Abstract
The concept of the Logarithm of the Odds (LogOdds) is frequently used in areas such as artificial neural networks, economics, and biology. Here, we utilize LogOdds for a shape representation that demonstrates desirable properties for medical imaging. For example, the representation encodes the shape of an anatomical structure as well as the variations within that structure. These variations are embedded in a vector space that relates to a probabilistic model.
We apply our representation to a voxel based segmentation algorithm. We do so by embedding the manifold of Signed Distance Maps (SDM) into the linear space of LogOdds. The LogOdds variant is superior to the SDM model in an experiment segmenting 20 subjects into subcortical structures.
We also use LogOdds in the non-convex interpolation between space conditioned distributions. We apply this model to a longitudinal schizophrenia study using quadratic splines. The resulting time-continuous simulation of the schizophrenic aging process has a higher accuracy then a model based on convex interpolation.
1 Introduction
Statistical shape representation in medical imaging applications is an important and challenging problem. The challenge arises from a number of issues including the need for a representation which lends itself to efficient statistical inference while simultaneously capturing the intrinsic properties of shapes. Signed distance maps (SDMs) are a popular shape representation that have recently been utilized in combination with principal components analysis (PCA) for statistical shape modeling. Examples of such approaches include [1,2,3,4,5] that, with some variation, essentially treat SDMs as elements of the vector space ℝN (N being the number of voxels), perform PCA over a set of example SDMs and then fit a statistical model to the resulting PCA coefficients. The resulting model is then useful for inference in a variety of tasks including segmentation and longitudinal studies of shape variation. The primary advantage of the approach is that it projects high-dimensional SDMs into a lower dimensional representation which lends itself to concise statistical modeling and inference. The utility of such approximate modeling has been demonstrated successfully in varying degrees. One drawback of this approach as noted by [5], however, is the implicit approximation of the manifold of SDMs as a linear vector space. Consequently, samples or compositions of samples from the inferred statistical models are in general not valid SDMs. This detail is usually dealt with by further approximation, necessitating an additional computational step, that projects samples from the distribution over PCA coefficients back onto the manifold of shapes. Additionally, like many shape models [6], SDMs do not capture space conditioned probabilities (SCP). SCPs are often used in voxel-based segmentation (such as [7,8,9,10]) to describe the anatomical variations within a population. Here we suggest an alternative representation, the log-odds maps (LogOdds), that explicitly addresses the issues above.
LogOdds maps form a vector space and as such can be composed via linear methods while simultaneously maintaining the intrinsic properties of shapes. This allows for computationally efficient approaches to modeling and inference without the need for projecting the representation back onto the manifold of shapes. There is a straightforward mapping of the LogOdds representation to SCPs as each entry in the LogOdds representation is the logarithm of the odds over an anatomical label versus the alternative. The specific parameterization of LogOdds maps presented here provides a notion of certainty about the boundary of the inferred shape. Thus, LogOdds not only describe the shape of a single structure but also capture some aspects of variations within a structure across populations and expert segmentations. SDMs can be used directly as LogOdds maps, consequently no additional computational burden is incurred by their use. For example, entries with zero correspond to the boundary of the structure under either interpretation. The computational burden of mathematical operations over SCPs is reduced by performing them in the vector space of LogOdds. Arsigny et al. [11] have made the same observation with respect to using the logarithm on tensor data, although the logarithms of tensor data are not comparable to LogOdds.
In Section 2, we review and establish some of the relevant properties of LogOdds maps. We also provide a probabilistic interpretation for mathematical operations of the vector space. In Section 3, we demonstrate the utility of LogOdds in two experiments. In the first experiment, a Bayesian classifier [12] segments 20 sets of MR images into subcortical structures. Each subject is automatically segmented using the deformable atlas based on both SDMs and the LogOdds representation, and the results are compared. The second experiment is based on a longitudinal schizophrenia study in which an atlas represents the aging process of that population. The atlas is comprised of three SCPs representing different time points in the development of the population. We generate a non-convex interpolation between the three time points within the space of LogOdds with superior results to a convex interpolation within the original space of SCPs.
2 LogOdds and Its Properties
In this section we review some properties of LogOdds representations. In so doing, we focus on the use of the LogOdds for statistical inference over shapes. Specifically, we establish mappings between various representations and the means by which one performs probabilistic computations on LogOdds representations.
2.1 An Introduction to LogOdds
The LogOdds of a probability p ∈ ℙ ≜ {p|p ∈ [0,1], i.e. p is a probability} is the logarithm of the odds between the probability p and its complement p̄ = 1− p:
In the classification problem of medical images, for example, p is the probability that a voxel is assigned to a particular anatomical structure, while the complement p̄ is the probability of assignment to any other structure. The function logit(p) is simply the logarithm of their ratio. The LogOdds space is defined as 𝕃 ≜ {logit(p)|p ∈ ℙ}. Consequently, 𝕃 = ℝ and 𝕃n defines a vector space through its equivalence to ℝn. The inverse of the log odds function logit(·) is the standard logistic function
𝒫(·) maps each element t ∈ 𝕃 to a unique probability p ∈ ℙ, thus, the function logit(·) and its inverse comprise a homomorphism between ℙ and 𝕃. Again, the equivalence of 𝕃n to ℙn is straightforward when 𝒫(·) is applied element-wise to 𝕃n.
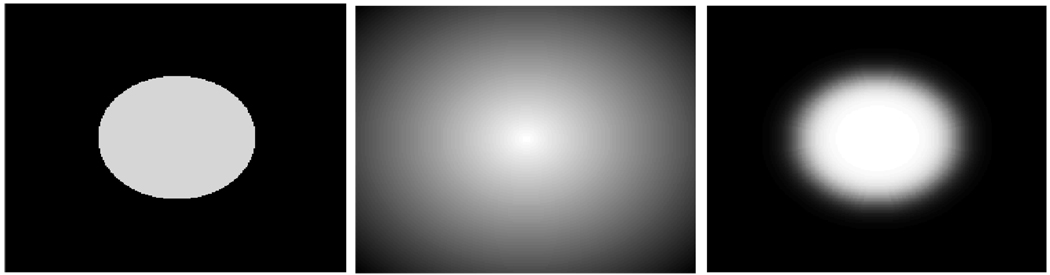
SDMs are elements of 𝕃n. The value of the SDM [𝒟]x at a voxel x is the distance from x to the closest point on the boundary. Figure 1(b) shows an example of a SDM corresponding to the binary map in Figure 1(a). It can be shown that any monotonic transformation 𝓥 (·) of 𝒟 is in 𝕃n. An obvious choice is the identity transformation [𝓥 (𝒟)]x ≜ [𝒟]x. In the LogOdds space, [𝒟]x is a representation for the probability px ≜ 𝒫([𝒟]x) ∈ ℙ that voxel x is assigned to the foreground. The corresponding vector p ≜ (p1, …, pn) therefore comprises a SCP of the foreground, where n is the number of voxels in the image. The relationship between SDMs and SCPs define a meaningful probabilistic model. The foreground is represented by the positive values of SDM corresponding to probabilities greater 0.5 in the SCP (see Figure 1 (c)). Conversely, negative values of the SDM define the background, which maps to probabilities less than 0.5.
Fig. 1.
The binary map of the circle in (a) corresponds to the SDM 𝒟 in (b). In 𝒟, positive values (bright) define the circle, while negative values (dark) represent the background. Panel (c) shows the corresponding SCP. Voxels inside the circle have probabilities greater 0.5.
2.2 The Relationship Between LogOdds and SCPs
We now make use of the Euclidean space 𝕃n to induce a vector space on ℙn. This results in a probabilistic model for addition and scalar multiplication in the log odds space 𝕃n. The derivations are performed with respect to 𝕃,ℙ for simplicity. It is easily extend to 𝕃n,ℙn by performing the operations individually for each vector entry.
Addition in ℙ
Based on the relationship between ℙ and 𝕃, the probabilistic addition ⊕ of p1, p2 ∈ ℙ is equivalent to the logistic function of the addition of the logarithm of odds logit(p1) and logit(p2):
| (1) |
As ℙ is closed under ⊕, (ℙ,⊕) forms an Abelian group with 0.5 as the neutral element and (1− p) the additive inverse of the element p. The complement of p1 ⊕ p2 is the probabilistic addition of the complements of p1 and p2, that is
Using suitably normalized likelihoods, the composition is equivalent to Bayes’ rule. Let the prior of the random variable X be p1 ≜ P(X)= 1−P(X̄) and the corresponding normalized likelihood be , then
Scalar Multiplication in ℙ
The probabilistic scalar multiplication ⊛ within the space of ℙ is equivalent to scalar multiplication, *, in the vector space 𝕃. Similar to ⊕, the multiplication between the scalar α ∈ ℝ and probability p ∈ ℙ is the logistic function of the product between α and the LogOdds logit(p)
(ℙ,⊕,⊛) defines a vector space, which is equivalent to (𝕃,+,*;). The complement of the probabilistic scalar multiplication α ⊛ p is equivalent to the scalar multiplication of the complement or negating α:
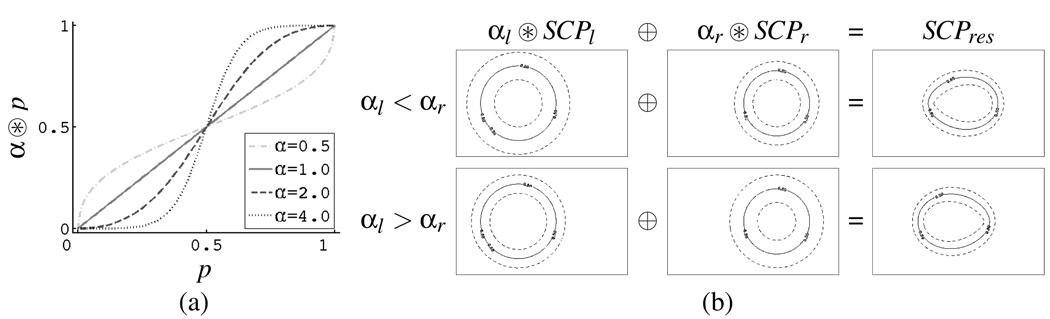
Figure 2(a) shows the influence of α on α ⊛ p. Changing α has the effect of increasing (α > 1) or decreasing (α < 1) the certainty of the boundary of a SCP. Probabilistic addition and scalar multiplication are illustrated in Figure 2 (b) using a synthetic shape description example. The figure depicts level sets of SCPl,SCPr ∈ ℙn that describe circles which differ in their means, radii, and boundary certainty (as captured by αl, αr). Figure 2 (b) shows how α effect the composition of the maps via the ⊕ operator. In the first row of Figure 2(b) αl = 20 (left circle) and αr = 30 (right circle). The distance between the level sets are seen to be closer for the right circle. Additionally, the composition of SCPs using ⊕ results in a combined SCP, which is biased towards the circle on the right. In the second row of Figure 2(b) the values of αl and αr are reversed resulting in a bias towards the circle on the left. Despite having different radii, if αl = αr the level sets of the combined SCP are ellipses, although the foci of the ellipse will depend on the relative radii.
Fig. 2.
(a) the effect of α on α ⊛ p. (b) Resulting SCP SCPres from combining 2 SCPs of circles (SCPl and SCPr) with different centers, radii, and α values.
In summary, we discussed basic properties of the vector space of log-odds 𝕃n relating them to the vector space of probabilities ℙn via the logistic function. This provided a probabilistic interpretation of scalar multiplication and vector addition in 𝕃n. We used LogOdds to describe binary maps of shapes. We note that the representation can be extended to multiple label maps.
3 Applications in Medical Imaging
We now apply LogOdds to two relevant applications in medical imaging. The first experiment measures the robustness of a shape based segmentation algorithm. In the second experiment, we build a temporal interpolation function from data of a longitudinal study. Both examples are based on MR images acquired by a 1.5-T MRI system (GE Medical System, three-dimensional Fourier transformation spoiled gradient-recalled acquisition, matrix=256×256×124, voxel dimension=0.9375×0.9375×1.5mm).
3.1 The Accuracy of a Segmenter with Respect to the Deformable Shape Atlas
We investigate the impact of our shape representation on the accuracy of a voxel-based segmenter [12]. The algorithm is guided by a deformable shape atlas that represents shape variation by PCA on SDMs. We analyze two implementations of the algorithm, which differ in the interpretation of the maps 𝒟 generated by the PCA atlas.
The first implementation (IMPL-𝓗) views 𝒟 as level set functions in a higher dimensional space. Each entry [𝒟]a,x of the SDM assigns the voxel x to the inside or outside of a structure a using the Heaviside function 𝓗 (·). 𝓗 (v) is one if v ≥ 0 and zero otherwise. A natural definition of the SCP is , where Σa′ 𝓗 ([𝒟]a′,x)) is the sum over all structures. In general, [SCP𝓗]a corresponds to a SCP with a steep slope along the structure’s boundary. Thus, the algorithm segments the structure according to [SCP𝓗]a, which only allows shapes within the PCA subspace.
The second implementation (IMPL-𝒫) interprets the maps 𝒟 as a vector of LogOdds 𝕃n. According to Section 2, the logistic function 𝒫([𝒟]a,x) defines the probability that voxel x is assigned to structure a. This approach is extended to multiple structures by defining the SCP as . [SCP𝒫]a is characterized by a gradual slope along the boundary of the structure. This provides the algorithm with some flexibility in determining the course of the contour – it allows the image evidence to nudge the resulting shape somewhat away from the PCA subspace.
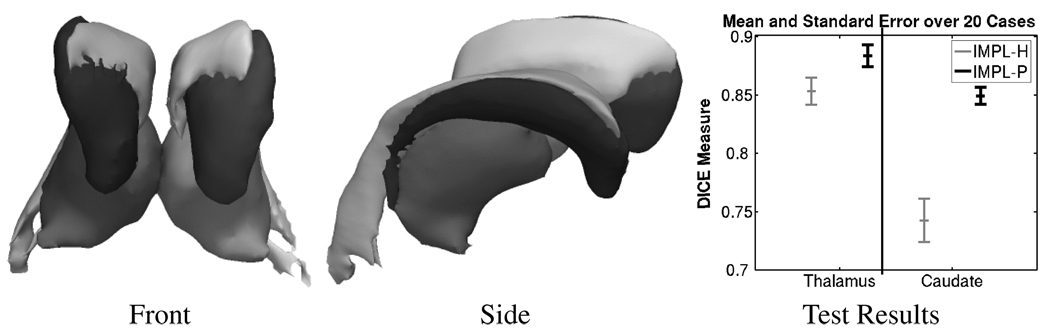
Both variations were used to segment 20 test cases into ventricles (light gray), the thalamus (gray), and caudate (black) (see Figure 3). Afterward, we compare the automatic segmentations of the thalamus and caudate to the manual segmentations via the Dice metric [13]. We focus on these two structures as they are characterized by different types of shapes (see Figure 3) and are partly invisible on MR images. Thus, the accuracy of the implementations is highly impacted by the shape model.
Fig. 3.
Different views of a 3D model of the thalamus (gray), the caudate (black), and the ventricles (light gray). The model is based on a segmentation generated by IMPL-𝒫. The graph to the right summarizes the results of our experiment. For both structures IMPL-𝒫 performs much better then IMPL-𝓗.
The graph in Figure 3 shows the result of the experiment for IMPL-𝓗 (thalamus: 85.3 ±1.2%, caudate: 74.3 ±1.6%; mean DICE score ± standard error) and IMPL-𝒫 (thalamus: 88.4 ±1.0%, caudate: 84.9 ±0.8%). For both structures, IMPL-𝒫 achieves a much higher average score than IMPL-𝓗. In addition, the segmentation results of IMPL-𝒫 are characterized by low standard error (in comparison to IMPL-𝓗) indicating high predictive power of the segmentation results. The differing accuracy of the implementations is mostly due to the different interpretations of the information in the shape atlas. The atlas underrepresents the shape variations within a population due to the limited amount of training data. The probabilistic model of IMPL-𝒫 addresses this issue by providing flexibility around the boundary, while IMPL-𝓗 in constrained to the limited-dimension PCA subspace.
This shortcoming of the atlas only slightly impacts the segmentation of the thalamus due its low shape variations within the healthy population. The bending of the horn shaped caudate, however, can vary substantially among subjects. This may explain the much lower score of IMPL-𝓗 in comparison to IMPL-𝒫 in case of this structure. We note that the input for the PCA was restricted to SDMs, a particular choice LogOdds function. The impact of different LogOdds classes on the segmentations would be interesting but outside the scope of this paper.
3.2 Defining an Interpolation Function from Time-Related Data Samples
Neuroscientists carry out longitudinal studies to better understand the aging process of a population. These studies are often defined by a set of subjects which have been scanned repeatedly at specific time points. The variations of the population at a specific point in time can be captured via SCPs. In this section, we explore the interpolation between the SCPs within the space of LogOdds, which results in continuous time atlases.
This experiment is based on a longitudinal data set consisting of eight Schizophrenic patients. Each patient was scanned three times with an average 14 month separation between first and second scan, and 23 month separation between the second and third scan. We generate the SCPs of each time point by first aligning the 24 MR images towards their central tendency using the population registration method by Zöllei et al. [14]. Afterward, we segment each aligned MR image into three brain tissue classes using Pohl et al. [15]. We then compute the SCP of an anatomical structure and time point based on the eight corresponding segmentations.
We can interpolate among atlases in either SCP or LogOdds space, however, with SCPs, to remain valid, we are limited to using convex combinations (CC), while we may use the broader class of linear combinations in the LogOdds vector space. In the following we will compare two temporal interpolation methods that are suited to the analysis of a small number of time points. One uses linear CCs of SCP, while the other uses a quadratic spline interpolation (that is not a CC) of LogOdds. It maps the SCPs to LogOdds’ space via the logistic function, applies the quadratic spline function and transfers the interpolated function back into the space of SCPs.
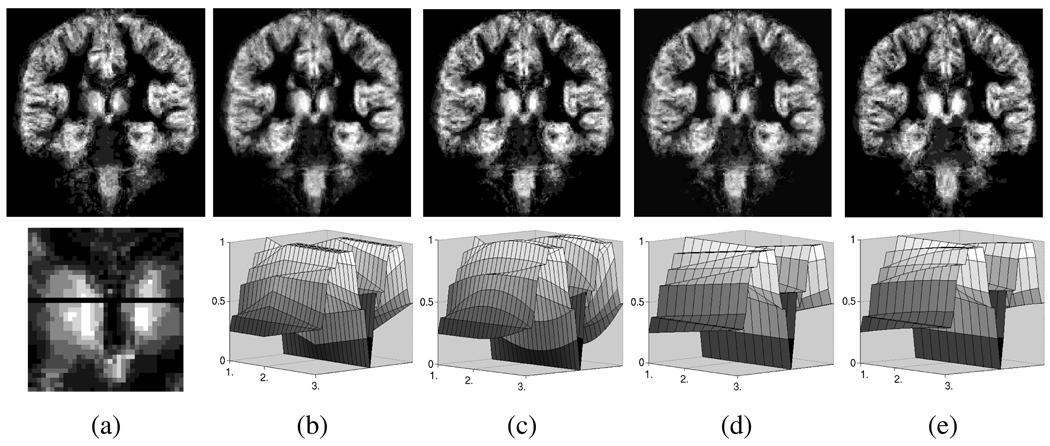
The first row of Figure 4 shows sample slices of the quadratic spline interpolation of the SCPs of gray matter. Bright indicates high and dark low probability of the gray matter. An area of particular interest is the region around the thalamus (in the center of the image) as the corresponding SCP changes substantially throughout the scans.
Fig. 4.
The first row shows a sample slice of the quadratic spline interpolation of a longitudinal schizophrenia study. Each image represents an SCP of the gray matter at a specific point in time of the study. Bright indicates high and dark low probability of the gray matter. The second row shows the SCP of the thalamus (a) with black indicating the voxels that are interpolated over three time point in (b)+(c) and two time points in (d) +(e). Graph (b)+(d) were produce by convex interpolation, while the smoother quadratic spline interpolation is representation (c)+(e).
The second row of Figure 4 shows a magnified version of the SCP of the thalamus region (a), and the corresponding CC (b)+(d) and quadratic spline interpolations (c)+(d). In the graphs, the z-axis symbolizes the probability, the x-axis is the time axis, and the y-axis represents the row of voxels highlighted by the black line in Figure 4 (a).
The graph of Figure 4 (b) is the CC of the SCPs from the three time points. The smoother interpolation in Figure 4 (c) is generated from quadratic splines within the LogOdds space. Unlike (b), this interpolation is differentiable over time.
In order to compare the accuracy of the interpolations, we repeat the experiment but now compute both functions just on the basis of the first and third scan. Samples of the results are shown the graphs of Figure 4(d)+(e). We then calculate the sum of squared errors of the two interpolations by comparing them to the SCP of the second scan. The quadratic spline interpolation achieves a 2% lower error for the gray matter and 5% lower error for the white matter in comparison to the CC. The quadratic spline interpolation may achieve this higher accuracy because it is smoother.
In this section, we applied our representation to the automatic segmentation of MR images as well as the interpolation of longitudinal data sets. We note that LogOdds maps can be used for a variety of other problems in medical imaging. For example, we have found the representation very helpful for capturing the variations between expert segmentations, though we do not discuss this topic here due to space limitations.
4 Conclusion
We presented a new shape representation called LogOdds. The representation can encode a single shape as well as its variations. A generalization of distance maps, this representation defines a vector space with a probabilistic interpretation.
We evaluate LogOdds within a voxel based segmentation algorithm. 20 subjects are automatically segmented into subcortical structures using the LogOdds and signed distance map model. The algorithm performs best with our new shape representation as LogOdds better capture the boundary constraints of anatomical structures.
In a second experiment, we use quadratic splines to interpolate SCPs from a longitudinal schizophrenia study. The splines are computed within the LogOdds space to simplify the calculations. We show that this models achieves a higher accuracy than the convex combination of SCPs in the original space.
Acknowledgments
This study was supported by the Department of Veterans Affairs Merit Awards, and grants from the NIH (R01 MH 40799, K05 MH 70047, R01 MH 50747, NIBIB NAMIC U54 EB005149, NCRR mBIRN U24-RR021382, NCRR NAC P41-RR13218, NINDS R01-NS051826) and NSF (JHU ERC CISST). We thank Mark Dreusicke from the Psychiatry Neuroimaging Laboratory, Harvard Medical School, for organizing the longitudinal study data and Torsten Rohlfing for his helpful comments.
Contributor Information
Kilian M. Pohl, Email: pohl@csail.mit.edu.
John Fisher, Email: fisher@csail.mit.edu.
Martha Shenton, Email: shenton@bwh.harvard.edu.
Robert W. McCarley, Email: mccarley@bwh.harvard.edu.
W. Eric L. Grimson, Email: welg@csail.mit.edu.
Ron Kikinis, Email: kikinis@bwh.harvard.edu.
William M. Wells, Email: sw@bwh.harvard.edu.
References
- 1.Leventon M, Grimson W, Faugeras O. CVPR. 2000. Statistical shape influence in geodesic active contours; pp. 1316–1323. [Google Scholar]
- 2.Tsai A, Yezzi A, Wells W, Tempany C, Tucker D, Fan A, Grimson W, Willsky A. A shape-based approach to the segmentation of medical imagery using level sets. TMI. 2003;vol. 22(no. 2):137–154. doi: 10.1109/TMI.2002.808355. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Staib LH, Duncan JS. Neighbor-constrained segmentation with level set based 3D deformable models. TMI. 2004;vol. 23(no. 8):940–948. doi: 10.1109/TMI.2004.830802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pohl K, Fisher J, Levitt J, Shenton M, Kikinis R, Grimson W, Wells W. A unifying approach to registration, segmentation, and intensity correction. MICCAI, LNCS. 2005:310–318. doi: 10.1007/11566465_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golland P, Grimson W, Shenton M, Kikinis R. Detection and analysis of statistical differences in anatomical shape. MIA. 2005;vol. 9:69–86. doi: 10.1016/j.media.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouix S. Medial Surfaces. PhD thesis. McGill University; 2003. [Google Scholar]
- 7.Collins D, Zijdenbos A, Barre W, Evans A. ANIMAL+INSECT: Improved cortical structure segmentation. IPMI. 1999;vol. 1613 [Google Scholar]
- 8.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. TMI. 1999;vol. 18(no. 10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 9.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Sgonne F, Salat D, Busa E, Seidman L, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale A. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;vol. 14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 10.Pohl KM, Fisher J, Grimson W, Kikinis R, Wells W. A bayesian model for joint segmentation and registration. NeuroImage. 2006;vol. 31(no. 1):228–239. doi: 10.1016/j.neuroimage.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 11.Arsigny V, Fillard P, Pennec X, Ayache N. Geometric means in a novel vector space structure on symmetric positive-definite matrices. SIAM JMAA. 2006 [Google Scholar]
- 12.Pohl K, Fisher J, Kikinis R, Grimson W, Wells W. ICCV, vol. 3765 of LNCS. Springer-Verlag; 2005. Shape based segmentation of anatomical structures in magnetic resonance images; pp. 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dice L. Measure of the amount of ecological association between species. Ecology. 1945;vol. 26(no. 3):297–302. [Google Scholar]
- 14.Zöllei L, Learned-Miller E, Grimson W, Wells W. Efficient population registration of 3D data. ICCV, vol. 3765 of LNCS. 2005:291–301. [Google Scholar]
- 15.Pohl K, Bouix S, Kikinis R, Grimson W. ISBI. Arlington, VA, USA: 2004. Anatomical guided segmentation with non-stationary tissue class distributions in an expectation-maximization framework; pp. 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]