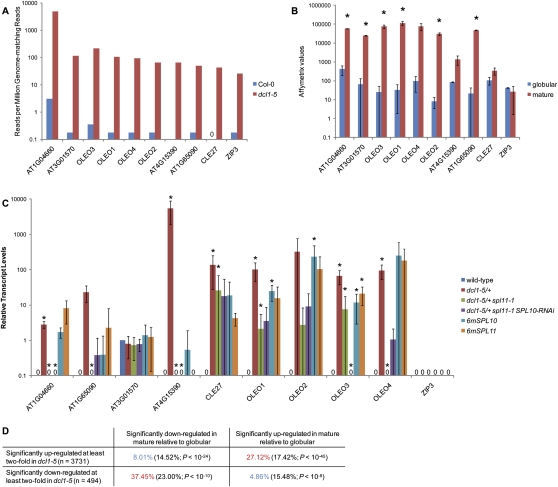
Because DCL1 is required for miRNA biogenesis (Park et al. 2002; Reinhart et al. 2002), the patterning defects that we observed in dcl1 embryos were likely due to the loss of specific miRNAs and the consequent up-regulation of their respective targets. To test which miRNA targets were up-regulated in dcl1 embryos relative to wild-type embryos, we performed genome-wide transcript profiling of wild-type and dcl1-5 early globular embryos using strand-specific mRNA-Seq. Of 205 annotated miRNA targets (see the Materials and Methods), 71 had at least 20 raw reads in dcl1, of which 68% (48 of 71), potentially targeted by 25 miRNA families, had at least twofold more reads in dcl1 relative to wild type (Supplemental Fig. 3; Supplemental Table 1). To focus on the miRNA targets most likely to be responsible for the defects observed in dcl1 embryos, predicted targets with at least 40 raw reads in dcl1 and at least fivefold more reads in dcl1 relative to wild type were selected for further analysis. Fifteen predicted targets of 10 miRNA families (miR156, miR159, miR160, miR166, miR168, miR319, miR393, miR400, miR778, and miR824) fulfilled these criteria (Fig. 2A). The most up-regulated target in dcl1 relative to wild type was the miR156 target SPL10, which increased ∼130-fold (Fig. 2A). Examination of SPL10-GFP translational fusions by confocal scanning laser microscopy (CSLM) confirmed that SPL10 increased in early globular dcl1-5 embryos (Fig. 2B). Signal corresponding to SPL10-GFP was mostly nuclear, which is consistent with its presumed role as a transcription factor.
Figure 2.
Many miRNA targets are overexpressed in dcl1-5 embryos. (A) Number of reads matching miRNA targets in libraries prepared from Col-0 (wild-type) and dcl1-5 early globular embryos. Only miRNA targets that had at least 40 raw reads in the dcl1-5 library and were increased at least fivefold in dcl1-5 embryos relative to Col-0 embryos are shown. Reads were normalized by dividing the number of reads that match each gene by the number of million genome-matching reads from each library. The corresponding miRNA is shown below each set of miRNA targets. See also Supplemental Figure 3. (B) CSLM images of early globular embryos expressing SPL10-GFP translational fusions under the control of upstream intergenic sequences (SPL10p∷SPL10-GFP). Genotypes are indicated above images. GFP and FM4-64 fluorescence are shown in green and red, respectively. Bars, 20 μm. (C) miRNA target transcript levels in eight-cell embryos from self-pollinated Col-0 and dcl1-5/+ plants based on qRT–PCR and normalized to Col-0 levels. The corresponding miRNA is shown below each set of miRNA targets. Error bars represent one standard deviation of the sample mean. Transcripts that were not detected are indicated by a zero. Asterisks indicate target transcripts that were significantly increased in dcl1-5/+ progeny compared with wild-type. Probabilities were calculated with heteroscedastic Student's t-tests using one-tailed distributions. Probabilities <0.05 were considered significant.
Because the earliest differentiation defect detected in dcl1 embryos occurred at the eight-cell stage (Fig. 1B), we hypothesized that the misregulation of one or more targets in dcl1 eight-cell embryos led to the widespread patterning defects observed. To test which miRNA target transcripts were increased in dcl1 eight-cell embryos, qRT–PCR was performed using RNA isolated from either wild-type eight-cell embryos or eight-cell embryos derived from selfed dcl1-5/+ plants. Of the 15 predicted targets analyzed, nine had significantly increased transcript levels in embryos derived from selfed dcl1-5/+ plants relative to wild type (Fig. 2C). All nine of the up-regulated targets encoded transcription factors that are predicted targets of either miR156, miR159, miR160, miR166, miR319, or miR824 (Fig. 2C). Therefore, multiple miRNAs appear to be repressing transcription factor production as early as the eight-cell stage of embryogenesis. Two known miR156 targets, SPL10 and SPL11 (Vazquez et al. 2004; Addo-Quaye et al. 2008; German et al. 2008), were the most up-regulated targets, with ≥150-fold increased transcript levels in progeny from self-fertilized dcl1-5/+ plants (Fig. 2C). When considering that only one-quarter of the embryos from selfed dcl1-5/+ plants were homozygous for dcl1-5, these results suggest that SPL10 and SPL11 transcript levels were elevated as much as ∼600-fold in dcl1-5 eight-cell embryos. Such dramatic elevations spurred us to test whether derepression of SPL10 or SPL11 was responsible for the defects observed in dcl1-5 embryos.
miR156 is localized and active throughout early embryogenesis
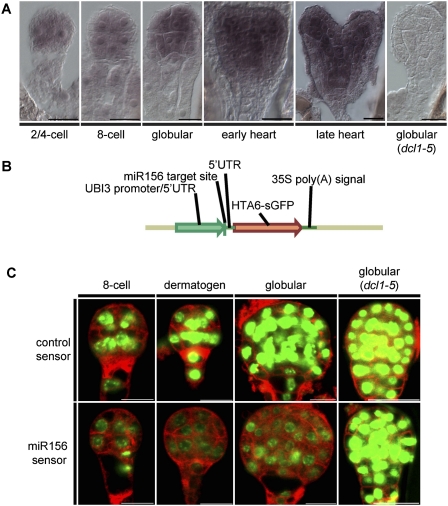
For the loss of miR156 to explain the observed up-regulation of SPL10 and SPL11 transcripts in dcl1 embryos, miR156 must be present and active at the relevant embryonic stages. To test for miR156 presence, we performed RNA in situ hybridizations on wild-type and dcl1-5 embryos using locked nucleic acid (LNA) probes antisense to miR156. Signal corresponding to miR156 was detected throughout two/four-cell and eight-cell embryos (Fig. 3A). Beginning at the globular stage, miR156 signal was increased in subprotodermal tissue layers, and this apparent differential accumulation of miR156 became more pronounced at the heart stage (Fig. 3A). Globular stage dcl1-5 embryos had undetectable signal, confirming both the probe specificity and miR156 loss in dcl1-5 embryos (Fig. 3A).
Figure 3.
miR156 is localized and active throughout early embryogenesis. (A) Representative micrographs of RNA in situ hybridizations performed with LNA probes antisense to miR156. All embryos are wild type except for the indicated dcl1-5 embryo. The stage of embryo development is listed below each image. (B) miR156 sensor construct used to detect miR156 activity. The UBI3 promoter/5′UTR, miR156 target site, translational fusion between Arabidopsis HISTONE 2A (HTA6) and sGFP, and Cauliflower Mosaic Virus 35S poly(A) site are indicated. (C) CSLM images of embryos expressing sensor constructs with either no site (control sensor; top) or miR156 target site (miR156 sensor; bottom). Embryo developmental stages are indicated above each column of images. All embryos are wild type except for the indicated dcl1-5 embryos. GFP and FM4-64 fluorescence are shown in green and red, respectively. Bars, 20 μm.
To test for miR156 activity, a sensor was constructed that contained a miR156 target site in the 5′ untranslated region (UTR) of a ubiquitously expressed nuclear-localized GFP (Fig. 3B). An identical construct without the miR156 target site served as a control. Transgenic plants expressing miR156 and control sensors were generated to determine the embryonic spatiotemporal activation domains of miR156. Compared with embryos with the control sensor, those with the miR156 sensor had reduced GFP signal throughout the eight-cell, dermatogen, and globular stages (Fig. 3C). Furthermore, GFP signal corresponding to the miR156 sensor was derepressed in dcl1-5 embryos. Together, these results demonstrate that miR156 is localized and active throughout early embryogenesis, and that miR156 activity is lost in dcl1-5 embryos.
SPL10 and SPL11 are partially responsible for dcl1 mutant phenotypes
The results presented so far indicate that miR156 is lost in dcl1 embryos and, as a result, SPL10 and SPL11 transcripts increase during early embryogenesis. To determine whether the up-regulation of SPL10 or SPL11 in dcl1 embryos was responsible for the Dcl1 phenotype, we tested whether spl10 or spl11 mutants suppressed dcl1 embryonic defects. Plant lines carrying T-DNA insertions in either SPL10 (spl10-1) or SPL11 (spl11-1) were obtained from the Arabidopsis thaliana Resource Center for Genomics at the French National Institute for Agricultural Research (Samson et al. 2002). Although SPL10 transcripts were detected in spl10-1 plants, SPL10 function was likely at least partially disrupted in spl10-1 plants due to a T-DNA insertion in the first coding sequence exon, which deletes 51 base pairs (bp) of adjacent genomic sequence (Supplemental Fig. 4; data not shown). Full-length transcripts corresponding to SPL11 could not be detected from spl11-1 plants (Supplemental Fig. 4). Embryos homozygous for spl10-1 or spl11-1 did not exhibit detectable morphological phenotypes.
Late globular and early heart stage embryos from self-pollinated dcl1-5/+ spl10-1, dcl1-5/+ spl11-1, and dcl1-10/+ spl11-1 plants were examined to test for a decrease in the frequency or severity of the Dcl1 phenotype. Selfed dcl1-5/+ spl10-1 plants produced ∼25% Dcl1 embryos, suggesting that SPL10 alone is not required for the phenotypes observed in dcl1 embryos (Table 1). In contrast, selfed dcl1-5/+ spl11-1 and selfed dcl1-10/+ spl11-1 plants yielded only ∼17% and ∼19% Dcl1 embryos, respectively (Table 1). Thus, spl11-1 suppressed the penetrance of the early embryonic Dcl1 phenotype.
Because SPL10 and SPL11 transcripts were both increased in dcl1-5 embryos and encode proteins that are ∼76% identical, we then tested whether SPL10 and SPL11 were redundantly required for the Dcl1 phenotype. dcl1/+ spl10 spl11 mutants could not be generated due to the tight linkage of the SPL10 and SPL11 loci (Supplemental Fig. 4). However, F1 embryos derived from crosses between dcl1-5/+ spl10-1 and dcl1-5/+ spl11-1 plants exhibited the Dcl1 phenotype at frequencies that were significantly less than those observed in embryos from self-pollinated dcl1-5/+ spl10-1 plants or crosses between dcl1-5/+ and dcl1-5/+ spl11-1 plants (Table 1). Therefore, simultaneously reducing the SPL10 and SPL11 wild-type alleles by one-half suppressed the penetrance of the Dcl1 phenotype. To further test whether SPL10 and SPL11 were redundantly required for the Dcl1 phenotype, we generated SPL10-RNAi constructs under the control of a ubiquitously expressed embryonic promoter, and transformed these SPL10-RNAi constructs into dcl1-5/+ spl11-1 plants. A strong knockdown of SPL10 transcripts was observed in embryos from dcl1-5/+ spl11-1 SPL10-RNAi plants relative to embryos from dcl1-5/+ spl11-1 plants, which demonstrated the functionality of this RNAi construct (Supplemental Fig. 5). Only a small percentage (1.5%; three of 203) of late globular and early heart embryos from selfed dcl1-5/+ spl11-1 SPL10-RNAi plants exhibited the Dcl1 phenotype. However, 8% (17 of 203) of embryos from these selfed plants exhibited a partial Dcl1 phenotype, characterized by defects in hypophysis but not subprotoderm cell division (Fig. 4A). With regard to later stages in development, ∼25% of late heart and torpedo stage embryos from selfed dcl1-5/+ spl11-1 plants (23.4%; 32 of 137) and selfed dcl1-5/+ spl11-1 SPL10-RNAi plants (22.1%; 35 of 159) resembled dcl1 embryos. More specifically, they failed to form cotyledons, exhibited aberrant cell divisions throughout the embryo and suspensor, and were arrested in development. Furthermore, seedlings homozygous for dcl1-5 were never recovered from the progeny of these plants. These results suggest that either the SPL10-RNAi construct is not effective at later stages of embryo development, or knockdown of other miRNA targets is also required for a full rescue of dcl1 embryos. Nevertheless, in globular and early heart stage dcl1 embryos, SPL10 and SPL11 are redundantly required for the observed morphological defects.
Figure 4.
SPL10 and SPL11 are redundantly required for dcl1 mutant phenotypes. (A) Representative Nomarski images of late globular embryos from self-pollinated wild-type, dcl1-5/+ spl11-1, and dcl1-5/+ spl11-1 SPL10-RNAi plants. Phenotypic classes are indicated directly above or below images and parental plant genotypes are listed directly above or below phenotypic classes. Phenotypic class frequencies for the given parental genotype and the total number of embryos examined are shown in the bottom left corner of each image. Asterisks indicate abnormal products of cell divisions, and carets designate positions where cell divisions did not take place in the mutants. (B) Schematic of base-pairing interactions between SPL10 and SPL11 (SPL10/11) and miR156, and between 6mSPL10/11 and miR156. Mutations introduced by site-directed mutagenesis are shown in red. Watson-Crick base-pairing (I), non-base-pairing (X), and G:U wobbles (O) for each pair are indicated. (C) Representative Nomarski images of late globular/transition stage embryos from self-pollinated gSPL10, 6mSPL10, gSPL11, and 6mSPL11 plants. Parental genotypes are listed directly above or below each image. Phenotypic frequencies and total number of embryos examined are indicated in the bottom left corner of each image. Asterisks indicate abnormal cell divisions. (D) Representative micrographs of RNA in situ hybridizations performed with either WOX2 antisense or sense probes on eight-cell embryos from self-pollinated wild-type and dcl1-5/+ spl11-1 SPL10-RNAi plants. Probes and parental genotypes are indicated directly above and below each image, respectively. Frequencies of embryos with detectable WOX2 transcripts and total number of embryos examined are shown at the bottom of each image. Bars, 20 μm.
We then tested whether the misregulation of SPL10 or SPL11 was sufficient to cause the Dcl1 phenotype. The miR156 target sites of SPL10 and SPL11 genes were disrupted with synonymous codon mutations within transformation constructs (6mSPL10 and 6mSPL11) (Fig. 4B). Upstream and downstream intergenic regions were included in these constructs to preserve other potential cis-regulatory elements that control SPL10 and SPL11 expression (Supplemental Fig. 4). Transgenic plants carrying nonmutated SPL10 and SPL11 loci (gSPL10 and gSPL11) were also generated as controls. Plants carrying either the 6mSPL10 or 6mSPL11 transgenes exhibited traits characteristic of precocious juvenile-to-adult vegetative-phase transitions (Supplemental Fig. 6), as reported for plants expressing miR156-resistant SPL10 or SPL11 transgenes (Wu et al. 2009). This, together with the observation that 6mSPL10 and 6mSPL11 embryos had significantly increased levels of SPL10 and SPL11 transcripts relative to wild type (Supplemental Fig. 5), indicated that the 6mSPL10 and 6mSPL11 transgenes used in our study were expressed and functional. Although they did not phenocopy dcl1 embryos, 6mSPL10 and 6mSPL11 embryos resembled dcl1 embryos in exhibiting division defects in cells at the position of the hypophysis derivatives (cf. Figs. 4C and 1A). Embryos expressing gSPL10 or gSPL11, the wild-type versions of these transgenes, resembled wild-type embryos, which suggested that disruption of miR156-mediated regulation of SPL10 or SPL11 was responsible for the observed phenotypes (Fig. 4C).
6mSPL10 and 6mSPL11 embryos had decreased levels of SPL10 and SPL11 transcripts relative to dcl1-5 embryos (Supplemental Fig. 5), which suggested the possibility that they did not phenocopy the dcl1 embryos because the 6mSPL10 and 6mSPL11 transgenic lines that expressed SPL10 and SPL11 at the levels observed in dcl1-5 embryos were not viable and could not be recovered because they did indeed phenocopy the dcl1 embryos. Alternatively, expression of SPL10 and SPL11 at the levels observed in dcl1-5 embryos still might not have been sufficient to cause the defects observed in dcl1 embryos. To help distinguish between these two possibilities, we examined embryos from self-pollinated 6mSPL10/_ 6mSPL11/_ plants, some of which had increased dosage of the miR156-resistant mRNAs because they were homozygous at both alleles. Approximately 16% (21 of 128) resembled the abnormal embryos observed in 6mSPL10 and 6mSPL11 embryos, but none phenocopied dcl1 embryos. Thus, disrupting miR156-mediated regulation of SPL10 and SPL11 caused defects during embryo morphogenesis but appears insufficient to phenocopy dcl1 embryos.
To test whether SPL10 and SPL11 were required for the loss of the apical cell lineage-specific transcript WOX2 in dcl1-5 preglobular embryos (Fig. 1B), we examined WOX2 transcripts in eight-cell and dermatogen stage embryos from selfed dcl1-5/+ spl11-1 SPL10-RNAi plants using RNA in situ hybridizations. The frequency of eight-cell and dermatogen stage embryos with detectable WOX2 transcripts significantly increased in the progeny of selfed dcl1-5/+ spl11-1 SPL10-RNAi plants relative to selfed dcl1-5/+ plants (88.5% [85 of 96] vs. 75.9% [66 of 87], respectively; P < 0.05) (Fig. 4D). Therefore, SPL10 and/or SPL11 appeared to be required for the reduction of WOX2 transcripts in dcl1-5 preglobular embryos. To test whether misregulated SPL10 or SPL11 were sufficient for WOX2 transcript reduction, we examined WOX2 transcript levels in eight-cell embryos from 6mSPL10 and 6mSPL11 plants. There was not a significant difference in transcript levels between wild-type, 6mSPL10, and 6mSPL11 eight-cell embryos (Supplemental Fig. 7). Collectively, our results indicate that derepression of SPL10 and SPL11 is required for a substantial part of the dcl1 patterning defects, but disrupting miR156-mediated regulation of these genes is not sufficient to fully recapitulate the dcl1 patterning defects.
SPL10 and SPL11 zygotic transcripts are repressed by miR156 during early embryogenesis
Because SPL10 and SPL11 transcripts were dramatically repressed by miR156 in eight-cell embryos (Fig. 2C; Supplemental Fig. 5), we tested whether miR156 clears maternal or paternal SPL10/SPL11 transcripts from early embryos. We did not detect significant parent-of-origin effects of the spl11-1 allele or the 6mSPL10 /6mSPL11 transgenes on zygotic SPL10/SPL11 transcript levels (Supplemental Fig. 8).
To assess the spatiotemporal expression patterns of SPL10 and SPL11 during embryogenesis, we examined the activity of SPL10 and SPL11 promoters transcriptionally fused to a nuclear-localized GFP (SPL10p∷NLS-GFP and SPL11p∷NLS-GFP). Weak signal corresponding to SPL10p∷NLS-GFP was detected in the subprotoderm and suspensors of dermatogen stage embryos and became more intense throughout the embryo during the globular stages (Fig. 5). Low levels of SPL11p∷NLS-GFP signal were detected throughout eight-cell embryos and, like SPL10p∷NLS-GFP signals, became progressively stronger during the globular stages (Fig. 5). Signal corresponding to SPL10p∷NLS-GFP and SPL11p∷NLS-GFP transgenes was more intense in the basal half of the embryo proper at the late globular stage, had peak levels at the heart stage, and were detected in torpedo stage embryos with increased levels in the shoot and root meristem precursors and vascular primordium (Fig. 5). These results were consistent with the Harada-Goldberg Arabidopsis seed development laser capture microdisection microarray data set (National Center for Biotechnology Gene Expression Omnibus [NCBI GEO]: GSE12404). We conclude that SPL10p∷NLS-GFP and SPL11p∷NLS-GFP are therefore expressed in overlapping patterns at the stages and in the cell types that exhibited patterning defects in dcl1, 6mSPL10, and 6mSPL11 embryos. Furthermore, they are expressed in the embryonic spatiotemporal domains where miR156 is active (cf. Figs. 3C and 5). Taken together, our results indicate that miR156 represses the zygotic expression of both SPL10 and SPL11 during early embryogenesis.
Figure 5.
Spatiotemporal activation domains of SPL10 and SPL11 promoters. Representative CSLM images of embryos from plants carrying SPL10p∷NLS-GFP or SPL11p∷NLS-GFP transgenes. Parental genotypes are indicated to the left of each row of images, and embryonic stages are designated above each column of images. The green signal corresponds to GFP, and the red signal corresponds to FM4-64. Bars, 20 μm.
Hundreds of genes are expressed prematurely in dcl1 embryos
In addition to the many miRNA targets up-regulated in dcl1-5 early globular embryos, we found many other genes that were also up-regulated. More specifically, 37.1% (4331 of 11,570) of genes with at least 20 raw reads in the dcl1-5 early globular library had at least twofold significantly more reads in dcl1-5 relative to wild type. Four of the 10 most up-regulated genes in dcl1-5 early globular embryos encoded OLEOSINS (Fig. 6A). Because OLEOSINS are found in the oil bodies of mature embryos (Frandsen et al. 2001), this suggested that OLEOSIN genes are expressed prematurely in dcl1-5 early globular embryos, and that the same might be true for other genes. Indeed, using the Harada-Goldberg data set (NCBI GEO: GSE12404), we found that nine out of the 10 most up-regulated genes in dcl1-5 early globular embryos had increased transcript levels in mature embryos relative to globular embryos (Fig. 6B). Moreover, significantly elevated transcript levels were detected for six out of nine of these genes in eight-cell embryos derived from selfed dcl1-5/+ plants relative to wild-type eight-cell embryos (Fig. 6C). In fact, transcripts corresponding to only one of the nine genes were detected in wild-type eight-cell embryos, consistent with their expression being mostly restricted to the maturation phase of embryogenesis. In summary, we observed that the most up-regulated genes in dcl1-5 early globular embryos are typically expressed at post-globular stages of embryo development, and are expressed prematurely as early as the eight-cell stage in dcl1-5 embryos.
Figure 6.
miR156 regulation of SPL10 and SPL11 prevents premature gene expression during early embryogenesis. (A) Normalized read numbers of the top 10 up-regulated genes in dcl1-5 early globular embryos relative to wild-type early globular embryos based on mRNA-Seq. Transcripts that were not detected are indicated by a zero. (B) Transcript levels of the top 10 up-regulated genes in dcl1 early globular embryos at globular and mature stages of embryo development based on the Harada-Goldberg data sets (NCBI GEO: GSE12404). Error bars represent one standard deviation of the sample. (C) Relative transcript levels of the top 10 up-regulated genes in eight-cell embryos from self-pollinated Col-0, dcl1-5/+, dcl1-5/+ spl11-1, dcl1-5/+ spl11-1 SPL10-RNAi, 6mSPL10, and 6mSPL11 plants based on qRT–PCR and normalized to Col-0 levels. Transcripts that were not detected are indicated by a zero. Error bars represent one standard deviation of the sample mean. Asterisks indicate either significantly increased transcript levels compared with wild-type (dcl1-5/+, 6mSPL10 and 6mSPL11 samples) or significantly decreased transcript levels compared with dcl1-5/+ (dcl1-5/+ spl11-1 and dcl1-5/+ spl11-1 SPL10-RNAi samples). Probabilities were calculated with heteroscedastic Student's t-tests. Probabilities <0.05 were considered significant. (D) Frequency of genes that are either significantly up-regulated or down-regulated in dcl1-5 early globular embryos (left column) that also have significantly different transcript levels in mature embryos relative to globular embryos (top row) based on publically available Affymetrix microarray data sets (NCBI GEO: GSE12404). The total number of genes that are either significantly up-regulated or down-regulated in dcl1-5 and that also have corresponding probes on the Affymetrix arrays are represented by n. Frequencies are color-coded in red or blue to indicate enrichment or depletion, respectively. Frequencies in parentheses are what is expected based on randomly selected control sets corrected for gene expression levels (see the Materials and Methods for more details). Probabilities (P) that differences between observed frequencies and expected frequencies (based on random control sets) are different due to chance are shown and were calculated using χ2 distributions.
To extend the analysis to the thousands of other genes up-regulated in dcl1-5 relative to wild-type early globular embryos, we tested whether these genes also tended to be expressed preferentially in mature embryos compared with globular embryos. Compared with control gene sets, genes with at least twofold significantly increased transcript levels in dcl1-5 early globular embryos were significantly enriched for genes with increased transcript levels in mature embryos relative to globular embryos, and were significantly depleted of genes with decreased transcript levels in mature embryos relative to globular embryos (Fig. 6D). Conversely, genes with transcript levels significantly decreased at least twofold in dcl1-5 early globular embryos were significantly enriched for genes that have decreased transcript levels in mature embryos relative to globular embryos, and were significantly depleted of genes that had increased transcript levels in mature embryos (Fig. 6D). Therefore, many genes are prematurely up-regulated or down-regulated in dcl1 early globular embryos relative to wild type.
Loss of miR156-mediated regulation of SPL transcripts is responsible for premature accumulation of transcripts in eight-cell embryos
The premature expression of many genes in dcl1-5 embryos could be a consequence of the up-regulation of miRNA targets (Fig. 2; Supplemental Table 1). Because SPL10 and SPL11 transcripts were dramatically up-regulated in eight-cell dcl1-5 embryos (Fig. 2C), we tested whether SPL10 and SPL11 function upstream of prematurely expressed genes. The levels of transcripts corresponding to the 10 most up-regulated genes in dcl1-5 early globular embryos were examined in eight-cell embryos from self-pollinated dcl1-5/+ spl11-1 plants to test whether spl11-1 suppressed the premature accumulation of transcripts observed in dcl1 embryos. Among the genes examined by qRT–PCR, transcript levels for all six that were significantly increased in dcl1-5 relative to wild-type eight-cell embryos were suppressed in eight-cell embryos from dcl1-5/+ spl11-1 plants (Fig. 6C). Transcript levels for these genes were also suppressed in eight-cell embryos from dcl1-5/+ spl11-1 SPL10-RNAi plants (Fig. 6C). These results indicate that SPL11 is necessary for the accumulation of transcripts corresponding to at least some of the genes expressed prematurely in dcl1-5 embryos.
To test whether disruption of miR156-mediated regulation of SPL10 or SPL11 is sufficient for the premature accumulation of transcripts in eight-cell embryos, we examined premature transcript levels in embryos expressing miR156-resistant 6mSPL10 or 6mSPL11 transgenes. Eight of the top 10 most up-regulated genes in dcl1-5 early globular embryos had both increased transcript levels in mature embryos relative to globular embryos and increased transcript levels in eight-cell embryos from selfed dcl1-5/+ plants (Fig. 6B,C). Seven of these eight genes also had at least twofold increased transcript levels in embryos expressing either the 6mSPL10 or 6mSPL11 transgene (Fig. 6C). These results suggest that, upon the disruption of miR156-mediated regulation, SPL10 and SPL11 are sufficient to promote the precocious accumulation of maturation-phase transcripts in eight-cell embryos.
Discussion
We found that DCL1 and, by implication, miRNAs were required for multiple embryonic patterning events beginning as early as the eight-cell stage. In fact, the only cell type that appeared to differentiate correctly in dcl1 embryos was the outermost protoderm layer. Furthermore, protoderm markers that are typically restricted to the embryo proper were ectopically expressed in dcl1 suspensors at the late globular and early heart stages (Fig. 1B; Supplemental Fig. 2), which helps explain a previously reported function of DCL1/SUS1 in maintaining the extraembryonic cell fate of the suspensor (Schwartz et al. 1994).
The requirement of miRNAs for proper embryonic patterning and cell differentiation might have suggested that embryonic miRNAs act to trigger developmental transitions, as was first reported for metazoan miRNAs (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000), where induced miRNAs act to down-regulate genes that contribute to the differentiation state of the precursor cells. Or they might act to sharpen developmental transitions, as observed in fish, frog, and, perhaps, mammalian embryos, where miR-430 or its orthologs clears maternally expressed messages to sharpen the transition from maternal to zygotic expression programs (Farh et al. 2005; Giraldez et al. 2006; Lund et al. 2009). Indeed, an analogous function in sharpening developmental transitions was proposed and has been observed for plant miRNAs at later developmental stages (Rhoades et al. 2002; Aukerman and Sakai 2003; Lauter et al. 2005; Wu et al. 2009). However, our molecular characterization of the Dcl1 phenotype shows that the earliest known developmental roles of miRNAs in plants is not to trigger or sharpen developmental transitions by repressing genes with developmental functions in precursor cell types, but instead is the opposite (Fig. 7). In eight-cell embryos, miRNAs repress genes that function in daughter cell types later in embryo development. Instead of attenuating pre-existing function, they prevent precocious function. Dicer and other miRNA biogenesis proteins are required for formation of the mammalian mesoderm and differentiation of murine embryonic stem cells (Bernstein et al. 2003; Kanellopoulou et al. 2005; O'Rourke et al. 2007; Wang et al. 2007). Perhaps as the roles of individual vertebrate miRNAs are examined in more detail, analogous functions in preventing precocious embryonic expression will help explain the requirement of miRNAs for mammalian embryonic development. Indeed, recent studies show that the extraembryonic lineage of mouse embryos uses miRNAs to prevent premature differentiation and maintain extraembryonic stem cell multipotency (Spruce et al. 2010).
Figure 7.
Opposite functions of animal and plant miRNAs during early embryogenesis. (A) In zebrafish embryos, miR-430 sharpens the maternal-to-zygotic transition by directing the destabilization of maternally derived target transcripts, which function earlier in development (Giraldez et al. 2006). MZdicer (maternal–zygotic dicer) embryos lack Dicer and miR-430 and exhibit delayed reduction of maternal miR-430 targets. (B) In Arabidopsis embryos, miR156 delays the production of maturation transcripts by directing the repression of SPL10/11. Early dcl1 mutant embryos lack miR156 and exhibit premature expression of miR156 targets SPL10 and SPL11, which in turn induces precocious expression of genes normally induced during the maturation phase of embryogenesis. We propose that additional plant miRNAs—including miR160, miR166, and miR319—also forestall expression of differentiation-promoting transcription factors such as ARF17, CNA, PHB, PHV, and TCP4. A link between delayed reduction of maternal transcripts and the morphogenesis phenotypes observed in MZdicer embryos awaits experimental confirmation. The same is true for the precocious expression of maturation transcripts and the patterning phenotypes observed in dcl1 embryos, although SPL10 and SPL11 have been experimentally linked to defects in both patterning and maturation gene expression.
One of the earliest perceptible roles of plant miRNAs is to repress transcription factors (Fig. 2C). We propose that, by repressing differentiation-promoting transcription factors (including SPL10, SPL11, ARF17, CNA, PHB, PHV, and TCP4), miRNAs maintain the potential of preglobular cells to generate diverse cell types at the subsequent globular stages. Since the reduction of WOX2 transcripts in dcl1 eight-cell embryos was the first detectable differentiation defect, and homeobox genes related to WOX2 have been implicated in maintaining stem cells during post-embryonic development (Laux et al. 1996; Mayer et al. 1998; Wu et al. 2005), we speculate that premature expression of miRNA targets leads to the reduction of WOX2 transcripts in dcl1 embryos and ultimately contributes to precocious differentiation and subsequent loss of developmental potential in preglobular cell types.
The two transcription factor genes that were most derepressed in eight-cell dcl1 embryos were miR156 targets SPL10 and SPL11, for which transcripts were up-regulated ≥150-fold (≥600-fold if only the homozygous mutant embryos contributed to the increase). These two genes were redundantly required for the embryonic patterning defects observed in dcl1 embryos (Table 1; Fig. 4A,D). The ability to suppress such a pleiotropic phenotype by knocking out/down only two miRNA targets was surprising when considering that dozens of miRNA targets were derepressed in dcl1 embryos. However, miR156-resistant SPL10 and SPL11 transgenes did not phenocopy dcl1 embryos, which suggests that the misregulation of additional targets contributes to the patterning defects observed. Consistent with this idea, at least seven miRNA target transcripts in addition to SPL10 and SPL11 have increased levels as early as the eight-cell stage, and all of these encode transcription factors.
Of these seven misregulated targets, five (ARF17, CNA, PHB, PHV, and TCP4) have reported embryonic functions after the preglobular stage (Palatnik et al. 2003; Mallory et al. 2005; Prigge et al. 2005). miR166-mediated regulation of PHB and PHV has also been reported to be important during early embryonic patterning (Grigg et al. 2009). However, phb phv double mutants do not suppress dcl1-null embryonic phenotypes (Grigg et al. 2009), suggesting that the misregulation of other miRNA targets is required for the dcl1 embryonic phenotypes. Perhaps the loss of both miR156-mediated regulation of SPL transcripts and miR166-mediated regulation of HD-ZIPIII transcripts in dcl1 embryos is responsible for the embryonic patterning phenotypes observed. Future characterization of how multiple miRNA/target interactions facilitate embryonic cell differentiation will contribute to a better understanding of how the basic plant body plan is established during embryo morphogenesis.
We also report previously uncharacterized roles of miR156-mediated SPL gene repression during embryogenesis. After morphogenesis, seed plant embryos transition to a maturation phase, when they accumulate storage proteins, undergo desiccation tolerance, and prepare to enter into a state of dormancy prior to germination. We found that miR156-mediated regulation of SPL10 and SPL11 prevents the transcription factor products of these genes from prematurely inducing seed maturation genes before the embryo has formed. To the extent that this regulation forestalls the morphogenesis-to-maturation-phase transition, this newly identified role for miR156-mediated repression of SPL genes resembles that observed for vegetative-, reproductive-, and meristem identity-phase transitions during post-embryonic development (Wu and Poethig 2006; Gandikota et al. 2007; Schwarz et al. 2008; Wang et al. 2009; Wu et al. 2009; Yamaguchi et al. 2009). This role for SPL10 and SPL11 repression might be tied to the requirement of these targets for the patterning defects observed in dcl1 embryos. Perhaps the premature induction of maturation-phase genes arrests morphogenesis before it is complete.
Later in development, the miR156-mediated repression of SPL10 and SPL11 must presumably be overcome in order for the embryo to undergo the morphogenesis-to-maturation transition. Although miR156 levels do not appear to decrease at later stages of morphogenesis (Fig. 3A), SPL10 and SPL11 promoter activities do increase at the same stages that the morphogenesis-to-maturation-phase transition occurs (Fig. 5). Therefore, at later stages of morphogenesis, miR156 may establish a threshold that SPL10 and SPL11 transcript levels must surpass in order to accumulate and promote maturation-phase gene expression programs. Despite the characterization of hormonal, metabolic, and genetic factors involved in embryo maturation (Nambara and Marion-Poll 2003; Weber et al. 2005; Braybrook and Harada 2008), the regulatory mechanisms that control the morphogenesis-to-maturation-phase transition are still not well understood. Multiple factors may influence SPL10 and SPL11 transcript levels and ultimately regulate the timing of the morphogenesis-to-maturation-phase transition. Further characterization of the embryonic functions of SPL transcription factors may provide a handle to acquire a better molecular understanding of the morphogenesis-to-maturation-phase transition.
Materials and methods
Growth conditions and genetic analyses
The dcl1-5 and dcl1-10 alleles were generated by McElver et al. (2001) and obtained from the Arabidopsis Biological Resource Center (ABRC). DR5rev∷GFP lines were obtained from the Nottingham Arabidopsis Stock Center (Scholl et al. 2000). Plants were grown at 20°C–24°C in a growth room with a 16-h light/8-h dark cycle.
Generation of transgenic lines
SPL10p∷SPL10-GFP constructs were generated by cloning genomic fragments, including 1.7 kb upstream of and including the SPL10 coding sequence (CDS), into pENTR/D-TOPO (Invitrogen), and then recombining these plasmids with Gateway-compatible pBIB-KAN-GWR-GFP plasmids using LR clonase (Invitrogen). The primers used in this study are listed in Supplemental Table 2.
Control sensor constructs were created by cloning the potato UBI3 promoter into pENTR/D-TOPO (Invitrogen). The resulting plasmids were then recombined with pCGTAG. A similar procedure was used to create miR156 sensor constructs, except that miR156 target sites were introduced into the construct by PCR.
SPL10p∷NLS-GFP and SPL11p∷NLS-GFP constructs were generated by PCR amplification of 1.9-kb and 2.2-kb genomic DNA, respectively. The resulting amplicons were then cloned into pCR8/GW-TOPO (Invitrogen) and recombined with pCGTAG.
6mSPL10 and 6mSPL11 constructs were created by cloning 4.9-kb (including 1.8 kb upstream of CDS, CDS, and 1.6 kb downstream from CDS) and 5.5-kb (including 2.7 kb upstream of CDS, CDS, and 1.3 kb downstream from CDS) genomic fragments in pENTR/D-TOPO (Invitrogen), respectively. The QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene) was used on the resulting plasmids to disrupt miR156-binding sites, and both nonmutagenized and mutagenized plasmids were recombined with pBIB-KAN-GW using LR clonase (Invitrogen).
SPL10-RNAi constructs were generated by amplifying 0.6 kb of the SPL10 CDS from cDNA prepared from inflorescence tissue total RNA isolated using TRIzol reagent (Invitrogen) and reverse-transcribed with oligo(dT) primers using SuperScript III Reverse Transcriptase (Invitrogen). The resulting amplicons were then cloned into pCR8/GW-TOPO (Invitrogen) and then recombined with UBI3p∷pFSPGW-HYGv02 to create an inverted repeat of the SPL10 cDNA fragment under the control of the UBI3 promoter. The UBI3p∷pFSPGW-HYGv02 vector was created by first adding AatII and XhoI sites on the 5′ and 3′ ends of the UBI3 promoter by PCR, cloning the amplicons into pENTR/D-TOPO (Invitrogen) to create UBI3F2R2∷pENTR, double-digesting UBI3F2R2∷pENTR and pFSPGW with AatII and XhoI, and ligating together the appropriate digestion fragments to create UBI3p∷pFSPGW. A hygromycin resistance cassette was then amplified from pCGTAG with primers that introduced Bsu36I and Acc65I sites on the 5′ and 3′ ends of the amplicons, respectively, and these were cloned into pCR8/GW-TOPO to create HYGF3R3∷pCR8. HYGF3R3∷pCR8 and UBI3p∷pFSPGW were then double-digested with Bsu36I and Acc65I and ligated together to create UBI3p∷pFSPGW-HYGv02.
All constructs were transformed into Col-0 and dcl1-5/+ plants via Agrobacterium-mediated transformation (Clough and Bent 1998). More than 20 embryos from at least five independent transgenic lines were examined for each construct.
Microscopy
Ovules were fixed and cleared as described previously (Ohad et al. 1996), and embryos were examined using Nomarski optics on a Nikon E800 upright microscope for morphological analysis. Images were collected with a Hamamatsu C4742-95 CCD camera and viewed with Openlab acquisition software (PerkinElmer).
For CSLM analysis, embryos were dissected in pH 7.2 potassium phosphate buffer, stained with 1 μg/mL FM4-64, and mounted in 5% glycerol. A 488-nm laser on a Zeiss LSM 510 confocal microscope was used to excite GFP, YFP, and FM4-64, and images were collected at 505–530 nm, 500–550 nm, and 644–719 nm, respectively.
RNA in situ hybridizations
WOX2, WOX8, RPS5A, eIF4A1, and PDF1 probes were generated by introducing T7 promoters with PCR and using the procedure outlined by Hejatko et al. (2006). miR156 LNA oligonucleotides were ordered directly from Integrated DNA Technologies. ATML1, PNH, and SCR probe generation; fixing, embedding, and sectioning of siliques; hybridization; washes; and immunological detection were performed as described previously (Nodine et al. 2007) except for miR156 LNA in situs, where 10 pmol of probe was used in each hybridization reaction.
mRNA sequencing and analysis
For RNA isolation, pools of 30 Col-0 (wild-type) and dcl1-5 early globular embryos were hand-dissected in water, immediately transferred to 30 μL of RNAlater (Ambion), incubated at 60°C with 500 μL of TRIzol reagent (Invitrogen) for 30 min, and then purified according to the TRIzol reagent protocol for RNA isolation from small quantities of tissue (Invitrogen). Two rounds of linear amplification of poly(A) RNA were performed using the Arcturus RiboAmp HS Plus kit (Molecular Devices). Strand-specific mRNA-Seq libraries were generated as described previously (Guo et al. 2010). After removing adapter sequences, sequences were mapped to the A. thaliana genome (TAIR9 assembly) using the Bowtie short-read aligner (Langmead et al. 2009), allowing up to two mismatches within a 25-nt “seed” sequence and retaining reads mapping uniquely to the genome. Wild-type and dcl1 libraries yielded 5.6 million and 4.5 million reads that uniquely matched the Arabidopsis genome, respectively. Since the Arcuturus RiboAmp HS Plus kit generates amplified RNAs in the antisense orientation, reads were counted as matching TAIR9 annotated genes if they overlapped the antisense strand. Approximately 90% of genome-matching reads (5,061,561 for wild-type and 3,973,657 for dcl1-5) matched annotated genes of the TAIR9 genome release (Swarbreck et al. 2008). To normalize for differences in library size, the number of reads matching each gene was divided by the total number of million genome-matching reads for each library. We then considered the number of normalized reads that matched miRNA targets listed in the Arabidopsis Small RNA Project (ASRP) database (Gustafson et al. 2005), comparing the read numbers in wild-type and dcl1-5 early globular embryo libraries (Supplemental Fig. 3). For comparisons between the set of genes up-regulated in dcl1-5 relative to wild-type embryos and the set of genes up-regulated in mature relative to globular embryos, we generated a control gene set. The control gene set was generated by first placing the genes significantly up-regulated at least twofold in dcl1-5 early globular embryos into 10 approximately equally sized bins based on expression levels. Genes expressed in Col-0 early globular embryos were then sorted into the established bins, and were randomly chosen uniformly from these bins to correct for associations between gene expression levels and the tendency for a gene to be up-regulated in either dcl1-5 early globular embryos or wild-type mature embryos. The mRNA-Seq data sets generated in this study have been deposited in NCBI GEO.
qRT–PCR analysis
Total RNA from pools of at least 20 embryos was isolated and linearly amplified as described above, and 100 ng of amplified RNA was used in reverse transcription reactions using Random Primers (Invitrogen) and SuperScript III Reverse Transcriptase (Invitrogen). Gene-specific and eIF4A1 (endogenous control) primers were used in the real-time PCR using a 7900HT Fast Real-Time PCR system from Applied Biosystems. ΔΔCt values were calculated for each gene, and relative transcript levels were derived from these values. For transcripts that were not detectable in wild type, test samples (from selfed dcl1-5/+ plants) were serially diluted with wild-type samples in order to determine the maximum cycle number within the exponential phase of the PCR for which fluorescence could be detected. The corresponding ΔCt value was then used to calculate the relative transcript levels in the test samples.
Acknowledgments
We thank J. Long and C. Zhang for providing the UBI3 promoter fragment and WOX5p∷NLS-YFP construct, respectively, and the Whitehead Genome Technology Core for sequencing. This work used the W.M. Keck Biological Imaging Facility at the Whitehead Institute and was supported by NIH grant GM067031 (to D.P.B.) and NIH Post-doctoral Fellowship F32GM084656 (to M.D.N). D.P.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1986710.
Supplemental material is available at http://www.genesdev.org.
References
- Abe M, Takahashi T, Komeda Y 1999. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol 40: 571–580 [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ 2008. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M 1997. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ 2008. LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW 1993. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet 241: 504–514 [DOI] [PubMed] [Google Scholar]
- Chen X 2009. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M 2005. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D 1991. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Carrington JC 2010. miRNA target prediction in plants. Methods Mol Biol 592: 51–57 [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL 2007. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- Frandsen GI, Mundy J, Tzen JT 2001. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112: 301–307 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P 2007. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. 2008. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Galinha C, Kornet N, Canales C, Scheres B, Tsiantis M 2009. Repression of apical homeobox genes is required for embryonic root development in Arabidopsis. Curr Biol 19: 1485–1490 [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD 2005. ASRP: The Arabidopsis small RNA project database. Nucleic Acids Res 33: D637–D640 doi: 10.1093/nar/gki127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C 2007. Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hejatko J, Blilou I, Brewer PB, Friml J, Scheres B, Benkova E 2006. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ 2008. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B 2006. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322 [DOI] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP 2005. MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci 102: 9412–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Laux T, Wurschum T, Breuninger H 2004. Genetic regulation of embryonic pattern formation. Plant Cell 16: S190–S202 doi: 10.1105/tpc.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J 2006. SERRATE: A new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Porat R, Nadeau JA, O'Neill SD 1996. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK 1999. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B 2004. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Misera S, Jurgens G 1991. Mutations affecting body organization in the Arabidopsis embryo. Nature 353: 402–407 [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A 2003. ABA action and interactions in seeds. Trends Plant Sci 8: 213–217 [DOI] [PubMed] [Google Scholar]
- Nodine MD, Yadegari R, Tax FE 2007. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev Cell 12: 943–956 [DOI] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL 1996. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci 93: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD 2007. Essential role for Dicer during skeletal muscle development. Dev Biol 311: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Harada JJ 2008. Arabidopsis embryogenesis. Methods Mol Biol 427: 3–16 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP 2002. MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP 2002. Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A 2002. FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A 2002. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH 2000. Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW 1994. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245 [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P 2008. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce T, Pernaute B, Di-Gregorio A, Cobb BS, Merkenschlager M, Manzanares M, Rodriguez TA 2010. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev Cell 19: 207–219 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. 2008. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 doi: 10.1093/nar/gkm965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD 2003. A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U 2005. Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jurgens G 2006. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]
- Wu G, Poethig RS 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D 2005. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15: 436–440 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D 2009. The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]