Abstract
Splicing regulatory networks are essential components of eukaryotic gene expression programs, yet little is known about how they are integrated with transcriptional regulatory networks into coherent gene expression programs. Here we define the MER1 splicing regulatory network and examine its role in the gene expression program during meiosis in budding yeast. Mer1p splicing factor promotes splicing of just four pre-mRNAs. All four Mer1p-responsive genes also require Nam8p for splicing activation by Mer1p; however, other genes require Nam8p but not Mer1p, exposing an overlapping meiotic splicing network controlled by Nam8p. MER1 mRNA and three of the four Mer1p substrate pre-mRNAs are induced by the transcriptional regulator Ume6p. This unusual arrangement delays expression of Mer1p-responsive genes relative to other genes under Ume6p control. Products of Mer1p-responsive genes are required for initiating and completing recombination and for activation of Ndt80p, the activator of the transcriptional network required for subsequent steps in the program. Thus, the MER1 splicing regulatory network mediates the dependent relationship between the UME6 and NDT80 transcriptional regulatory networks in the meiotic gene expression program. This study reveals how splicing regulatory networks can be interlaced with transcriptional regulatory networks in eukaryotic gene expression programs.
Keywords: Regulated splicing, regulons, splicing-sensitive microarray, epistasis
Cell identities and functional states arise from distinctive sets of expressed genes. Transitions from one state to another are achieved through activation of gene expression programs that lead to stable changes in the set of expressed genes. Programs are composed of regulatory networks, or regulons (Ben-Tabou de-Leon and Davidson 2007), that ensure coordinated expression of required groups of genes. Defining gene regulatory networks and obtaining insight into their relationships with each other is essential for understanding any developmental program.
Much work in this area has focused on transcription factors and the signaling pathways that activate them to promote coordinate transcription of groups of genes in a defined transcriptional regulon. Splicing regulatory networks may function in a parallel manner whereby splicing factors activate the coordinate splicing of specific transcripts, leading to changes in protein function important to progression of the gene expression program. A widely known cascade of splicing regulation occurs during sex determination in Drosophila, where Sex lethal (Sxl) promotes the productive splicing of transformer (tra) pre-mRNA. Tra protein (with Tra-2) then controls whether the male (no Tra) or the female (with Tra) form of the doublesex transcription factor is produced (Baker 1989; Lopez 1998; Black 2003). With the exception of this one example, little is known about how splicing and transcriptional regulators might control each other in complex programs of eukaryotic gene expression.
Meiosis in the budding yeast Saccharomyces cerevisiae is accompanied by a well-studied developmental gene expression program associated with transcriptional regulons (Chu et al. 1998; Primig et al. 2000). The program includes a transcriptional cascade that can be separated into at least three components: early meiotic genes regulated by Ume6p/Ime1p (Strich et al. 1994; Williams et al. 2002), middle meiotic genes activated by Ndt80p (Xu et al. 1995; Chu and Herskowitz 1998; Hepworth et al. 1998), and late meiotic genes (Mitchell 1994; Kassir et al. 2003). As meiotic events such as chromosome synapsis and recombination take place, checkpoints mediated by phosphorylation of regulatory kinases ensure event completion and allow progression through meiosis (Hochwagen and Amon 2006). In the absence of progress, checkpoint activation causes a delay in the transcriptional program to coordinate meiotic cellular events with gene expression.
In addition to transcription, splicing is regulated during meiosis in yeast. Best understood is the activation of a small set of introns by the KH domain RNA-binding protein Mer1p (Nandabalan and Roeder 1995; Spingola and Ares 2000). MER1 was first identified genetically by its contribution to spore viability, meiotic recombination, and synaptonemal complex (SC) formation (Engebrecht and Roeder 1989, 1990; Engebrecht et al. 1990), but turned out to be a splicing factor (Engebrecht et al. 1991). Its expression is induced during meiosis (Engebrecht and Roeder 1990) to activate the splicing of MER2/REC107 (Engebrecht et al. 1991), MER3/HFM1 (Nakagawa and Ogawa 1999), and SPO70/AMA1 (Cooper et al. 2000; Davis et al. 2000) through an interaction with a conserved intronic enhancer sequence (5′-AYACCCYU-3′) (Spingola and Ares 2000). NAM8/MRE2, a component of the U1 snRNP, contributes to 5′ splice site recognition (Gottschalk et al. 1998; Puig et al. 1999) and is required for meiosis (Nakagawa and Ogawa 1997), in part through its role in splicing activation of Mer1p-responsive transcripts (Spingola and Ares 2000). Consistent with this, Mer1p also binds to the U1 snRNP (Spingola and Ares 2000) and its interactions with other spliceosome components have been enumerated (Spingola and Ares 2000; Spingola et al. 2004; Balzer and Henry 2008), but its mechanism of action remains unclear.
Despite increasing ability to define splicing regulatory networks (Ule et al. 2003; Zhang et al. 2008; Du et al. 2010), little is known about how transcriptional regulation is coordinated with splicing regulation and other cellular events in eukaryotic gene expression programs. In this study, we address two intimately connected problems. First, we want to understand how the Mer1p splicing regulatory network is connected to the transcriptional regulatory networks that operate in meiosis. Second, we want to understand the specific contributions of the genes in the Mer1p splicing network to the progress of meiosis and the meiotic gene expression program. Using splicing-sensitive microarrays, we compared splicing and mRNA levels in wild-type and mer1Δ cells after initiation of the meiotic program. In addition to observing inhibited splicing of the three known Mer1p-activated introns, we identified only one additional gene (SPO22/ZIP4) whose splicing is inhibited in mer1Δ cells. Surprisingly, both MER1 and three of its four targets are under the control of Ume6p, the activator of the early meiotic genes (Strich et al. 1994; Steber and Esposito 1995; Williams et al. 2002). Proper function of Mer1p is necessary (through its contributions to the expression of Mer1p-responsive genes) for full activation of Ndt80p, the activator of the middle meiotic genes (Hepworth et al. 1998; Tung et al. 2000), suggesting a model in which the MER1 splicing regulon bridges two major transcriptional regulons during meiosis.
Results
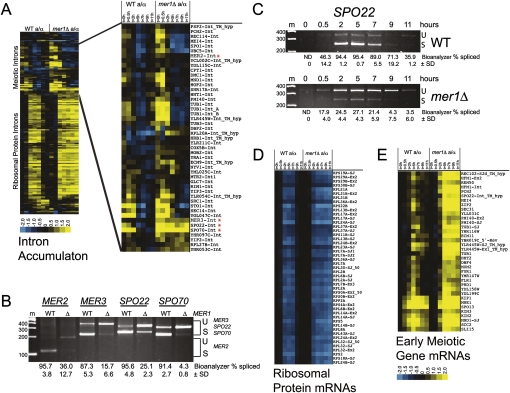
Deletion of MER1 inhibits splicing of four introns in the yeast genome
Mer1p is required for splicing of three pre-mRNAs (Engebrecht et al. 1991; Nakagawa and Ogawa 1999; Davis et al. 2000), but it is unclear how many more Mer1p-responsive introns might be lurking in the yeast genome. To observe the contribution of MER1 to the meiotic gene expression program, we compared the global changes in mRNA levels and splicing during meiosis in synchronized wild-type yeast (SK1) to those of isogenic mer1Δ yeast using whole-genome splicing-sensitive microarrays (Fig. 1). As judged by their intron accumulation indexes (IAI) (see the Materials and Methods), only four meiotic genes (Fig. 1A, asterisks) show reduced splicing efficiency in mer1Δ as compared with wild-type cells. This is confirmed by RT–PCR (Fig. 1B) using RNA from the 5-h meiotic time point. The splicing efficiency of MER2/REC107, MER3/HFM1, SPO22/ZIP4, and SPO70/AMA1 is substantially reduced in the absence of MER1. MER2, MER3, and SPO70 pre-mRNAs are known to require Mer1p (Engebrecht et al. 1991; Nakagawa and Ogawa 1999; Davis et al. 2000), and here we show that SPO22/ZIP4 pre-mRNA splicing efficiency also depends on Mer1p (Fig. 1C). The SPO22 intron sequence contains a Mer1p intronic enhancer sequence (5′-AUACCCUU-3′) that closely matches the consensus 5′-AYACCCUY-3′ (Spingola and Ares 2000) 21 nucleotides downstream from the noncanonical 5′ splice site (GUAUAU instead of the canonical GUAUGU). We also tested several meiotically expressed intron-containing genes that appeared to have reasonable matches to the Mer1p enhancer near their 5′ splice sites using RT–PCR and found that none appeared to depend on Mer1p (Supplemental Table S1). Because we cannot strictly exclude the possibility that another Mer1p-responsive intron remains undetected in the genome, we tentatively conclude that the four known Mer1p-responsive genes identified thus far constitute the complete MER1 splicing regulatory network.
Figure 1.
Meiotic gene expression in the absence of the Mer1p splicing factor. (A) Splicing changes as represented by intron accumulation indexes (Clark et al. 2002) during the time course of wild-type (left panel) and mer1Δ (right panel) meiosis. The asterisk (*) indicates introns whose splicing efficiency during meiosis is reduced in mer1Δ cells compared with wild type. Yellow represents an increase in the intron accumulation index, and thus a decrease in splicing efficiency. Blue represents an increase in splicing efficiency. (B) RT–PCR validation of MER2, MER3, SPO22, and SPO70 splicing efficiencies in wild-type (WT) and mer1Δ yeast 5 h after induction of meiosis. (C) Expression and splicing of SPO22 mRNA during meiosis in wild-type (WT) and mer1Δ strains. (D) RP gene expression during meiosis in wild-type (left panel) and mer1Δ (right panel) cells. Blue represents decrease in expression. (E) Expression of early meiotic genes in wild-type (left panel) and mer1Δ (right) cells. Yellow represents increase in expression. For B and C, “U” indicates unspliced pre-mRNA and “S” indicates spliced mRNA. Marker sizes are in base pairs. Splicing efficiency was calculated as described in the Materials and Methods.
Late meiotic gene expression is delayed by deletion of MER1
Mer1p is a splicing factor; thus, the direct effect of loss of MER1 is the inhibition of efficient splicing of Mer1p enhancer containing pre-mRNAs. Although Mer1p could have yet-unknown functions, most downstream (indirect) effects of loss of MER1 would presumably be due to compromised expression of the four Mer1p-responsive transcripts. To determine the indirect effects that loss of MER1 has on the meiotic gene expression program, we compared total gene expression profiles of wild-type SK1 cells and isogenic mer1Δ cells during meiosis. The major differences in mRNA expression profiles affect the genes in two classes: the ribosomal protein (RP) transcripts (Fig. 1D) and the meiotic transcripts (Fig. 1E). Both the transcriptional repression of RP transcripts (Fig. 1D) and the transcriptional induction of early meiotic genes (Fig. 1E) remain unperturbed in cells lacking MER1 compared with wild type. However, a block to progression through meiosis in mer1Δ cells is evident by 9 h based on RP and meiotic gene expression profiles. RP gene expression fails to be activated by 9 h in mer1Δ cells (Fig. 1D, right panel). In addition, meiosis-specific transcripts remain high in late meiosis in the mer1Δ cells relative to wild type (Fig. 1E, right panel). We conclude that deletion of MER1 affects the meiotic gene expression program by causing a delay in the reduction of meiotic transcript levels as well as a failure to activate RP expression in late meiosis. Because Mer1p is a splicing factor that promotes splicing of just four genes (Fig. 1A,B), we infer that the global delay in the gene expression program arises as an indirect consequence of failure to express adequately one or more of the Mer1p-responsive genes.
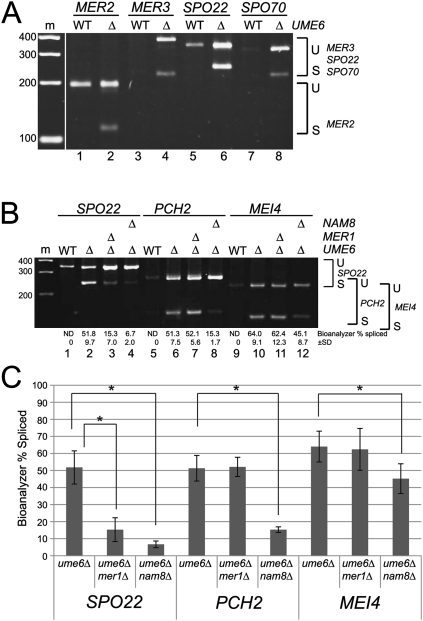
MER1 and three of four Mer1p-responsive genes are activated by Ume6p
Nutrient signals trigger the expression of early meiotic genes that convert the Ume6p transcription factor from its repressor form in vegetative cells to an activator of early meiotic genes (Mitchell 1994). The expression of MER1 and its responsive genes (Fig. 1A) increases during the early wave of transcription. To determine whether Ume6p activates MER1 and its responsive genes, we searched their promoters for the Ume6p-binding site (URS1) (Buckingham et al. 1990; Strich et al. 1994; Steber and Esposito 1995). We found the URS1 in the promoters of MER1, MER3, SPO22, and SPO70, but not MER2 (Supplemental Fig. S1; Harbison et al. 2004). Consistent with this, MER2 is not repressed during vegetative growth; its pre-mRNA is spliced only during meiosis when Mer1p is present (Engebrecht et al. 1991). Deletion of UME6 in vegetative cells leads to derepression of meiotic genes during vegetative growth (Strich et al. 1994). We exploited this fact to test the role of Ume6p in expression of the MER1 regulon as well as other meiotic intron-containing genes (Fig. 2; Table 1). A splicing-sensitive microarray experiment comparing vegetatively growing ume6Δ cells to wild type confirms derepression of SPO22 (Williams et al. 2002) and also reveals new Ume6p-activated genes, MER1 and SPO70 (Table 1, shaded). To validate the array results, and to test MER3 (for which array signals were not robust), we performed RT–PCR using RNA from ume6Δ and wild-type strains (Fig. 2A). MER3, SPO22, and SPO70 transcription is derepressed in ume6Δ vegetative cells. Spliced transcripts from these genes as well as MER2 are greatly increased in ume6Δ vegetative cells (Fig. 2A, lanes 2,4,6,8), indicating expression of MER1. Thus, we conclude that transcription of MER1 and three of the four Mer1p-responsive genes is repressed by Ume6p in vegetative cells and is activated by Ume6p during meiosis. This means that the MER1 splicing regulatory network is largely under the control of the Ume6p transcription factor.
Figure 2.
Derepression of meiotic genes in vegetative cells reveals splicing factor requirement for meiosis. (A) Expression and splicing of MER2, MER3, SPO22, and SPO70 in wild-type (WT) and ume6Δ vegetative cells. (B) Expression and splicing of SPO22, PCH2, and MEI4 in wild-type (WT), ume6Δ, mer1Δume6Δ, and nam8Δume6Δ vegetative cells. (C) Splicing factor dependence for efficient splicing of SPO22, PCH2, and MEI4. The asterisk (*) indicates a statistically significant difference in splicing efficiency (α = 0.05) using a t-test (see the Materials and Methods). (U) Unspliced pre-mRNA; (S) spliced mRNA. Marker sizes are in base pairs. Splicing efficiency was calculated as described in the Materials and Methods.
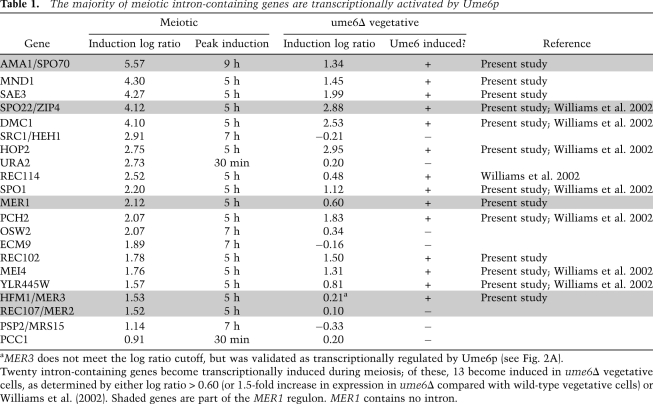
Table 1.
The majority of meiotic intron-containing genes are transcriptionally activated by Ume6p
aMER3 does not meet the log ratio cutoff, but was validated as transcriptionally regulated by Ume6p (see Fig. 2A).
Twenty intron-containing genes become transcriptionally induced during meiosis; of these, 13 become induced in ume6Δ vegetative cells, as determined by either log ratio > 0.60 (or 1.5-fold increase in expression in ume6Δ compared with wild-type vegetative cells) or Williams et al. (2002). Shaded genes are part of the MER1 regulon. MER1 contains no intron.
An overlapping meiotic splicing regulon is controlled by Nam8p
The three previously identified Mer1p-responsive pre-mRNAs require both Mer1p and the U1snRNP protein Nam8p for splicing activation (Spingola and Ares 2000). To test whether splicing activation of SPO22 also requires NAM8, we used vegetative ume6Δ cells containing or lacking either MER1 or NAM8, and measured SPO22 splicing efficiency (Fig. 2B, lanes 1–4). Splicing efficiency of SPO22 is significantly reduced (Fig. 2C) in both mer1Δume6Δ and nam8Δume6Δ cells, indicating that, like the other Mer1p-responsive genes, SPO22 splicing activation requires both NAM8 and MER1.
We also tested other Ume6p-activated meiotic intron-containing genes using the mer1Δume6Δ and nam8Δume6Δ strains. We found that splicing efficiency of PCH2 (Fig. 2B [lanes 5–8], C) is strongly dependent on NAM8, as is MEI4, albeit to a lesser but still statistically significant degree (Fig. 2B [lanes 9–12], C). Neither intron is affected by loss of MER1, because they lack the Mer1p enhancer. Transcriptional control of NAM8 is distinct from that of MER1, since NAM8 is expressed in both vegetative and meiotic cells and is not under Ume6p control (Ekwall et al. 1992). We conclude that a second meiotic splicing regulatory network is controlled by NAM8, and that this network overlaps with the Mer1p network but includes splicing events that do not require Mer1p.
A previous report described 13 meiosis-specific intron-containing genes based on tiling arrays (Juneau et al. 2007). We found additional genes whose expression is up-regulated during meiosis, and determined which of these are under Ume6p repression in vegetative cells (Table 1). The array experiment confirmed seven out of eight previously identified Ume6p-activated genes (Williams et al. 2002), including SPO22, and identified three new meiotic intron-containing genes regulated by Ume6p (MND1, REC102, and SAE3) (Table 1). Together with this new recognition that MER3 and SPO70 are under Ume6p control, we counted a total of 13 of 20 meiosis-induced intron-containing genes regulated by Ume6p.
Expression of the Mer1p-responsive genes is delayed relative to other Ume6p-activated genes
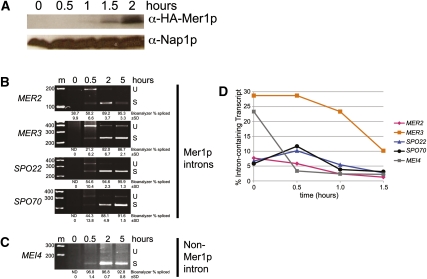
Induction of expression of Mer1p by Ume6p simultaneously with its responsive pre-mRNAs seems unusual, since the time needed for Mer1p translation would produce a delay in splicing and expression of the responsive genes. If true, for a period of time after Ume6p induction, Mer1p-responsive pre-mRNAs should accumulate while Mer1p protein is being produced. We measured Mer1p induction early in meiosis, and were first able to detect Mer1p 1 h after transfer to sporulation medium, increasing up to 2 h after induction of meiosis (Fig. 3A). Efficient Mer1p-dependent splicing was observed 2 h after transfer to sporulation medium, while unspliced transcripts were detected within 30 min (Fig. 3B). Furthermore, MER2, which is not under Ume6p control and is transcribed during both vegetative growth and meiosis (Engebrecht et al. 1991), exhibited a similar delay in splicing efficiency. A splicing delay was not evident for other intron-containing transcripts not under Mer1p control, such as MEI4 (Fig. 3C). To examine this more closely, we performed RT–qPCR on RNA isolated at 30-min intervals after transfer to sporulation medium (Fig. 3D). Each of the Mer1p-responsive genes displayed a higher percentage of intron-containing transcript 30 min after onset of meiosis as compared with 1 h, when Mer1p first became evident. This experiment revealed a splicing-dependent timing mechanism that separates expression of genes induced by a common transcription factor into two temporal components: those immediately expressed, and those delayed by the time necessary to translate sufficient splicing factor. This suggests that one contribution of the MER1 splicing regulatory network to the gene expression program might be to promote appropriate timing of expression of a subset of meiotic genes.
Figure 3.
Accumulation of mRNA for Mer1p-responsive genes is delayed relative to other Ume6p-activated genes. (A) Western blot measuring Mer1p expression in wild-type cells early in meiosis. Nap1p was used as a loading control. (B) Measurement of expression and splicing of the Mer1p-responsive MER2, MER3, SPO22, and SPO70 genes in wild-type cells at the indicated times after induction of meiosis. (C) Measurement of expression and splicing of the Mer1p-independent gene MEI4 in wild-type cells at the indicated times after induction of meiosis. (D) Measurement of percent of intron-containing transcript (calculated as described in the Materials and Methods) as determined by RT–qPCR of MER2, MER3, SPO22, SPO70, and MEI4 in wild-type cells at the indicated times after induction of meiosis. (U) Unspliced pre-mRNA; (S) spliced mRNA. Marker sizes are in base pairs. Splicing efficiency was calculated as described in the Materials and Methods.
Deletion of MER3 and SPO22 delays NDT80 transcriptional induction
After expression of Ume6p-induced genes, transcription of a second meiotic wave was triggered (Chu et al. 1998; Primig et al. 2000). This wave is regulated by the transcription factor Ndt80p and allows expression of middle meiotic genes, leading to exit from pachytene and entry into Meiosis I (Xu et al. 1995; Chu and Herskowitz 1998). Because the MER1 splicing regulon is expressed as a consequence of the UME6 transcriptional regulon, we wanted to ask how expression of the MER1 regulon contributes (directly or indirectly) to the succeeding cellular events and the progress of the gene expression program. Functions of all four Mer1p-responsive genes have been studied, but their contributions to the meiotic gene expression program are unknown. Three of the four Mer1p-responsive gene products function during meiotic prophase. Mer2p/Rec107p is required for formation of double-stranded breaks (DSBs) to initiate recombination (Keeney 2001; Li et al. 2006); loss of MER2 allows a rapid aberrant meiosis that bypasses the recombination pathway (Malone et al. 2004). Mer3p/Hfm1p is a recombination-specific DNA helicase (Nakagawa and Ogawa 1999; Nakagawa and Kolodner 2002; Mazina et al. 2004); in the absence of MER3, cells arrest in prophase due to the inability to resolve DSB intermediates (Nakagawa and Ogawa 1999). Spo22p/Zip4p promotes SC formation (Tsubouchi et al. 2006; Lynn et al. 2007); strains mutant for SPO22 exhibit delayed progression through meiosis due to the defect in SC formation (Tsubouchi et al. 2006). The fourth gene product, Spo70p/Ama1p, is a meiosis-specific anaphase-promoting complex (APC) regulatory subunit that functions during chromosome segregation and spore formation (Oelschlaegel et al. 2005; Penkner et al. 2005; Diamond et al. 2009); spo70Δ cells arrest with segregated chromosomes but without spore formation (Rabitsch et al. 2001). Although Spo70p is absolutely required for spore formation (Rabitsch et al. 2001; Coluccio et al. 2004), its function in chromosome segregation is redundant with other APC regulatory subunits (Oelschlaegel et al. 2005; Penkner et al. 2005).
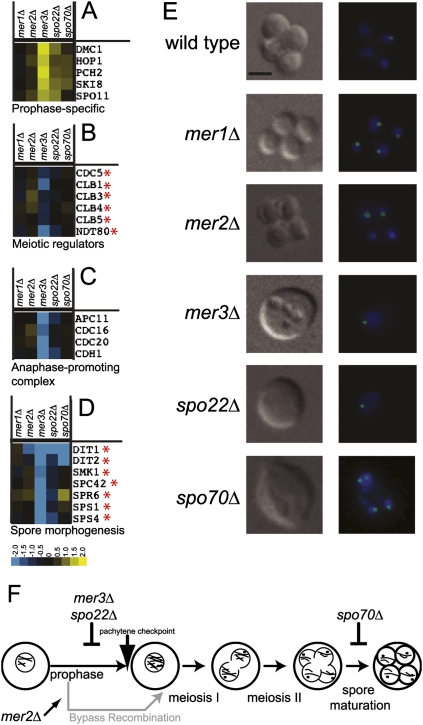
The phenotype of the mer1Δ strain is complex because loss of MER1 results in the simultaneous reduction of expression of MER2, MER3, SPO22, and SPO70, each of which has distinct meiotic functions. To address this, we obtained deletions of each gene and separately assessed the contribution of each to the meiotic gene expression program by measuring mRNA levels of each mutant relative to wild type at 9 h after transfer to sporulation medium. Prophase-specific genes (Fig. 4A) have increased expression relative to wild type in each of the mutants, especially mer3Δ and spo22Δ, indicating blocked or delayed reduction of the Ume6p-activated transcripts in these strains. Furthermore, expression of NDT80, the transcriptional activator of the middle genes (Fig. 4B), as well as important middle genes such as the B-type cyclins (CLB1, CLB3, CLB4, and CLB5) (Chu and Herskowitz 1998) and polo-like kinase CDC5 (Clyne et al. 2003), is decreased in mer3Δ and spo22Δ cells (Fig. 4B). Other genes that function following the NDT80 transcriptional wave (Chu et al. 1998), such as those required for active APC (Fig. 4C) or those involved in spore morphogenesis (Fig. 4D), display lower levels of gene expression compared with wild type in mer3Δ and spo22Δ strains. Consistent with Mer3p and Spo22p function in prophase, strains lacking these proteins do not enter the meiotic divisions and arrest before chromosome segregation at the pachytene checkpoint (Fig. 4E,F; for review, see Hochwagen and Amon 2006). Expression of genes required for completion of spore formation, such as DIT1 and DIT2 (Briza et al. 1994; Coluccio et al. 2004), is strongly reduced in the spo70Δ strain (Fig. 4D; see also Coluccio et al. 2004), indicating a delay or block in late gene expression. This block must occur after segregation but before spore formation, since spo70Δ cells arrest in meiosis with segregated chromosomes but no spores (Fig. 4E,F; see also Rabitsch et al. 2001; Coluccio et al. 2004).
Figure 4.
MER1 regulon expression is required for induction of NDT80 and Ndt80p-regulated genes. (A) Expression of genes whose products function during prophase in mer1Δ, mer2Δ, mer3Δ, spo22Δ, and spo70Δ compared with wild type 9 h after induction of meiosis. (B) Same as A for genes whose products are regulators of the meiotic divisions. (C) Same as A for genes whose products function in the APC. (D) Same as A for genes whose products function in spore morphogenesis. Yellow represents increase in expression, while blue represents decrease in expression relative to wild type. The asterisk (*) indicates genes shown by Chu and Herskowitz (1998) or Clyne et al. (2003) to be regulated by Ndt80p. (E) Sample pictures of major phenotypes of wild-type, mer1Δ, mer2Δ, mer3Δ, spo22Δ, and spo70Δ cells 9 h after induction of meiosis. On the left are differential interference contrast micrographs, and on the right are superimposed fluorescence micrographs of DNA stained with DAPI (blue) and CenV-GFP (green). Bar, 2 μm. (F) Diagram of meiotic events and execution points of Mer1p-responsive gene deletions.
Deletion of MER2 does not block meiotic progression, but an aberrant meiosis takes place in which no DSBs form and aneuploid spores are produced at high frequency (Engebrecht et al. 1990; Cool and Malone 1992; Malone et al. 2004). We counted cell phenotypes in the mer1Δ strain at 9 h and found the majority (70.3%, 147 of 209) of mer1Δ cells resemble the mer2Δ phenotype and complete the meiotic gene expression program. A detectable fraction of mer1Δ cells arrests at positions similar to the arrest points of mer3Δ and spo22Δ (prophase) (11.0%, 23 of 209) or spo70Δ (segregated chromosomes but no spores) (18.7%, 39 of 209), suggesting that the phenotype of individual mer1Δ tetrads is influenced by stochastic events, such as whether a threshold level of Mer2p is produced through leaky splicing (Fig. 1B). Decreased expression of NDT80 and Ndt80p-regulated genes in mer3Δ and spo22Δ cells shows that the gene expression program is halted in the absence of sufficient Mer3p or Spo22p. We conclude that the MER1 splicing regulon is interposed between the UME6 and NDT80 transcriptional regulons.
Loss of Mer1p generates heterotypic effects on meiotic progression that are resolved by epistasis
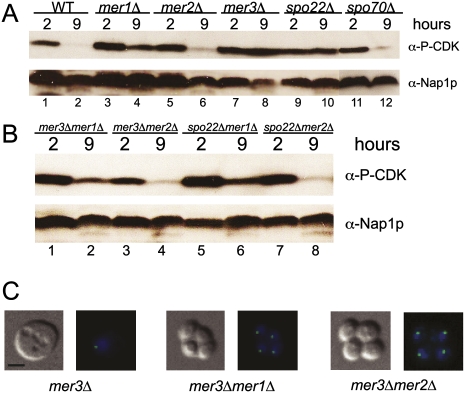
Loss of Mer1p splicing factor leads to reduced expression of genes whose loss produces heterotypic block points in meiosis (Fig. 4). For example, reduced levels of either Mer2p or Spo70p would not be expected to trigger the pachytene checkpoint, whereas reduced levels of Mer3p or Spo22p would. To confirm this and evaluate checkpoint activation in the mer1Δ strain, we assayed the activation state of CDK (Cdc28p) by detecting inhibitory phosphorylation at Y19 (Leu and Roeder 1999) using a phospho-specific antibody. We observe strong, persistent Cdc28p phosphorylation at Y19 late in meiosis in mer3Δ and spo22Δ strains, and, to a lesser extent, in mer1Δ (Fig. 5A, lanes 4,8,10). In wild-type, mer2Δ, and spo70Δ strains, CDK is mostly unphosphorylated by 9 h into meiosis, indicating that these cells progress past pachytene (Fig. 5A, lanes 2,6,12). Presumably, the partial activation of the pachytene checkpoint in the mer1Δ strain is due to residual splicing of Mer1p-responsive transcripts in the absence of Mer1p (Fig. 1B; Engebrecht et al. 1991; Davis et al. 2000; Spingola and Ares 2000), eventually allowing slow progression past the checkpoint and explaining the delayed gene expression program. This partial activation of the checkpoint likely occurs in subpopulations of mer1Δ cells that lack adequate Mer3p or Spo22p but produce sufficient Mer2p to initiate DSBs. Other subpopulations that produce inadequate Mer2p would immediately bypass the checkpoint because DSBs would not form in those cells.
Figure 5.
Pachytene checkpoint activation persists in mer3Δ and spo22Δ, and, to a lesser degree, in the mer1Δ strain. (A) Western blot measuring phosphorylation state of CDK on Y19 in wild-type (WT), mer1Δ, mer2Δ, mer3Δ, spo22Δ, and spo70Δ strains 2 h and 9 h after induction of meiosis. (B) Same as A using mer3Δmer1Δ, mer3Δmer2Δ, spo22Δmer1Δ, and spo22Δmer2Δ strains. Nap1p was used as a loading control in A and B. (C) Sample pictures of major phenotypes of mer3Δ, mer3Δmer1Δ, and mer3Δmer2Δ strains at 9 h after induction of meiosis. On the left are differential interference contrast micrographs, and on the right are superimposed fluorescence micrographs of DNA stained with DAPI (blue) and CenV-GFP (green). Bar, 2 μm.
To test this idea, we constructed double deletions within the MER1 regulon to assess epistasis (Fig. 5B). Double mutants of mer3Δ or spo22Δ with mer1Δ showed reduced activation of the checkpoint (Fig. 5B, lanes 2,6), the same as the mer1Δ single mutant (Fig. 5A, lane 4), rather than the strong checkpoint activation observed in the mer3Δ or spo22Δ single mutants. Double mutants of mer3Δ or spo22Δ with mer2Δ showed little or no detectable pachytene checkpoint activation (Fig. 5B, lanes 4,8), the same as the mer2Δ single mutant (Fig. 5A, lane 6). The meiotic arrest points of the mer1Δ strain resemble the mer2Δ mutant (Fig. 4E). Likewise, the arrest points of the mer3Δmer1Δ and mer3Δmer2Δ resemble the mer2Δ single mutant, rather than mer3Δ (Fig. 5C). These results indicate that both mer1Δ and mer2Δ are epistatic to mer3Δ and spo22Δ with respect to pachytene checkpoint activation. Thus, loss of Mer1p leads primarily to meiotic events that arise as a consequence of limited expression of Mer2p. Furthermore, this experiment shows that the successful expression of Mer3p and Spo22p is monitored by the pachytene checkpoint, ensuring that the activity of the MER1 regulon leads to NDT80 expression.
Discussion
In this study, we define the MER1 splicing regulatory network as consisting of Mer1p splicing factor and Mer1p-responsive pre-mRNA transcripts from four genes: MER2/REC107, MER3/HFM1, SPO22/ZIP4, and SPO70/AMA1 (Fig. 1). Deletion of MER1 reduces splicing efficiency of these four pre-mRNAs and causes a cascade of defects in the transcriptional program, including prolonged high levels of Ume6p-activated gene transcripts and a delay in induction of middle and late gene transcripts. Surprisingly, MER1 and all but one of its responsive genes are under the control of the Ume6p transcription factor (Fig. 2). This arrangement divides Ume6p-controlled genes into two waves, one of which (including the Mer1p-responsive genes) is delayed in mRNA expression by the amount of time necessary to accumulate Mer1p after Ume6p-mediated activation (Fig. 3). Function of the MER1 splicing regulon is necessary, in turn, for the expression of the NDT80 transcriptional regulon (Fig. 4). Specifically, compromising expression of either of two Mer1p-responsive genes, MER3 and SPO22, blocks NDT80 expression (Fig. 4) and triggers the activation of the pachytene checkpoint (Fig. 5), resulting in prophase arrest. Although loss of MER1 reduces expression of all four genes, it appears that the consequent loss of Mer2p in the mer1Δ mutant accounts for much of the phenotype (Fig. 4). We show that the MER1 splicing regulon is primarily under the control of one transcription factor (Ume6p) and is required for the activation of another (Ndt80p), and thus bridges two key transcriptional regulons during the meiotic gene expression program (Fig. 6).
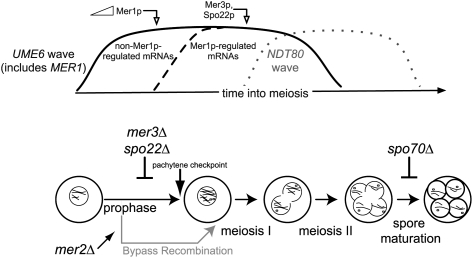
Figure 6.
The MER1 regulatory network and the meiotic gene expression program. The UME6 expression wave is divided into two temporal components: an early component including non-Mer1p-regulated mRNAs (including Mer1p mRNA itself), and a later component including the mRNAs whose splicing is dependent on Mer1p. Loss of expression of either of two Mer1p-responsive genes (MER3 and SPO22) arrests the cells at the pachytene checkpoint, which must be passed in order for induction of the subsequent NDT80 expression wave to proceed. However, loss of Mer1p-responsive gene MER2 bypasses recombination and the pachytene checkpoint due to the absence of DSBs. This allows completion of meiosis, even in the absence of MER3 or SPO22. Loss of Mer1p-responsive gene SPO70 arrests the cells after chromosome segregation but before spore formation, later in the meiotic gene expression program. Note that the timing of peak RNA expression precedes the execution points for several of the proteins, presumably due to regulatory events at other levels.
How does splicing regulation contribute to meiotic gene expression?
Although only ∼300 yeast genes have introns, the presence of introns is strongly associated with gene functional class. After cytoplasmic RP genes (103 introns in 100 genes), the largest functional class of yeast intron-containing genes are meiotically induced genes, most of which are under Ume6p transcriptional control (13 of 20 meiotic intron-containing genes) (Table 1). One explanation for this might be that introns help keep meiotic genes from being expressed in vegetative cells, adding an additional layer of protection in the event of incomplete transcriptional repression (Juneau et al. 2007). Two findings suggest that such effects may be more subtle or only enforced on evolutionary time scales. First, ume6Δ cells grow reasonably well, given the loss of nonmeiotic functions of Ume6p, while actively transcribing and splicing early meiotic introns (Fig. 2). Second, only one meiosis-specific splicing factor has been found (Mer1p) (Engebrecht et al. 1991), and it activates the splicing of only four pre-mRNAs (Fig. 1; Engebrecht et al. 1991; Nakagawa and Ogawa 1999; Davis et al. 2000). It is possible that Mer2p and Spo22p represent the key regulatory subunits of their respective protein complexes, and that the function of these complexes is critically dependent on Mer1p-activated expression of MER2 and SPO22, but additional experiments would be required to demonstrate this.
We favor the hypothesis that introns in meiotic genes allow for temporal expression regulation during meiosis. As cells shift into the meiotic gene expression program, there appears to be a general increase in splicing efficiency for meiotic genes (Fig. 1; Juneau et al. 2007) as well as for intron-containing genes that are expressed in both vegetative growth and meiosis (Fig 1). The mechanism of this increase in splicing efficiency during meiosis is unknown, but must be independent of MER1 function, since the residual splicing of Mer1p-responsive genes observed in the absence of MER1 also increases at this time (Fig. 1).
What is the specific value of the MER1 regulon to the timing of the meiotic gene expression program? By inducing transcription of MER1 and its responsive genes with the same transcriptional regulator, the cell creates a timed delay in expression of all Mer1p-responsive genes relative to other Ume6p-induced genes. This is distinct from the NAM8 splicing regulon, which includes Ume6p-induced PCH2 and MEI4 (Table 1). NAM8 is transcribed during both vegetative growth and meiosis and is not regulated by Ume6p (Ekwall et al. 1992). Although Nam8p function is essential for meiosis (Nakagawa and Ogawa 1997), the NAM8 splicing network produces no delay in expression of PCH2 or MEI4 (Fig. 3). The special nature of the MER1 splicing regulon divides the expression of coinduced genes into two components: an early wave that is independent of the splicing factor, and a delayed wave that is dependent on the splicing factor. Thus, the existence of a splicing regulatory network can contribute to the coordination of gene expression in time by creating secondary waves of splicing-dependent expression within large waves of transcriptional regulation (Fig. 6).
Transcriptional regulons are interlaced with splicing regulons
The meiotic gene expression program requires both transcriptional (UME6 and NDT80) and splicing (MER1) networks for progression, as failed expression of these program regulators blocks meiosis (Engebrecht and Roeder 1990; Steber and Esposito 1995; Xu et al. 1995). We asked how the splicing regulatory network is integrated with each transcriptional regulatory network. Using genomics and genetics, we found that the UME6 transcriptional network activates the expression of the MER1 splicing network, which in turn is required for activation of the subsequent NDT80 transcriptional network.
An intriguing characteristic of the MER1 regulon is that it has evolved a complex relationship with the NDT80 transcriptional regulon. Loss of Mer1p expression does not completely block NDT80 induction or inhibit meiotic progress in the same way in every cell. Splicing of Mer1p-responsive transcripts occurs, but is much less efficient in mer1Δ cells, allowing some mRNA from each of the four Mer1p-responsive genes to be made, presumably resulting in partially inadequate levels of Mer2p, Mer3p, Spo22p, and Spo70p. Depending on stochastic events, these proteins may be limiting in different cells attempting the meiotic program. If Mer2p is limiting, no DSBs will be made, and thus neither Mer3p nor Spo22p will be required, leading to recombination bypass, NDT80 induction, and mostly successful chromosome segregation (except that spore viability suffers due to increased nondisjunction in the absence of recombination) (Roeder 1997).
In those cells where Mer2p is not limiting, DSBs are formed, but limiting amounts of Mer3p or Spo22p (or both) result in delays at the pachytene checkpoint and delayed NDT80 induction (Tung et al. 2000) until adequate levels of the missing protein can accumulate to pass the checkpoint. This explains the leaky, mixed phenotype of mer1Δ tetrads, and insinuates splicing regulation into both the initiation and resolution steps of recombination, the key checkpoint-regulated step in meiosis. The evolutionary importance of this is underscored by the limited number of genes in yeast that still require splicing—not to mention regulated splicing—for their expression, but why it is important seems obscure. Nonetheless, the nature and function of the Mer1p-responsive genes ensure that correct regulated splicing must occur for NDT80 induction and efficient, accurate meiosis to take place.
Implications
Developmental programs progress through tightly coordinated gene regulatory networks. Completely defining a gene regulatory network in complex systems is challenging, since the main experimental approach involves determining the effect of loss of function of the master regulator. Such experiments produce complex phenotypes comprised of direct effects and a cascade of indirect effects that must be distinguished. Even for the well-studied Drosophila sex determination pathway, in which expression of the master regulator Sxl ultimately leads to a male or female form of the transcription factor Dsx (Baker 1989; Lopez 1998; Black 2003), we cannot begin to explain the integration of observed sex-specific transcription and splicing (Robida et al. 2007; Telonis-Scott et al. 2009). Part of this is due to Sxl regulation of translation as well as splicing (Penalva and Sanchez 2003), and another part is due to incomplete understanding of the sets of genes that respond to Tra and Dsx, and what the effects of those might be on sex-specific transcription and splicing.
Our study shows that, even for the relatively simple MER1 splicing regulatory network, such downstream effects can be at cross-purposes and difficult to dissect. Sorting true responsive genes from indirectly activated genes will require comparison of large sets of perturbations, as well as the identification of sequence features that mediate action of the master regulator(s). Finally, more effort is needed to relate transcription and splicing regulatory networks to each other. Discerning higher-level dependence relationships will help identify and attribute many secondary events to specific primary events. We will need to know which transcription factors regulate the expression of which splicing factor genes, which splicing factors regulate expression of which other splicing factors, and how alternative splicing of transcription factor mRNAs affect transcription factor function.
Materials and methods
Strains
A complete list of strains is in Supplemental Table S2. Briefly, all vegetatively grown haploid strains were derived from the yeast deletion set background (Winzeler et al. 1999). All experiments involving meiosis used the high-meiotic synchrony strains with the SK1 background (Primig et al. 2000). Diploid single-mutant SK1 strains were constructed by cassette-based gene replacement, followed by sporulation and verification of the deletion by PCR. Diploid double-mutant SK1 strains were constructed by cassette-based gene replacement in the heterozygous knockout of each single deletion, followed by sporulation and verification of the double deletion by PCR. HA3-MER1 was constructed by N-terminally tagging MER1 under its native promoter with three copies of the HA epitope (HA3) marked by TRP1 (Wach et al. 1997; Longtine et al. 1998). All derivatives in this study harbor a tet operator array near the centromere of chromosome V and express the tet repressor-GFP fusion protein to allow for fluorescence detection of chromosome V segregation (Michaelis et al. 1997).
Media, culture conditions, and induction of meiosis
Standard methods for yeast culture were used (Sherman 1991) at 30°C. SK1 cells were induced for synchronous meiosis as described in Padmore et al. (1991). Briefly, cells were streaked from frozen stocks to YP-glycerol (3%) plates, and then single colonies from glycerol were streaked to YPD plates. After 2 d on YPD, a single colony was inoculated into 5 mL of YPD, and, 30 h later, 50 mL of YPA (1% potassium acetate, 1% yeast extract, 2% peptone) was inoculated to OD600 = 0.25 and shaken for 14 h. After growth in YPA, cells were washed with water and suspended in SPM (1% potassium acetate, 0.02% raffinose), defined as time 0 of meiosis. Aliquots were taken at 30 min, 2 h, 5 h, 7 h, 9 h, and 11 h for RNA or protein preparation; spun down at room temperature; flash-frozen in liquid nitrogen; and stored at −80°C.
RNA isolation
RNA was isolated as described in Rio et al. (2010). Total meiotic RNA was extracted according to method 2 to ensure uniform RNA extraction from late spore stages. Total vegetative RNA was prepared according to method 1.
Microarray analysis
Microarray analysis was done as described previously (Clark et al. 2002; Burckin et al. 2005). Our arrays are printed in-house and contain ∼20,000 spots containing oligonucleotides for all yeast genes (in duplicate) and intron, splice junction, and second exon probes for all intron-containing genes (in quadruplicate) (Burckin et al. 2005). Data from four independent meiotic time courses were combined as follows. Each time point from each replicate meiotic time course was hybridized to a pair of dye-swapped arrays using an arbitrary reference pool of RNA comprised of 50% time 0 RNA plus 10% each of time 2 h, 5 h, 7 h, 9 h, and 11 h. After normalization and removal of outlier arrays, the data was zero-subtracted (to eliminate variation derived from the reference pools) and averaged. To evaluate splicing changes, we used the IAI, which is derived by subtracting the log ratios of the second exon signals from the intron signals in order to normalize for changes in transcript level on a gene-by-gene basis (Clark et al. 2002). The data presented in Table 1 came from an experiment in which RNA from the ume6Δ strain was compared with wild type grown in YPD, and represent the average of a dye-swapped pair of arrays. For the experiment comparing the 9-h time expression pattern of deletion of each member gene in the MER1 regulon (Fig. 4), we compared 9-h RNA from each mutant to 9-h RNA from wild-type cells, again as the average of dye-swapped pairs. To produce the images in Figures 1 and 4, we used Gene Cluster 3.0 (de Hoon et al. 2004) and Java Treeview (Saldanha 2004). Array data was released through the Gene Expression Omnibus under accession number GSE24686.
RT–PCR and qPCR
RNA was extracted from at least three biological replicates. Reverse-transcribed RNA (cDNA) was amplified using the primers in Supplemental Table S3. Semiquantitative RT–PCR was carried out by limiting cycle numbers to 20 and using cDNA derived from 300 ng of total RNA. PCR products were first analyzed by agarose gel electrophoresis. To obtain estimates of splicing efficiency, we used the Agilent 2100 Bioanalyzer to determine molar amounts of each PCR product and estimated splicing efficiency as follows: percent spliced = [(molarity of spliced peak)/(molarity of unspliced peak + molarity of spliced peak)] * 100. Bioanalyzer percent spliced values from triplicate biological replicates were averaged and the standard deviations are shown. To test for significant differences in splicing between samples (as in Fig. 2C), a paired, two-tailed t-test was performed and P < 0.05 was considered significant. qPCR was performed using a commercially available master mix (Fermentas) and qPCR primers described in Supplemental Table S3. The graph shown in Figure 3D is a measure of percent of intron-containing RNA from 0 h to 1.5 h every 30 min after the onset of meiosis. This analysis used two primer sets for each gene: one pair for intron-containing pre-mRNA (spanning the 3′ splice site) and one set for total RNA (within the second exon). Primer pair amplification efficiencies were confirmed to be >1.95. Threshold cycles were determined using reactions containing the same amount of cDNA and the percent of intron-containing 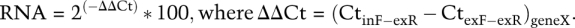
Western blotting
Frozen cell pellet aliquots from the 0-h, 0.5-h, 1-h, and 1.5-h time points (Fig. 3A) or from the 2-h and 9-h time points (Fig. 5) were prepared as in Rudner et al. (2000). After electrophoresis on SDS-containing 15% acrylamide gels, samples were transferred to nitrocellulose membrane. The blot in Figure 3A was blocked in 3% milk in PBST buffer containing 387 mM NaCl total and was incubated overnight at 4°C in blocking buffer containing 1:1000 α-HA.11 monoclonal antibody (Covance). The blots in Figure 5 were blocked in TBST containing 5% BSA and incubated overnight at 4°C in blocking buffer containing 1:1000 α-phospho-cdc2 (Tyr 15) (Cell Signaling Technology) for Cdc28p-Y19 detection, visualized (see below), then stripped and reprobed overnight with 1:2000 α-Nap1 (affinity-purified rabbit polyclonal raised against Nap1p; a gift from Doug Kellogg, University of California at Santa Cruz) as a loading control. Primary antibody was detected with HRP-conjugated sheep anti-mouse secondary antibody (for HA detection) (GE Healthcare) or donkey anti-rabbit secondary antibody (for phospho CDK and Nap1p detection) (Santa Cruz Biotechnology) and was visualized with ECL Plus (GE Healthcare).
Microscopy
Aliquots (100 μL) from the 9-h time point were fixed with formaldehyde for 1 h at room temperature. Samples were DAPI-stained and visualized with a Leica DM5500 microscope (Leica Microsystems) using DIC, as well as GFP and DAPI channels.
Acknowledgments
We thank Rhonda Perriman for critical reading of the manuscript, Doug Kellogg and Stacy Harvey for generosity with antibodies and Western blotting materials, Gloria Brar (University of California at San Francisco) and Needhi Bhalla (University of California at Santa Cruz) for insightful meiosis advice, and Dr. Yu-Chen Hwang (University of California at Santa Cruz Life Sciences Microscopy Facilities) for technical support with microscopy. M.A. and H.I. thank Tracy Johnson (University of California at San Diego) for her hospitality during a sabbatical year, when part of this work was done. This work was supported primarily by GM040478 from the National Institutes of Health to M.A. L.S. was supported by GM084317. E.M. and K.D. were supported by National Institutes of Health Training Grant T32 GM008646. L.T. was supported by R25 GM5890.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1977410.
Supplemental material is available at http://www.genesdev.org.
References
- Baker BS 1989. Sex in flies: The splice of life. Nature 340: 521–524 [DOI] [PubMed] [Google Scholar]
- Balzer RJ, Henry MF 2008. Snu56p is required for Mer1p-activated meiotic splicing. Mol Cell Biol 28: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH 2007. Gene regulation: Gene control network in development. Annu Rev Biophys Biomol Struct 36: 191–212 [DOI] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Briza P, Eckerstorfer M, Breitenbach M 1994. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci 91: 4524–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham LE, Wang HT, Elder RT, McCarroll RM, Slater MR, Esposito RE 1990. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci 87: 9406–9410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M Jr 2005. Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol 12: 175–182 [DOI] [PubMed] [Google Scholar]
- Chu S, Herskowitz I 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1: 685–696 [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell 3: 1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool M, Malone RE 1992. Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol Cell Biol 12: 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci 97: 14548–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Grate L, Spingola M, Ares M Jr 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res 28: 1700–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol Biol Cell 20: 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol 17: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Kermorgant M, Dujardin G, Groudinsky O, Slonimski PP 1992. The NAM8 gene in Saccharomyces cerevisiae encodes a protein with putative RNA binding motifs and acts as a suppressor of mitochondrial splicing deficiencies when overexpressed. Mol Gen Genet 233: 136–144 [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Roeder GS 1989. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics 121: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Roeder GS 1990. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol 10: 2379–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Hirsch J, Roeder GS 1990. Meiotic gene conversion and crossing over: Their relationship to each other and to chromosome synapsis and segregation. Cell 62: 927–937 [DOI] [PubMed] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS 1991. Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Seraphin B, Rosbash M, Luhrmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393 [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 18: 5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Amon A 2006. Checking your breaks: Surveillance mechanisms of meiotic recombination. Curr Biol 16: R217–R228 doi: 10.1016/j.cub.2006.03.009 [DOI] [PubMed] [Google Scholar]
- Juneau K, Palm C, Miranda M, Davis RW 2007. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci 104: 1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G 2003. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol 224: 111–171 [DOI] [PubMed] [Google Scholar]
- Keeney S 2001. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Leu JY, Roeder GS 1999. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol Cell 4: 805–814 [DOI] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS 2006. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lopez AJ 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu Rev Genet 32: 279–305 [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV 2007. ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15: 591–605 [DOI] [PubMed] [Google Scholar]
- Malone RE, Haring SJ, Foreman KE, Pansegrau ML, Smith SM, Houdek DR, Carpp L, Shah B, Lee KE 2004. The signal from the initiation of meiotic recombination to the first division of meiosis. Eukaryot Cell 3: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117: 47–56 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Mitchell AP 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev 58: 56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kolodner RD 2002. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol Cell Biol 22: 3281–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H 1997. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells 2: 65–79 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J 18: 5714–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandabalan K, Roeder GS 1995. Binding of a cell-type-specific RNA splicing factor to its target regulatory sequence. Mol Cell Biol 15: 1953–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W 2005. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120: 773–788 [DOI] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256 [DOI] [PubMed] [Google Scholar]
- Penalva LO, Sanchez L 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev 67: 343–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner AM, Prinz S, Ferscha S, Klein F 2005. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell 120: 789–801 [DOI] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE 2000. The core meiotic transcriptome in budding yeasts. Nat Genet 26: 415–423 [DOI] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Seraphin B 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev 13: 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol 11: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M, Hannon GJ, Nilsen TW 2010. RNA: A laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Robida MD, Rahn A, Singh R 2007. Genome-wide identification of alternatively spliced mRNA targets of specific RNA-binding proteins. PLoS ONE 2: e520 doi: 10.1371/journal.pone.0000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS 1997. Meiotic chromosomes: It takes two to tango. Genes Dev 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Rudner AD, Hardwick KG, Murray AW 2000. Cdc28 activates exit from mitosis in budding yeast. J Cell Biol 149: 1361–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—Extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Sherman F 1991. Getting started with yeast. In Guide to yeast genetics and molecular biology. (ed. Guthrie C, Fink GR), pp. 3–21 Academic Press, San Diego, CA [Google Scholar]
- Spingola M, Ares M Jr 2000. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell 6: 329–338 [DOI] [PubMed] [Google Scholar]
- Spingola M, Armisen J, Ares M Jr 2004. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res 32: 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Esposito RE 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc Natl Acad Sci 92: 12490–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev 8: 796–810 [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Kopp A, Wayne ML, Nuzhdin SV, McIntyre LM 2009. Sex-specific splicing in Drosophila: Widespread occurrence, tissue specificity and evolutionary conservation. Genetics 181: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS 2006. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell 10: 809–819 [DOI] [PubMed] [Google Scholar]
- Tung KS, Hong EJ, Roeder GS 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc Natl Acad Sci 97: 12187–12192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Williams RM, Primig M, Washburn BK, Winzeler EA, Bellis M, Sarrauste de Menthiere C, Davis RW, Esposito RE 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci 99: 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15: 6572–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]