Abstract
Nucleotide synthesis is a universal response to DNA damage, but how this response facilitates DNA repair and cell survival is unclear. Here we establish a role for DNA damage-induced nucleotide synthesis in homologous recombination (HR) repair in fission yeast. Using a genetic screen, we found the Ddb1–Cul4Cdt2 ubiquitin ligase complex and ribonucleotide reductase (RNR) to be required for HR repair of a DNA double-strand break (DSB). The Ddb1–Cul4Cdt2 ubiquitin ligase complex is required for degradation of Spd1, an inhibitor of RNR in fission yeast. Accordingly, deleting spd1+ suppressed the DNA damage sensitivity and the reduced HR efficiency associated with loss of ddb1+ or cdt2+. Furthermore, we demonstrate a role for nucleotide synthesis in postsynaptic gap filling of resected ssDNA ends during HR repair. Finally, we define a role for Rad3 (ATR) in nucleotide synthesis and HR through increasing Cdt2 nuclear levels in response to DNA damage. Our findings support a model in which break-induced Rad3 and Ddb1–Cul4Cdt2 ubiquitin ligase-dependent Spd1 degradation and RNR activation promotes postsynaptic ssDNA gap filling during HR repair.
Keywords: Rad3, Ddb1–Cul4Cdt2 ubiquitin ligase, Spd1, ribonucleotide reductase, homologous recombination repair, fission yeast
DNA double-strand breaks (DSBs) are potentially lethal lesions that, if left undetected or repaired incorrectly, can threaten the integrity of the genome. DSBs arise at a low frequency during normal cell metabolism and can also arise from exposure to DNA-damaging agents such as ionizing radiation (IR) (Shrivastav et al. 2008), potentially leading to chromosomal rearrangements, cancer, or cell death (Pfeiffer et al. 2000). Consequently, an intricate network of cellular responses for detection and accurate repair of such lesions exists within the cell (Jackson and Bartek 2009).
Cells have evolved two distinct repair pathways to maintain genome integrity following a DSB: nonhomologous end-joining (NHEJ), in which DNA ends are directly ligated, and homologous recombination (HR) (Shrivastav et al. 2008). In yeast, HR involves the RAD52 epistasis group (Krogh and Symington 2004) and uses a homologous sequence as a template for repair—typically the sister chromatid or, less frequently, the homologous chromosome (Kadyk and Hartwell 1992). Repair is initiated by 5′–3′ resection of the broken ends to form a 3′ ssDNA overhang. This is a two-step process in which MRX/MRN (Mre11–Rad50–Xrs2 in Saccharomyces cerevisiae [Sc]; Mre11–Rad50–Nbs1 in Schizosaccharomyces pombe [Sp]) and Sae2Sc/Ctp1Sp initiate resection, while Exo1 together with Sgs1Sc and Dna2Sc facilitates extensive resection (Mimitou and Symington 2008; Zhu et al. 2008). Replication protein A (RPA) binds the ssDNA before being displaced by Rad51Sc/Rhp51Sp, creating a nucleoprotein filament in a process requiring Rad52Sc/Rad22Sp and the heterodimer Rad55Sc–Rad57Sc (Rhp55Sp–Rhp57Sp) (Sung 1997; Sugawara et al. 2003; Wolner et al. 2003; Haruta et al. 2008). Stabilization of the nucleoprotein filament is achieved through interaction between Rad51 and Rad54; strand invasion is then initiated, resulting in the formation of a displacement (D) loop (Petukhova et al. 1998; Mazin et al. 2003; Sugawara et al. 2003). RPA further functions postsynaptically to stabilize DNA pairing and possibly the displaced ssDNA (Eggler et al. 2002; Wang and Haber 2004). HR can then proceed through a choice of genetically distinct subpathways: During synthesis-dependent strand annealing, the invading strand is expelled following DNA synthesis and reanneals to the second end. Alternatively, the second broken end can anneal to the D-loop following DNA synthesis, resulting in the formation of a double-Holliday junction structure, which can be resolved with or without crossovers in a process involving GEN1/Yen1Sc, Mus81Sc–Mms4Sc (Mus81Sp–Eme1Sp), and SLX1–SLX4Hs/Sc resolvases or dissolved through the activities of Sgs1–Top3 (for review, see Svendsen and Harper 2010). It is anticipated that dNTPs will be required for efficient HR to facilitate both DNA synthesis within the D-loop and gap filling synthesis following strand annealing prior to ligation.
The DNA damage checkpoint is activated in response to DSBs and acts to both facilitate DSB repair and delay the cell cycle, thus preventing cell cycle progression prior to completion of repair (Lazzaro et al. 2009). Central to the DNA damage checkpoint are the phosphatidylinositol 3′ kinase-like kinases ATM (Tel1Sc/Sp) and ATR (Rad3Sp/Mec1Sc), which localize to DNA damage sites and act as the initial sensor kinases (Abraham 2001). Following damage detection in fission yeast, the signal is transduced through Rad4Sp (TOPBP1) and Crb2Sp (53BP1) to the effector kinase Chk1Sp/Sc, which in turn acts to delay the cell cycle and promote repair (Carr 2002). Thus, the DNA damage checkpoint pathway plays a crucial role in maintaining genome stability in response to a DSB.
Synthesis of dNTPs, catalyzed by ribonucleotide reductase (RNR), is required for both DNA replication and DNA repair (Elledge et al. 1992; Chabes et al. 2003). RNR is a heterotetramer composed of two large catalytic subunits (termed R1) encoded by cdc22+ in fission yeast, and two small regulatory subunits (R2) encoded by suc22+. Tight regulation of dNTP pools is essential, as excessive levels can cause genetic mutation (Bebenek et al. 1992) and insufficient levels can affect genome stability (Chabes et al. 2003; Holmberg et al. 2005; Mathews 2006). Consequently, multiple mechanisms for regulation of RNR activity exist within the cell. Allosteric regulation is achieved in a positive manner by ATP and negatively by dATP (Nordlund and Reichard 2006); RNR protein levels are also regulated via transcription (Elledge et al. 1993). Additionally in both S. cerevisiae and S. pombe, regulation by subcellular compartmentalization effectively restrains RNR activity outside of S phase and in the absence of DNA damage (Liu et al. 2003; Yao et al. 2003). This compartmentalization in S. pombe is achieved in part by Spd1, a small inhibitor protein. Preparation for DNA synthesis or the presence of DNA damage results in degradation of Spd1 and relocalization of the small subunits of RNR from the nucleus to the cytoplasm, whereby, through interaction with the large subunits, active RNR is constituted (Liu et al. 2003; Håkansson et al. 2006). Degradation of Spd1 is achieved through the activity of a Ddb1–Cul4Cdt2 ubiquitin ligase complex consisting of components of the COP9 signalosome complex (CSN), a Cullin-4 ubiquitin ligase (Pcu4), the Ddb1 protein, and the adapter protein Cdt2 (Liu et al. 2003, 2005; Holmberg et al. 2005). Activation of this complex is achieved through fluctuations in Cdt2 protein levels during the cell cycle, peaking at S phase and reducing following DNA synthesis (Rustici et al. 2004; Liu et al. 2005). Additionally, DNA damage in G2 cells induces Cdt2 levels in a manner that is dependent on the DNA damage checkpoint (Watson et al. 2004; Liu et al. 2005).
Here we identify roles for the Ddb1–Cul4Cdt2 ubiquitin ligase complex and RNR in HR. Our data support a model in which Rad3-dependent activation of the Ddb1–Cul4Cdt2 ubiquitin ligase complex promotes HR through degradation of Spd1 in response to DNA damage. Such RNR-induced nucleotide synthesis facilitates efficient postsynaptic ssDNA gap filling following resection during HR.
Results
Ddb1 and Cdt2 are required for minichromosome maintenance following a DSB
To screen for genes required to maintain genome stability following a DSB, a colony sectoring assay was adapted to allow rapid visualization of mutants defective in repair and maintenance of a broken nonessential minichromosome (Ch16) following site-specific DSB induction. We previously adapted Ch16, an experimentally derived nonessential minichromosome (Niwa et al. 1986), to screen for suppressors of break-induced loss of heterozygosity (LOH) (Tinline-Purvis et al. 2009). This minichromosome was further adapted here to carry the HO endonuclease gene on the left arm to form Ch16-LMYAU (Fig. 1A). Following removal of thiamine (T) from the media, the HO endonuclease is expressed, inducing a unique site-specific DSB at the MATa recognition site located on the right arm. This newly created minichromosome was introduced into a subset of 205 mutants from the S. pombe Bioneer haploid deletion library (Kim et al. 2010) exhibiting sensitivity to the alkylating agent methylmethane sulfonate (MMS) and/or the radiomimetic bleomycin (Deshpande et al. 2009; our unpublished results). Ch16 encodes an ade6-M216 point mutation that, when present with an ade6-M210 heteroallele on ChIII, results in an ade+ (white) phenotype through intragenic complementation (Leupold and Gutz 1964). Mutants were thus assayed for those that exhibited increased loss of the minichromosome ade6-M216 heteroallele located centromere-distal from the MATa site following break induction, resulting in ade− cells that could be detected as red sectors within colonies on plates containing low levels of adenine (Materials and Methods).
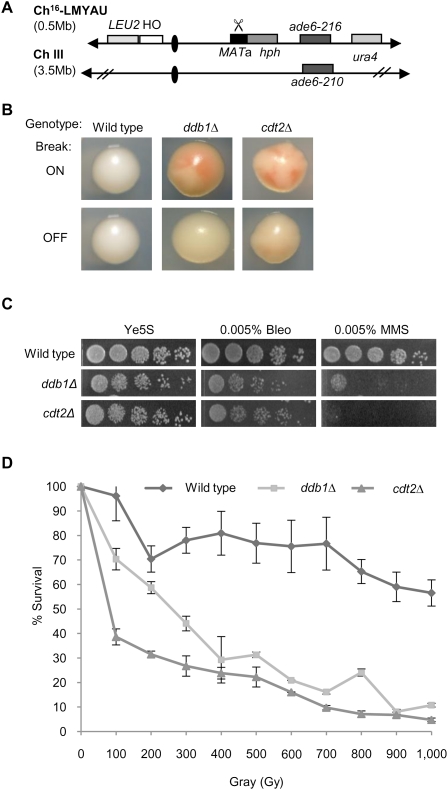
Figure 1.
Ddb1 and Cdt2 are required for break-induced minichromosome maintenance and resistance to DNA-damaging agents. (A) Schematic of the minichromosome Ch16-LMYAU. Ch16-LMYAU, a 530-kb fragment of ChIII, contains the MATa site with an adjacent hygromycin resistance marker gene (hph) and the ade6-M216 ade heteroallele, complemented by ade6-M210, present on ChIII, as described previously (Tinline-Purvis et al. 2009). Ch16-LMYAU additionally carries a ura4 marker ∼50 kb centromere-distal to ade6-M216 at the cid2 locus. The HO endonuclease gene, under control of nmt promoter, with adjacent LEU2 marker is integrated into SPCC1795.09 on the left arm of the minichromosome. Derepression of HO endonuclease (by removal of thiamine) generates a DSB at the MATa target site (indicated by scissors). (B) Colony coloration of wild-type, ddb1Δ, and cdt2Δ colonies grown on EMM plus leucine, uracil, histidine, arginine, and low adenine (5 mg/L) in the presence (break off) or absence (break on) of thiamine. (C) Fivefold serial dilutions of wild-type, ddb1Δ, and cdt2Δ strains on Ye5S, Ye5S + 0.005% bleomycin, and Ye5S + 0.005% MMS. (D) IR survival curve for wild-type, ddb1Δ, and cdt2Δ strains. Means ± standard errors of three experiments are shown.
Deletion mutants of ddb1+ and cdt2+ were identified as exhibiting striking break-induced sectoring (Fig. 1B). When tested for sensitivity to damaging agents, ddb1Δ and cdt2Δ both exhibited exquisite sensitivity to MMS, bleomycin, and IR (Fig. 1C,D), suggesting a role for Ddb1 and Cdt2 in response to a DSB. Furthermore, ddb1Δ and cdt2Δ mutations exhibited an epistatic relationship in response to DNA-damaging agents (Supplemental Fig. 1).
Ddb1 and Cdt2 are required for HR
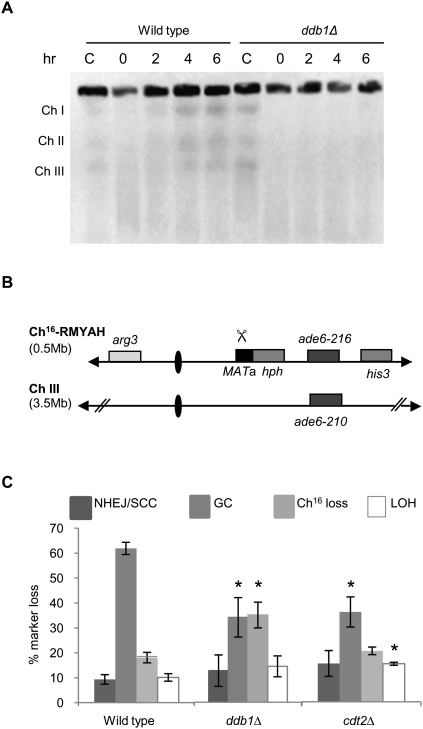
The suggested role of ddb1Δ in DSB repair was further investigated by assaying for the ability to repair DSBs caused by exposure to bleomycin. Following bleomycin treatment, broken chromosomes are visible by pulsed-field gel electrophoresis (PFGE) as a low-molecular-weight smear. In wild-type cells, reformation of distinct chromosomes can be seen after 2 h, with complete reconstitution of all three chromosomes being visible 4 h following damage (Fig. 2A). In contrast, the chromosomes in ddb1Δ mutant cells remain unrepaired 6 h following treatment (Fig. 2A). To quantify DSB repair in ddb1Δ and cdt2Δ backgrounds, DSB-induced marker loss was assessed using a DSB assay in which a previously described minichromosome (Ch16-RMYAH) was cleaved uniquely at the MATa target site following HO endonuclease derepression from a plasmid (pREP81X-HO) (Fig. 2B; Tinline-Purvis et al. 2009). Following break induction, wild-type cells predominantly repair the break by HR repair (61%), resulting in arg+ hygS ade+ his+ colonies in which the MATa break site and adjacent HygR marker are lost through gene conversion (GC) using the endogenous ChIII as a repair template (Prudden et al. 2003). In contrast to wild type, significantly reduced levels of GC were observed in both ddb1Δ (34%, P = 0.005) and cdt2Δ (33%, P = 0.02) backgrounds following break induction, an outcome indicative of inefficient HR. Failed DSB repair results in Ch16 loss, resulting in arg− hygS ade− his− colonies, seen in 17% of repair events in wild-type cells. Consistent with inefficient DSB repair by GC, ddb1Δ exhibited significantly increased levels of Ch16 loss (34%, P = 0.01) compared with wild type. High levels of minichromosome loss were also observed in a cdt2Δ background (37%); however, a large proportion of this, 18%, was spontaneous (i.e., HO break-independent) (data not shown). An increase in arg+ hygS ade− his− colonies, consistent with increased LOH, was observed in cdt2Δ (14%, P = 0.03) and ddb1Δ (14%), although not significantly different from wild type in the latter (P = 0.2). Analysis of LOH events by PFGE revealed predominantly isochromosomes, and some de novo telomere addition in both ddb1Δ and cdt2Δ backgrounds (Supplemental Fig. 2). No significant difference in levels of arg+ hygR ade+ his+ colonies, arising from NHEJ/sister chromatid conversion (SCC), was observed (Fig. 2C). These data identify roles for both Ddb1 and Cdt2 in promoting efficient HR repair and maintaining genome stability.
Figure 2.
Ddb1 and Cdt2 are required for efficient HR repair. (A) PFGE analyses of chromosomes (Ch) from bleomycin-treated (5 μg/mL) wild-type and ddb1Δ cells. Cells were either untreated (C) or treated with bleomycin for 1 h, washed to remove bleomycin, and harvested at the times indicated following bleomycin removal. (B) Schematic of the minichromosome Ch16-RMYAH. Ch16-RMYAH, a 530-kb fragment of ChIII, contains the MATa site with an adjacent hygromycin resistance marker gene (hph) and the ade6-M216 ade heteroallele, complemented by ade6-M210, present on ChIII, in addition to a his3 and arg3 markers, as shown previously (Tinline-Purvis et al. 2009). Derepression of pREP81X-HO (not shown) generates a DSB at the MATa target site (indicated by scissors). (C) Percentage of DSB-induced marker loss in wild-type, ddb1Δ, and cdt2Δ backgrounds. Means ± standard errors of three experiments are shown. The asterisk (*) represents significant difference compared with wild type.
Deletion of spd1+ suppresses the DSB repair defect of ddb1Δ and cdt2Δ
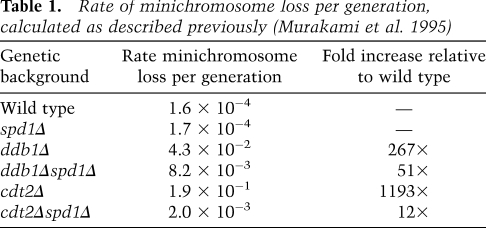
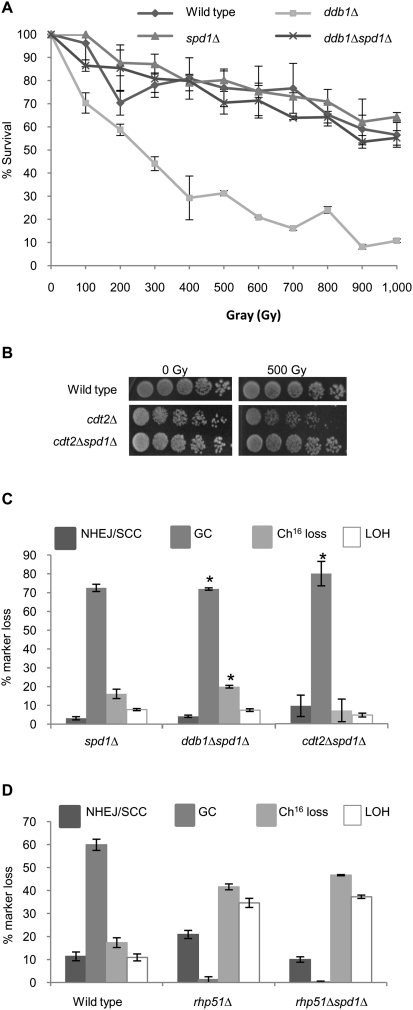
Ddb1 and Cdt2 function as part of the Ddb1–Cul4Cdt2 ubiquitin ligase complex. One function of this complex in S. pombe is to ubiquitylate and promote the degradation of Spd1, a small inhibitor protein of RNR (Bondar et al. 2004; Holmberg et al. 2005). To establish whether loss of Spd1 degradation in ddb1Δ and cdt2Δ leads to the observed repair defects, spd1+ was deleted in ddb1Δ and cdt2Δ backgrounds, thus bypassing the requirement for a functional Ddb1–Cul4Cdt2 ubiquitin ligase complex to degrade Spd1. Deletion of spd1+ suppressed the increased spontaneous minichromosome loss observed in ddb1Δ and cdt2Δ single mutants (Table 1). In addition, deletion of spd1+ was found to suppress the IR sensitivity of both ddb1Δ and cdt2Δ single mutants, restoring wild-type levels of resistance (Fig. 3A,B). These findings are in accordance with the radiation sensitivity observed in the ddb1Δ and cdt2Δ mutants, resulting solely from their inability to degrade Spd1. Furthermore, quantification of DSB repair in the double mutants revealed that deletion of spd1 restored wild-type levels of GC in both ddb1Δ (69%, P = 0.001) and cdt2Δ (78%, P = 0.006) backgrounds (cf. Figs. 2C and 3C). The observed increase in GC levels was associated with decreased Ch16 loss in both ddb1Δspd1Δ (19%, P = 0.007) and cdt2Δspd1Δ (8%, P = 0.17) backgrounds compared with individual ddb1Δ or cdt2Δ mutants (Figs. 2C, 3C), thus restoring DSB repair to wild-type levels. In contrast, deletion of spd1+ was unable to suppress the repair defect observed in an rhp51Δ or rhp51E345K hypomorphic point mutant background (Fig. 3D; Supplemental Fig. 3, respectively). Hence, deletion of spd1+ did not promote HR repair in general, but specifically suppressed the DSB repair defects of ddb1Δ and cdt2Δ mutants. Together, these data suggest that the inefficient GC observed in the ddb1Δ and cdt2Δ mutants results solely from their inability to degrade Spd1, and thus efficient HR repair and radiation resistance requires degradation of Spd1 through the action of the Ddb1–Cul4Cdt2 ubiquitin ligase complex.
Table 1.
Rate of minichromosome loss per generation, calculated as described previously (Murakami et al. 1995)
Figure 3.
spd1Δ suppresses the repair defect of ddb1Δ and cdt2Δ. (A) IR survival curves for wild type, spd1Δ, ddb1Δ, and ddb1Δspd1Δ. Means ± standard errors of three experiments are shown. (B) Fivefold serial dilutions of wild type, cdt2Δ, and cdt2Δspd1Δ strains grown on Ye5S in response to 0 or 500 Gy of IR. (C) Percentage of DSB-induced marker loss in spd1Δ, ddb1Δspd1Δ, and cdt2Δspd1Δ backgrounds. Means ± standard errors of three experiments are shown. The asterisk (*) represents significant difference compared with ddb1Δ and cdt2Δ single mutants, respectively (see Fig. 2C). (D) Percentage of DSB-induced marker loss in wild-type, rhp51Δ, and rhp51Δspd1Δ backgrounds. Means ± standard errors of three experiments are shown.
dNTP production following a DSB is required for efficient HR
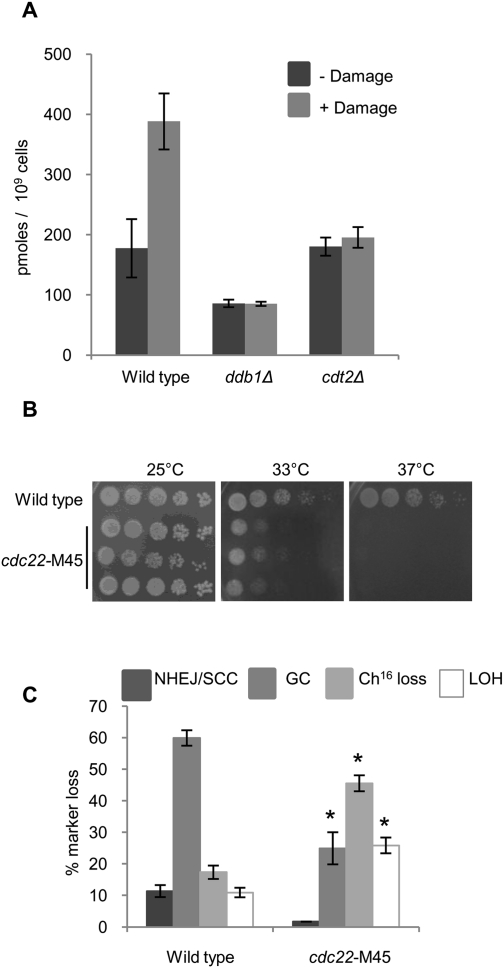
Following DNA damage, dNTPs are produced, creating a measurable fold increase in dNTP pools within the cell (Håkansson et al. 2006). To assess dNTP production following damage in ddb1Δ and cdt2Δ strains, bleomycin was used to induce DNA breaks and levels of dTTP were subsequently measured. Wild-type cells exhibit a 2.5-fold increase in dTTP levels following damage (Fig. 4A), seemingly in preparation for the increased demand for nucleotides experienced during repair. In contrast, neither ddb1Δ nor cdt2Δ cells exhibit increased dTTP pool levels (Fig. 4A), consistent with their inability to degrade the RNR inhibitor Spd1 following damage (Holmberg et al. 2005; Liu et al. 2005). To investigate whether reduced RNR activity and, thus, reduced nucleotide pools have a detrimental effect on levels of HR in general, rather than in ddb1Δ and cdt2Δ mutants specifically, levels of break-induced marker loss were quantified in a cdc22-M45 mutant. cdc22-M45, a temperature-sensitive mutant of the large subunit of RNR (Dickinson 1981), can be grown at a semipermissive temperature of 33°C (Fig. 4B); growth at this temperature partially impairs cdc22 gene function in this mutant (Singer and Johnston 1985). Performing the DSB assay in a cdc22-M45 background at 33°C revealed a significant reduction in GC (25%, P = 0.03) and an increase in minichromosome loss (46%, P = 0.02) compared with wild type, an outcome consistent with failed HR repair (Fig. 4C). Levels of break-induced LOH were also increased (26%, P = 0.05), and were found to result from both isochromosome formation and de novo telomere addition (Supplemental Fig. 2). FACS analysis indicated that cdc22-M45 cells exhibited a 2C peak, and were thus predominantly in G2 at the time of DSB induction during the DSB assay (Supplemental Fig. 4; Prudden et al. 2003).
Figure 4.
RNR and dNTP production following a DSB is required for efficient HR repair. (A) dTTP levels measured in wild-type, ddb1Δ, and cdt2Δ strains 100 min following 5 μg/mL bleomycin treatment. Means ± standard errors of three experiments are shown. (B) Fivefold serial dilutions of three independently derived cdc22-M45 strains grown on Ye5S. Plates at 33°C and 37°C were incubated for 2 d; the plate at 25°C was incubated for 4 d. (C) Percentage of DSB-induced marker loss in cdc22-M45 grown for 72 h at 33°C. Means ± standard errors of three experiments are shown. The asterisk (*) represents significant difference compared with wild type.
A role for the Ddb1–Cul4Cdt2 ubiquitin ligase complex in gap filling of break-induced resected ssDNA
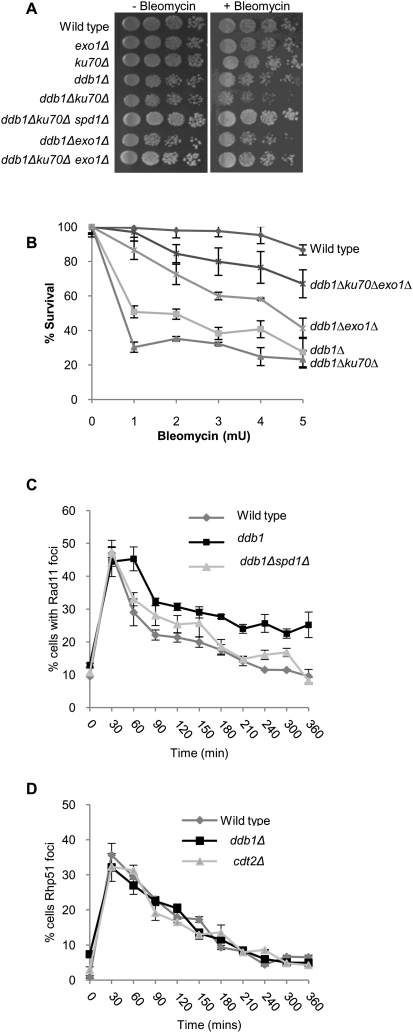
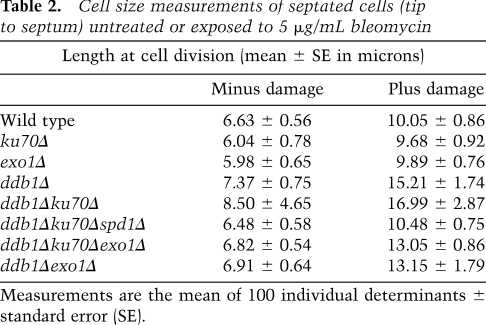
Reduced nucleotide pools observed in a ddb1Δ background would be expected to affect DNA synthesis steps required for gap filling and efficient GC following a DSB. The extent of ssDNA generated through resection would dictate the length of new DNA synthesis required to fill the gaps following annealing of ssDNA ends. Ku70, a DNA end-binding protein, protects the DNA ends following a DSB, and loss of Ku70 consequently results in increased resection at the break site (Lee et al. 1998). We hypothesized that the increased resection in a ku70Δ strain would create a larger “gap” to be filled in by new DNA synthesis. This in turn would compromise DSB repair in a ddb1Δ mutant further due to an increased requirement for nucleotides to fill these extended gaps. Accordingly, ddb1Δku70Δ showed an increased sensitivity to bleomycin (Fig. 5A,B), an effect not seen in ddb1Δlig4Δ (Supplemental Fig. 5), indicating that increased sensitivity in ddb1Δku70Δ is not a result of impaired NHEJ. Furthermore, ddb1Δku70Δ cells were increased in length compared with the single mutants after DNA damage, consistent with the checkpoint-dependent cell cycle delay associated with reduced repair (Table 2; Supplemental Fig. 6). Importantly, deletion of spd1+ in the ddb1Δku70Δ double mutant suppressed this enhanced sensitivity (Fig. 5A) and the increase in cell size (Table 2; Supplemental Fig. 6), consistent with increased nucleotide pools restoring DNA synthesis and gap filling in the ddb1Δku70Δ background. Exo1 is a 5′–3′ exonuclease required for efficient resection at a DSB (Mimitou and Symington 2008; Zhu et al. 2008). To confirm that the increased sensitivity of the ddb1Δku70Δ double mutant resulted from the inability to repair the extensively resected broken ends, we tested whether this sensitivity could be suppressed by deletion of exo1+ in which resection is reduced (Tomita et al. 2003; Limbo et al. 2007). We found bleomycin sensitivity in a ddb1Δku70Δexo1Δ triple mutant was substantially reduced compared with that of the ddb1Δku70Δ double mutant (Fig. 5A,B). The increased cell length observed in the ddb1Δku70Δ double mutant was also suppressed by deletion of exo1+ (Table 2; Supplemental Fig. 6). Deletion of exo1+ also reduced the sensitivity and cell length in a ddb1Δ background (Fig. 5B; Table 2; Supplemental Fig. 6). To further assess the kinetics of ssDNA metabolism following DNA damage in wild-type and ddb1Δ cells, RPA-coated ssDNA was measured following exposure to 50 Gy of IR using a GFP-tagged RPA subunit, Rad11-GFP (Kibe et al. 2007). Formation of Rad11-GFP foci did not differ between wild-type and ddb1Δ cells. In contrast, Rad11-GFP foci were retained for longer in ddb1Δ cells compared with wild type in response to IR (Fig. 5C). In accordance with the DNA damage sensitivity of ddb1Δ being suppressed by deletion of spd1+, the retention of Rad11-GFP foci observed in a ddb1Δ background was reduced by deletion of spd1+ following IR (Fig. 5C). These findings indicate that Spd1 degradation and, thus, RNR activation are required for efficient removal of RPA-coated ssDNA following exposure to IR. To determine whether RNR functioned presynaptically or postsynaptically to promote HR, the kinetics of Rad51-CFP foci in response to IR were measured in wild-type, ddb1Δ, and cdt2Δ cells (Osman et al. 2005). All strains exhibited very similar profiles of Rad51-CFP foci appearance and disappearance following exposure to IR (Fig. 5D), consistent with a postsynaptic role for Ddb1–Cul4Cdt2 and RNR in facilitating ssDNA and RPA removal during HR. Our findings further support a postsynaptic role for RPA in checkpoint signaling.
Figure 5.
A role for the Ddb1–Cul4Cdt2 ubiquitin ligase complex in gap filling of break-induced resected ssDNA. (A) Fivefold serial dilutions of wild-type, exo1Δ, ku70Δ, ddb1Δ, ddb1Δku70Δ, ddb1Δku70Δspd1Δ, ddb1Δexo1Δ, and ddb1Δku70Δexo1Δ strains grown on Ye5S and Ye5S + 0.0025% bleomycin. (B) Bleomycin survival curve for wild-type, ddb1Δ, ddb1Δku70Δ, ddb1Δexo1Δ, and ddb1Δku70Δexo1Δ strains exposed to the indicated doses of bleomycin for 1 h. Means ± standard errors of three experiments are shown. (C) Quantification of Rpa1(Rad11)-GFP foci in wild-type, ddb1Δ, and ddb1Δspd1Δ strains following exposure to 50 Gy of IR treatment. (D) Quantification of Rhp51-CFP foci in wild-type, ddb1Δ, and cdt2Δ strains following exposure to 50 Gy of IR.
Table 2.
Cell size measurements of septated cells (tip to septum) untreated or exposed to 5 μg/mL bleomycin
Measurements are the mean of 100 individual determinants ± standard error (SE).
Rad3 facilitates efficient HR by regulating Cdt2 and dNTP levels
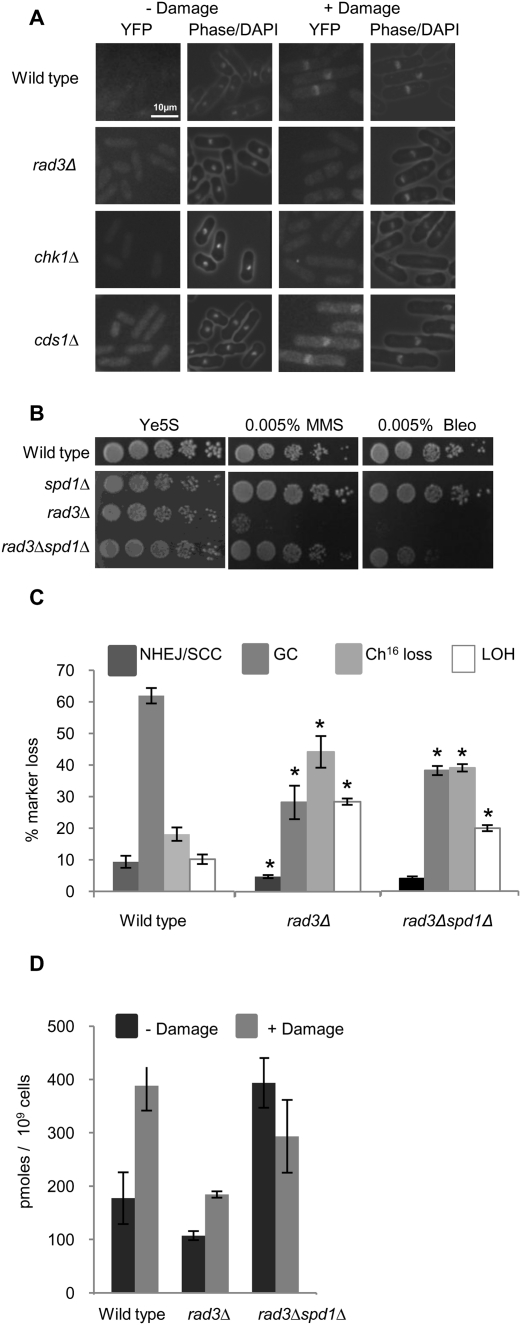
We wished to determine the mechanism by which the Ddb1–Cul4Cdt2 ubiquitin ligase might be regulated in response to a DSB. We and others previously identified a role for Rad3 in regulating Cdt2 expression levels in response to IR (Watson et al. 2004; Liu et al. 2005). In accordance with this, we found that exposure to bleomycin resulted in a strong increase in nuclear Cdt2-YFP levels (Fig. 6A). Increased nuclear levels required cycloheximide, consistent with Cdt2 localization requiring gene expression. Localization was not observed in the presence of the nuclear export inhibitor leptomycin B, suggesting nuclear accumulation was not a result of altered nuclear export (Supplemental Fig. 7). Furthermore, Cdt2-YFP appearance was DNA damage checkpoint-dependent, as, while Cdt2-YFP levels were observed in a cds1Δ background, no Cdt2-YFP was observed in a rad3Δ or chk1Δ background following exposure to bleomycin (Fig. 6A). In contrast to Cdt2, Ddb1 levels were not altered in the presence or absence of DNA damage (our unpublished results). Consistent with an incomplete damage response, rad3Δ cells were found to be sensitive to DNA--damaging agents (MMS and bleomycin) (Fig. 6B). These findings supported a role for Rad3 in regulating Cdt2 nuclear expression and, thus, Ddb1–Cul4 ubiquitin ligase activity in response to DNA damage. If correct, deletion of spd1+ in rad3Δ should suppress, in part, the repair defect of rad3Δ by overcoming the need for Cdt2 up-regulation. Consistent with this, suppression of sensitivity to damaging agents (MMS and bleomycin) was observed following deletion of spd1+ in a rad3Δ background (Fig. 6B). Quantitative analysis of break repair in rad3Δ revealed a defect in HR repair, resulting in a significantly reduced level of GC (26%, P = 0.001) and a concomitant increase in Ch16 loss (42%, P = 0.03) and LOH (27%, P = 0.0003) compared with wild type (Fig. 6C; Prudden et al. 2003), reminiscent of the repair profile observed in ddb1Δ and cdt2Δ. Furthermore, analysis of break repair following deletion of spd1+ in a rad3Δ background partially restored GC levels (37%, P = 0.05) and reduced levels of minichromosome loss (38%, P = 0.5) and LOH (20%, P = 0.01) compared with rad3Δ (Fig. 6C). As GC levels in a rad3Δ background were only partially restored compared with those of ddb1Δ following deletion of spd1+, this suggests additional roles for Rad3 in facilitating efficient HR repair, including expression of RNR genes in response to DNA-damaging agents (Harris et al. 1996; Watson et al. 2004). Deletion of spd1+ was unable to suppress the DNA damage sensitivity observed in a chk1Δ background (our unpublished results), suggesting additional roles for Chk1 in the DNA damage response.
Figure 6.
A role for Rad3 in RNR regulation. (A) Methanol-fixed samples of asynchronous Cdt2-YFP wild type, rad3Δ, chk1Δ, and cds1Δ cells imaged following their growth in the presence or absence of 5 μg/mL bleomycin for 3 h. (B) Fivefold serial dilutions of wild-type, spd1Δ, rad3Δ, and rad3Δspd1Δ strains on Ye5S, Ye5S + 0.005% MMS, and Ye5S + 0.005% bleomycin. (C) Percentage of DSB-induced marker loss in wild type, rad3Δ, and rad3Δspd1Δ backgrounds. Means ± standard errors of three experiments are shown. The asterisk (*) represents significant difference of rad3Δ compared with wild type and rad3Δspd1Δ compared with rad3Δ single mutant. (D) dTTP levels measured in wild-type, rad3Δ, and rad3Δspd1Δ strains 100 min following 5 μg/mL bleomycin treatment. Means ± standard errors of three experiments are shown.
To investigate whether failed GC resulted from misregulation of dNTP levels following a DSB in a rad3Δ background, levels of dTTP were quantitated in wild-type and rad3Δ mutants following exposure to bleomycin. While in a wild-type strain a 2.5-fold increase in dTTP levels was observed, a significantly lower increase in dTTP levels was observed in rad3Δ cells (1.8-fold, P = 0.04) following exposure to bleomycin (Fig. 6D). The improved repair profile of the double mutant correlated with a higher level of dTTP present in a rad3Δspd1Δ background compared with rad3Δ (Fig. 6D), indicating the deficient repair in rad3Δ is, at least in part, due to impaired dNTP production following a DSB. Together, these data support a role for Rad3 in facilitating efficient HR repair by increasing nuclear Cdt2 levels, which in turn leads to Ddb1–Cul4Cdt2 ubiquitin ligase-dependent Spd1 degradation and subsequently elevated dNTP levels following a DSB.
Discussion
Regulation of RNR and efficient HR
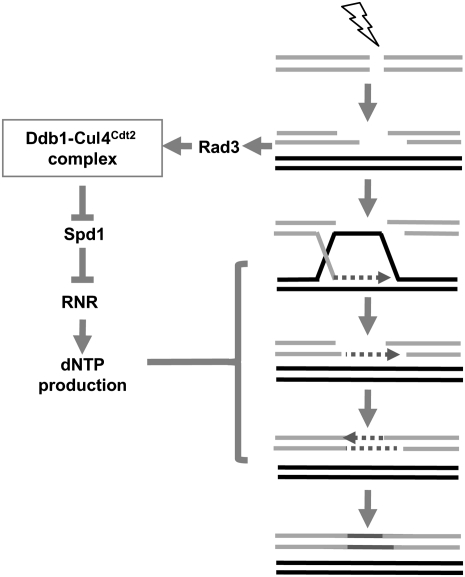
The present study provides insights into the role and regulation of nucleotide synthesis during HR repair of DSBs. We show that regulation of RNR through Ddb1–Cul4Cdt2 ubiquitin ligase-dependent degradation of the RNR inhibitor Spd1 is critical for efficient DSB repair by HR and for resistance to DNA-damaging agents. We found that deletion of ddb1+ or cdt2+ results in reduced GC, lowered dNTP pools, and subsequently increased sensitivity to DNA-damaging agents. Conversely, the deletion of spd1+ in a ddb1Δ or cdt2Δ deletion background results in efficient GC and subsequently increased resistance to DNA-damaging agents. Consistent with this, we found that a mutant in the R1 subunit, Cdc22, exhibits reduced GC, thus identifying a role for RNR in promoting efficient HR. Furthermore, we show that the DNA damage sensitivity in a ddb1Δ background is exacerbated by deletion of ku70+, and this increased DNA damage sensitivity was suppressed by deletion of exo1+. These findings support a role for the Ddb1–Cul4Cdt2 ubiquitin ligase complex in gap filling of resected ssDNA ends. Accordingly, increased accumulation of RPA-coated ssDNA was observed in a ddb1Δ background following irradiation, which was suppressed by deletion of spd1+. As the kinetics of Rhp51-CFP foci were unaffected in ddb1Δ or cdt2Δ backgrounds, this suggests that the Ddb1–Cul4Cdt2 ubiquitin ligase complex and, thus, RNR activity promote HR postsynaptically. Together, these findings support a role for the Ddb1–Cul4Cdt2 complex in facilitating postsynaptic repair of resected ssDNA ends during HR by promoting Spd1 degradation, leading to RNR activation and nucleotide pool increase (Fig. 7).
Figure 7.
Model for the role and regulation of nucleotide synthesis during HR repair. Rad3 activation in response to a DSB leads to activation of the Ddb1–Cul4Cdt2 ubiquitin ligase complex. Complex activation results in Spd1 degradation, RNR activation, and subsequent increase in dNTP pools. This dNTP pool increase is required for postsynaptic gap filling during HR repair.
Spd1 plays a key role in regulating RNR activity in fission yeast, where it can interact directly with Cdc22 to mediate biochemical inhibition of RNR (Håkansson et al. 2006). Spd1 also regulates the subcellular localization of the R2 subunit Suc22, which is localized primarily to the nucleus in non-S-phase cells and is relocalized to the cytoplasm during S phase or following DNA damage (Liu et al. 2003). In this respect, Spd1 has been shown recently to function as a nuclear import factor for the Suc22 R2 subunit, where its degradation facilitates cytoplasmic R2 localization, thus functioning analogously to Dif1 in S. cerevisiae (Liu et al. 2003; Lee et al. 2008; Nestoras et al. 2010). However, detailed mutagenic analysis of Spd1 indicated that its ability to restrain RNR activity is not a consequence of its ability to sequester R2 to the nucleus (Nestoras et al. 2010). Our results suggest that increased DSB repair and resistance to DNA-damaging agents in response to Spd1 degradation would necessitate increased nuclear dNTP pools in response to DNA damage. In this respect, RNR may be linked physically to the sites of DSB repair, as a role for the histone acetyltransferase Tip60 in recruiting RNR to DNA damage sites during G1 has been reported recently (Niida et al. 2010).
A number of anti-cancer drugs function to inhibit RNR activity, including hydroxyurea (HU), gemcitabine 5′-diphosphate, 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3AP), [Tris(1,10-phenanthroline)-lanthanum(III)] trithiocyanate (KP772), motexafin gadolinium (MGd), triapine, and others (for review, see Shao et al. 2006). Our findings indicate that these drugs are likely to not only inhibit DNA replication, but are expected also to block efficient HR, and therefore sensitize cells to the types of DNA damage and damaging agents that require HR repair. In accordance with this, HU treatment has been shown recently to sensitize NHEJ-deficient cells to IR-induced chromosome breaks (Burkhalter et al. 2009).
We found recently that the extensive resection associated with failed or inefficient HR resulted in chromosomal rearrangements and extensive LOH (Tinline-Purvis et al. 2009). Consistent with this, we observed elevated levels of break-induced LOH arising predominantly through isochromosome formation, in which the entire distal arm of the minichromosome was lost and the intact arm was duplicated in ddb1Δ, cdt2Δ, and cdc22-M45 backgrounds in which HR is compromised. These observations support the generality of this model, and suggest a mechanism by which the Ddb1–Cul4Cdt2 complex or other factors involved in RNR function may act as tumor suppressors. Furthermore, low levels of LOH arising from de novo telomere addition were also detected in these mutants, again consistent with disrupted HR (Cullen et al. 2007).
The Ddb1–Cul4Cdt2 complex and DNA repair
A role for DDB1 was originally described in DNA repair, where it was found to bind UV-induced damage as a heterodimer with DDB2, thus recruiting the nucleotide excision repair machinery (Tang et al. 2000). More recently, a role for DDB1 in the DDB1–CUL4 ubiquitin ligase has been documented that targets a variety of substrates that regulate a broad spectrum of cellular processes, including cell proliferation, survival, DNA repair, and genomic integrity. In humans, this range of functions is achieved through its interaction with up to 60 DDB1–CUL4-associated factors (DCAFs), which target specific proteins for degradation (for review, see Lee and Zhou 2007).
Cdt2 in humans performs a number of functions related to DNA repair and genome stability, where it is required as an E3 ubiquitin ligase to monoubiquitinate PCNA to promote translesion DNA synthesis (Terai et al. 2010); it also functions as a DCAF to promote p21 proteolysis when treatments such as UV irradiation trigger replication fork stress (Soria and Gottifredi 2010). In both humans and fission yeast, Cdt2 functions as a DCAF required for the ubiquitylation and degradation of Cdt1, which functions together with Cdc18 to license DNA replication (for review, see Feng and Kipreos 2003). In fission yeast, Cdt2 is required for damage-induced proteolysis of Cdt1 and Spd1 (Liu et al. 2005; Ralph et al. 2006). Our data indicate that cdt2Δ has increased spontaneous levels of minichromosome loss compared with ddb1Δ, yet spd1+ deletion effectively suppressed both spontaneous and DNA damage-induced minichromosome loss in both strains. This suggests that cdt2Δ may incur additional lesions that require dNTP synthesis for genome stability. Thus, while Cdt2 may target other proteins for ubiquitin-dependent degradation, a major role for Cdt2 during HR repair is to facilitate Spd1 degradation and the activation of RNR. While it is possible that Spd1 has additional functions, it is likely that the suppressive effects of Spd1 degradation on HR repair are mediated through dNTP synthesis, as we also found RNR activity to be required for HR.
Checkpoint-dependent ssDNA regulation and DSB repair
The DNA damage checkpoint signal is generated in response to ssDNA detection by the ATR–ATRIP and 9-1-1 complexes (Lydall and Weinert 1995; Melo et al. 2001; Zou and Elledge 2003). The DNA damage checkpoint has been shown to be required for efficient DSB repair where Rad24Sc is required for resection and, thus, ssDNA production (Aylon and Kupiec 2003), while Rad9Sc has been shown to limit ssDNA production associated with DSB repair (Lazzaro et al. 2008). This function appears to be conserved, as the human Rad9 homolog 53BP1 has been shown to block extensive resection and suppress the repair defect of BRCA1 (Bunting et al. 2010). In S. cerevisiae, lethality associated with deletion of checkpoint kinase Mec1 can be suppressed by deletion of the RNR inhibitor Sml1; this suppression is thought to arise by facilitating efficient dNTP synthesis required for DNA replication (Zhao et al. 1998). In S. pombe, the Mec1 ortholog Rad3 is viable when deleted, thus facilitating comparison of DSB repair in both rad3Δ and rad3Δspd1Δ backgrounds. Viability in a rad3Δ background is likely to reflect a second checkpoint-independent pathway for Spd1 degradation during S phase in fission yeast (Liu et al. 2003; Ralph et al. 2006). Following DNA damage, our data support a new role for checkpoint-dependent RNR-induced ssDNA gap repair and efficient HR. This functions through Rad3-dependent nuclear expression of Cdt2 and Ddb1–Cul4Cdt2 ubiquitin ligase-dependent Spd1 degradation, leading to RNR activation and increased nucleotide pools. These findings together support a central role for the DNA damage checkpoint in regulating ssDNA metabolism, where, in addition to its detection, it regulates ssDNA generation through resection and, finally, its removal by RNR-dependent gap filling, thus facilitating efficient DSB repair and genome stability (Fig. 7). Such a mechanism implies the existence of a negative feedback loop, which promotes recovery after damage: DSB end processing generates ssDNA, which in turn activates Rad3. This leads to increased nucleotide pools, which helps fill the ssDNA gaps and hence switches off the checkpoint.
Materials and methods
Yeast strains, media, and genetic methods
All S. pombe strains were cultured, manipulated, and stored as described previously (Prudden et al. 2003). All strain genotypes are listed in Supplemental Table 1.
Colony sectoring assay
Following growth on selective media with a low concentration of thiamine (2 μM), cells were diluted and plated (∼100 cells per plate) onto sectoring plates: EMM + arginine (15 mg/L), histidine (15 mg/L), uracil (15 mg/L), leucine (15 mg/L), and adenine (5 mg/L) with and without thiamine (break off/on). Plates were incubated for 56 h at 32°C and were stored for 48 h at 4°C before being scored for the presence of sectored colonies. For strains exhibiting break-induced sectoring, this procedure was repeated two further times to ensure reproducibility of results.
Site-specific DSB assay
The DSB assay was performed as described previously (Prudden et al. 2003). The percentage of colonies undergoing NHEJ/SCC (arg+ HygR ade+ his+), GC (arg+ HygS ade+ his+), minichromosome loss (arg− HygS ade− his−), or LOH (arg+ HygS ade− his−) was calculated. To determine levels of break-induced minichromosome loss, background minichromosome loss at 48 h −T in blank vector assays was subtracted from break-induced minichromosome loss at 48 h −T in cells transformed with pREP81X-HO. More than 1000 colonies were scored for each time point, and each experiment was performed three times using three independently derived strains for all mutants tested.
HPLC analysis of nucleotides
Cells were harvested by centrifugation, and pellets were stored at −80°C pending TCA extraction. Thawed cells were disrupted with glass beads in 10% TCA, and were immediately frozen at −80°C. Defrosted cell extracts (15 μL) were mixed with 85 μL milli Q water (final concentration of 1.5% TCA). The nucleotides were neutralized for analysis by extracting the samples with an equal volume (100 μL) of freon (1,1,2 trichlorotrifluorethane):trioctylamine (4:1). The aqueous upper layer was removed and 40 μL was injected for analysis. Chromatography was carried out based on a previous method (Pires et al. 2010) on a Waters 2695 system with diode array detection (Waters 2996), monitoring at 254 nm. The column was an Ace (3 μm, 3 × 125 mm) column maintained at 30°C. Samples were maintained at 10°C. Separation was achieved with eluent A (10 mM potassium dihydrogen phosphate, 10 mM tetrabutylammonium hydroxide, 10% methanol at pH 6.9) and eluent B (50 mM potassium dihydrogen phosphate, 6 mM tetrabutylammonium hydroxide, 30% methanol at pH 7), using a flow rate of 0.6 mL per minute and a gradient of 25%–80% B over 20 min, with a run time of 25 min. Nucleotides were identified and quantitated against commercially available dNTPs.
Fluorescence microscopy
Asynchronous cultures were treated with 5 μg/mL bleomycin (1 h at 26°C), before being fixed in methanol then acetone. Samples were rehydrated and stained with 6′-diamidino-2-phenylindole (DAPI) before examination using a Zeiss Axioplan 2ie microscope, Hamamatsu Orca ER camera, and micromanager software. For visualization of Rad11-GFP and Rhp51-CFP foci, cells were irradiated with 50 Gy of IR using a 137Cs source with a dose rate of 2.8 Gy per min, before being fixed and visualized as above.
Serial dilution assay
A dilution series for the indicated mutants was spotted onto Ye5S plates and Ye5S with the indicated concentration of MMS or bleomycin. Plates were incubated for 2 d at 32°C before analysis.
IR survival curve
Logarithmically growing cells were irradiated by using a 60Co source at a dose rate of 31 Gy per min. Irradiated and nonirradiated cells were plated on Ye5S and incubated for 4 d at 32°C before colonies were counted.
PFGE
PFGE was carried out as described previously (Kearsey et al. 2007).
Acknowledgments
We thank Tony Carr, Miguel Ferreira, and Matthew Whitby for strains and reagents. This research was funded by the Medical Research Council (United Kingdom) (J.M., H.T.P., C.W., and T.C.H.), Cancer Research-UK (J.H., L.F., S.K., and M.S.), the Danish Cancer Society (C.H. and O.N.), the North West Cancer Research Fund (O.F.), the intramural research program of KRIBB (Mission 2007) (D.K. and K.H.), the Chemical Genomics Research Program from MOEST of Korea (K.H.), and the Bioneer Corporation (H.P.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1970810.
Supplemental material is available at http://www.genesdev.org.
References
- Abraham RT 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M 2003. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol Cell Biol 23: 6585–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Roberts JD, Kunkel TA 1992. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem 267: 3589–3596 [PubMed] [Google Scholar]
- Bondar T, Ponomarev A, Raychaudhuri P 2004. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J Biol Chem 279: 9937–9943 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhalter MD, Roberts SA, Havener JM, Ramsden DA 2009. Activity of ribonucleotide reductase helps determine how cells repair DNA double strand breaks. DNA Repair 8: 1258–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1: 983–994 [DOI] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401 [DOI] [PubMed] [Google Scholar]
- Cullen JK, Hussey SP, Walker C, Prudden J, Wee BY, Dave A, Findlay JS, Savory AP, Humphrey TC 2007. Break-induced loss of heterozygosity in fission yeast: Dual roles for homologous recombination in promoting translocations and preventing de novo telomere addition. Mol Cell Biol 27: 7745–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande GP, Hayles J, Hoe KL, Kim DU, Park HO, Hartsuiker E 2009. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair 8: 672–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JR 1981. The cdc22 mutation by Schizosaccharomyces pombe is a temperature-sensitive defect in nucleoside diphosphokinase. Eur J Biochem 119: 341–345 [DOI] [PubMed] [Google Scholar]
- Eggler AL, Inman RB, Cox MM 2002. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J Biol Chem 277: 39280–39288 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB 1992. Ribonucleotide reductase: Regulation, regulation, regulation. Trends Biochem Sci 17: 119–123 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15: 333–339 [DOI] [PubMed] [Google Scholar]
- Feng H, Kipreos ET 2003. Preventing DNA re-replication-divergent safeguards in yeast and metazoa. Cell Cycle 2: 431–434 [PubMed] [Google Scholar]
- Håkansson P, Dahl L, Chilkova O, Domkin V, Thelander L 2006. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem 281: 1778–1783 [DOI] [PubMed] [Google Scholar]
- Harris P, Kersey PJ, McInerny CJ, Fantes PA 1996. Cell cycle, DNA damage and heat shock regulate suc22+ expression in fission yeast. Mol Gen Genet 252: 284–291 [DOI] [PubMed] [Google Scholar]
- Haruta N, Akamatsu Y, Tsutsui Y, Kurokawa Y, Murayama Y, Arcangioli B, Iwasaki H 2008. Fission yeast Swi5 protein, a novel DNA recombination mediator. DNA Repair 7: 1–9 [DOI] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey SE, Stevenson AL, Toda T, Wang SW 2007. Fission yeast Cut8 is required for the repair of DNA double-strand breaks, ribosomal DNA maintenance, and cell survival in the absence of Rqh1 helicase. Mol Cell Biol 27: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T, Ono Y, Sato K, Ueno M 2007. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol Biol Cell 18: 2378–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS 2004. Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair 8: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P 2007. DCAFs, the missing link of the CUL4–DDB1 ubiquitin ligase. Mol Cell 26: 775–780 [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U, Gutz H 1964. Genetic fine structure in Schizosaccharomyces pombe. Proc XI Intl Congr Genet 2: 31–35 [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM 2005. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J 24: 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T 1995. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science 270: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Mathews CK 2006. DNA precursor metabolism and genomic stability. FASEB J 20: 1300–1314 [DOI] [PubMed] [Google Scholar]
- Mazin AV, Alexeev AA, Kowalczykowski SC 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J Biol Chem 278: 14029–14036 [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Yanagida M, Niwa O 1995. A large circular minichromosome of Schizosaccharomyces pombe requires a high dose of type II DNA topoisomerase for its stabilization. Mol Gen Genet 246: 671–679 [DOI] [PubMed] [Google Scholar]
- Nestoras K, Mohammed AH, Schreurs AS, Fleck O, Watson AT, Poitelea M, O'Shea C, Chahwan C, Holmberg C, Kragelund BB, et al. 2010. Regulation of ribonucleotide reductase by Spd1 involves multiple mechanisms. Genes Dev 24: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, et al. 2010. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev 24: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M 1986. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet 203: 397–405 [Google Scholar]
- Nordlund P, Reichard P 2006. Ribonucleotide reductases. Annu Rev Biochem 75: 681–706 [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC 2005. The F-box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol 25: 8084–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393: 91–94 [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, Goedecke W, Obe G 2000. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15: 289–302 [DOI] [PubMed] [Google Scholar]
- Pires IM, Bencokova Z, Milani M, Folkes LK, Li JL, Stratford MR, Harris AL, Hammond EM 2010. Effects of acute versus chronic hypoxia on DNA damage responses and genomic instability. Cancer Res 70: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Evans JS, Hussey SP, Deans B, O'Neill P, Thacker J, Humphrey T 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J 22: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE 2006. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep 7: 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J 2004. Periodic gene expression program of the fission yeast cell cycle. Nat Genet 36: 809–817 [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Chu B, Yen Y 2006. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets 6: 409–431 [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Singer RA, Johnston GC 1985. Indirect suppression of the wee1 mutant phenotype in Schizosaccharomyces pombe. Exp Cell Res 158: 533–543 [DOI] [PubMed] [Google Scholar]
- Soria G, Gottifredi V 2010. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA Repair 9: 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sung P 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Harper JW 2010. GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev 24: 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell 5: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K, Abbas T, Jazaeri AA, Dutta A 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell 37: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinline-Purvis H, Savory AP, Cullen JK, Dave A, Moss J, Bridge WL, Marguerat S, Bahler J, Ragoussis J, Mott R, et al. 2009. Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast. EMBO J 28: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Haber JE 2004. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol 2: E21 doi: 10.1371/journal.pbio.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A, Mata J, Bahler J, Carr A, Humphrey T 2004. Global gene expression responses of fission yeast to ionizing radiation. Mol Biol Cell 15: 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B, van Komen S, Sung P, Peterson CL 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell 12: 221–232 [DOI] [PubMed] [Google Scholar]
- Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci 100: 6628–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]