Abstract
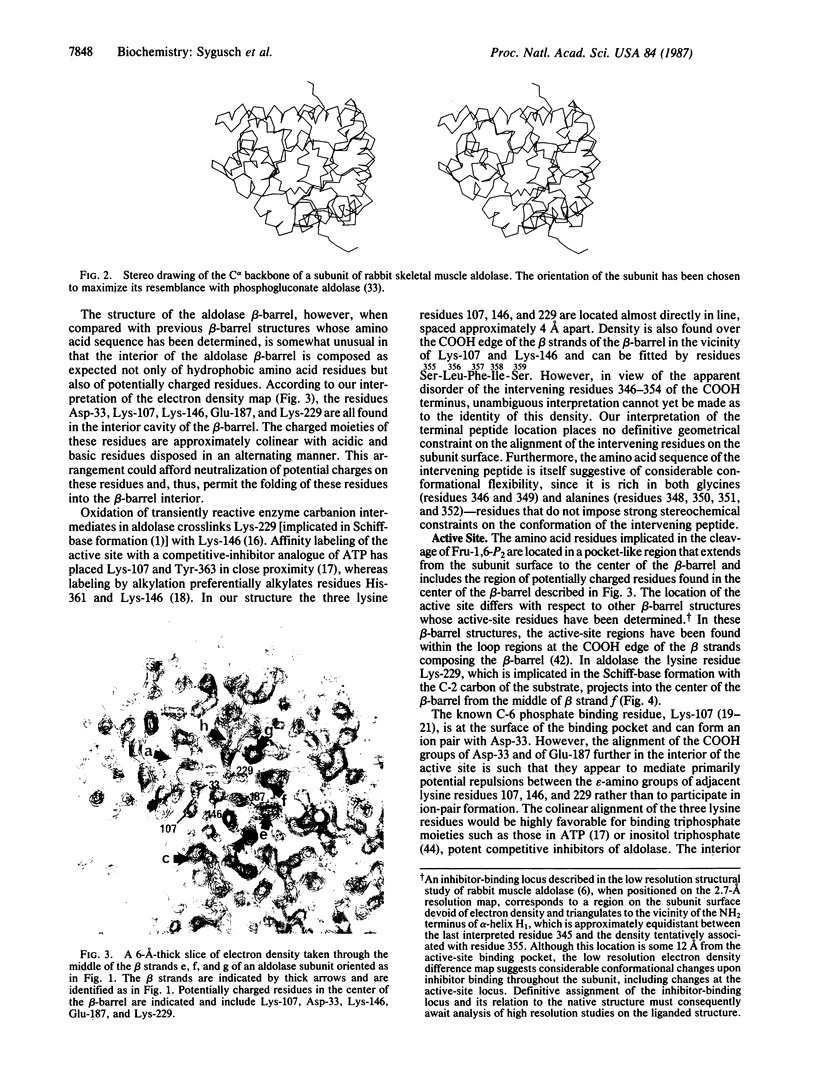
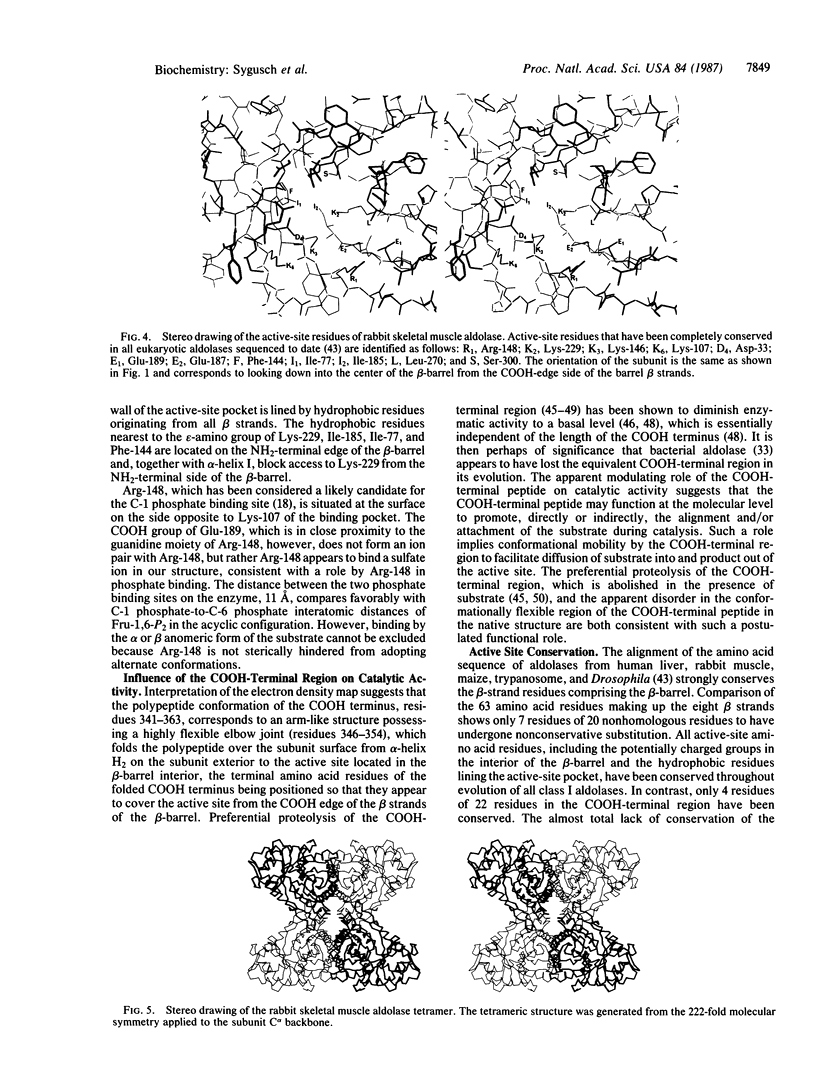
The molecular architecture of the rabbit skeletal muscle aldolase (D-fructose-1,6-bisphosphate D-glyceraldehyde-3-phosphate-lyase, EC 4.1.2.13) tetramer has been determined to 2.7-A resolution. Solution of the three-dimensional structure of rabbit muscle aldolase utilized phase information from a single isomorphous Pt(CN)4(2-) derivative, which was combined with iterative-phase refinement based upon the noncrystallographic 222-fold symmetry exhibited by the tetramer subunits. The electron-density map calculated from the refined phases (mf = 0.72) was interpreted on the basis of the known amino acid sequence (363 amino acids per subunit). The molecular architecture of the aldolase subunit corresponds to a singly wound beta-barrel of the parallel alpha/beta class structures as has been observed in triose phosphate isomerase, pyruvate kinase, phosphogluconate aldolase, as well as others. Close contacts between tetramer subunits are virtually all between regions of hydrophobic residues. Contrary to other beta-barrel structures, the known active-site residues are located in the center of the beta-barrel and are accessible to substrate from the COOH side of the beta-barrel. Biochemical and crystallographic data suggest that the COOH-terminal region of aldolase covers the active-site pocket from the COOH side of the beta-barrel and mediates access to the active site. On the basis of sequence studies, active-site residues as well as residues lining the active-site pocket have been totally conserved throughout evolution. By comparison, homology in the COOH-terminal region is minimal. It is suggested that the amino acid sequence of the COOH-terminal region may be, in part, the basis for the variable specific activities aldolases exhibit toward their substrates.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anai M., Lai C. Y., Horecker B. L. The pyridoxal phosphate-binding site of rabbit muscle aldolase. Arch Biochem Biophys. 1973 Jun;156(2):712–719. doi: 10.1016/0003-9861(73)90324-x. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., Corran P. H., Furth A. J., Milman J. D., Offord R. E. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975 Jun 19;255(5510):609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Benfield P. A., Forcina B. G., Gibbons I., Perham R. N. Extended amino acid sequences around the active-site lysine residue of class-I fructose 1,6-bisphosphate aldolases from rabbit muscle, sturgeon muscle, trout muscle and ox liver. Biochem J. 1979 Nov 1;183(2):429–444. doi: 10.1042/bj1830429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J. S., Barrett A. J. Degradation of fructose-1,6-bisphosphate aldolase by cathepsin B. Biochem J. 1980 Jul 1;189(1):17–25. doi: 10.1042/bj1890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner-Holzach O., Smit J. D. Crystallization and preliminary crystallographic data for fructose-1,6-bisphosphate aldolase from Drosophila melanogaster. J Biol Chem. 1982 Oct 10;257(19):11747–11749. [PubMed] [Google Scholar]
- Carrell H. L., Rubin B. H., Hurley T. J., Glusker J. P. X-ray crystal structure of D-xylose isomerase at 4-A resolution. J Biol Chem. 1984 Mar 10;259(5):3230–3236. [PubMed] [Google Scholar]
- Eagles P. A., Johnson L. N., Joynson M. A., McMurray C. H., Gutfreund H. Subunit structure of aldolase: chemical and crystallographic evidence. J Mol Biol. 1969 Nov 14;45(3):533–544. doi: 10.1016/0022-2836(69)90310-6. [DOI] [PubMed] [Google Scholar]
- GRAZI E., CHENG T., HORECKER B. L. The formation of a stable aldolase-dihydroxyacetone phosphate complex. Biochem Biophys Res Commun. 1962 Apr 20;7:250–253. doi: 10.1016/0006-291x(62)90184-5. [DOI] [PubMed] [Google Scholar]
- Hannappel E., MacGregor J. S., Davoust S., Horecker B. L. Limited proteolysis of liver and muscle aldolases: effects of subtilisin, cathepsin B, and Staphylococcus aureus protease. Arch Biochem Biophys. 1982 Mar;214(1):293–298. doi: 10.1016/0003-9861(82)90033-9. [DOI] [PubMed] [Google Scholar]
- Hartman F. C., Brown J. P. Affinity labeling of a previously undetected essential lysyl residue in class I fructose bisphosphate aldolase. J Biol Chem. 1976 May 25;251(10):3057–3062. [PubMed] [Google Scholar]
- Healy M. J., Christen P. Mechanistic probes for enzymatic reactions. Oxidation-reduction indicators as oxidants of intermediary carbanions (studies with aldolase, aspartate aminotransferase, pyruvate decarboxylase, and 6-phosphogluconate dehydrogenase). Biochemistry. 1973 Jan 2;12(1):35–41. doi: 10.1021/bi00725a006. [DOI] [PubMed] [Google Scholar]
- Heidner E. G., Weber B. H., Eisenberg D. Subunit structure of aldolase. Science. 1971 Feb 19;171(3972):677–679. doi: 10.1126/science.171.3972.677. [DOI] [PubMed] [Google Scholar]
- Humphreys L., Reid S., Masters C. Evidence for the spatial separation of the binding sites for substrate and for cytoskeletal proteins on the enzyme aldolase. Int J Biochem. 1986;18(1):7–13. doi: 10.1016/0020-711x(86)90003-0. [DOI] [PubMed] [Google Scholar]
- Kasprzak A. A., Kochman M. Interaction of fructose-1,6-bisphosphate aldolase with adenine nucleotides. Binding of 5'-mononucleotides and phosphates to rabbit muscle aldolase. Eur J Biochem. 1980 Mar;104(2):443–450. doi: 10.1111/j.1432-1033.1980.tb04446.x. [DOI] [PubMed] [Google Scholar]
- Kelley P. M., Tolan D. R. The complete amino Acid sequence for the anaerobically induced aldolase from maize derived from cDNA clones. Plant Physiol. 1986 Dec;82(4):1076–1080. doi: 10.1104/pp.82.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppitz B., Vogel F., Mayr G. W. Mammalian aldolases are isomer-selective high-affinity inositol polyphosphate binders. Eur J Biochem. 1986 Dec 1;161(2):421–433. doi: 10.1111/j.1432-1033.1986.tb10462.x. [DOI] [PubMed] [Google Scholar]
- LAI C. Y., TCHOLA O., CHENG T., HORECKER B. L. THE MECHANISM OF ACTION OF ALDOLASES. 8. THE NUMBER OF COMBINING SITES IN FRUCTOSE DIPHOSPHATE ALDOLASE. J Biol Chem. 1965 Mar;240:1347–1350. [PubMed] [Google Scholar]
- Lai C. Y., Nakai N., Chang D. Amino acid sequence of rabbit muscle aldolase and the structure of the active center. Science. 1974 Mar;183(130):1204–1206. doi: 10.1126/science.183.4130.1204. [DOI] [PubMed] [Google Scholar]
- Lebioda L., Hatada M. H., Tulinsky A., Mavridis I. M. Comparison of the folding of 2-keto-3-deoxy-6-phosphogluconate aldolase, triosephosphate isomerase and pyruvate kinase. Implications in molecular evolution. J Mol Biol. 1982 Dec 5;162(2):445–458. doi: 10.1016/0022-2836(82)90537-x. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y., Brändén C. I. Structure of glycolate oxidase from spinach. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6855–6859. doi: 10.1073/pnas.82.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubini D. G., Christen P. Paracatalytic modification of aldolase: a side reaction of the catalytic cycle resulting in irreversible blocking of two active-site lysyl residues. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2527–2531. doi: 10.1073/pnas.76.6.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midelfort C. F., Mehler A. H. A chymotrypsin-catalyzed modification of rabbit muscle aldolase. J Biol Chem. 1972 Jun 10;247(11):3618–3621. [PubMed] [Google Scholar]
- Millar J. R., Shaw P. J., Stammers D. K., Watson H. C. The low-resolution structure of human muscle aldolase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):209–214. doi: 10.1098/rstb.1981.0074. [DOI] [PubMed] [Google Scholar]
- Model P., Ponticorvo L., Rittenberg D. Catalysis of an oxygen-exchange reaction of fructose 1,6-diphosphate and fructose 1-phosphate with water by rabbit muscle aldolase. Biochemistry. 1968 Apr;7(4):1339–1347. doi: 10.1021/bi00844a014. [DOI] [PubMed] [Google Scholar]
- Offermann M. K., McKay M. J., Marsh M. W., Bond J. S. Glutathione disulfide inactivates, destabilizes, and enhances proteolytic susceptibility of fructose-1,6-bisphosphate aldolase. J Biol Chem. 1984 Jul 25;259(14):8886–8891. [PubMed] [Google Scholar]
- Palczewski K., Hargrave P. A., Folta E. J., Kochman M. Affinity labeling of rabbit muscle fructose-1,6-bisphosphate aldolase with 5'-[p-(fluorosulfonyl)benzoyl]-1,N6-ethenoadenosine. Eur J Biochem. 1985 Jan 15;146(2):309–314. doi: 10.1111/j.1432-1033.1985.tb08654.x. [DOI] [PubMed] [Google Scholar]
- Penhoet E. E., Kochman M., Rutter W. J. Molecular and catalytic properties of aldolase C. Biochemistry. 1969 Nov;8(11):4396–4402. doi: 10.1021/bi00839a026. [DOI] [PubMed] [Google Scholar]
- Penhoet E., Kochman M., Valentine R., Rutter W. J. The subunit structure of mammalian fructose diphosphate aldolase. Biochemistry. 1967 Sep;6(9):2940–2949. doi: 10.1021/bi00861a039. [DOI] [PubMed] [Google Scholar]
- Penhoet E., Rajkumar T., Rutter W. J. Multiple forms of fructose diphosphate aldolase in mammalian tissues. Proc Natl Acad Sci U S A. 1966 Oct;56(4):1275–1282. doi: 10.1073/pnas.56.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSE I. A., O'CONNELL E. L., MEHLER A. H. MECHANISM OF THE ALDOLASE REACTION. J Biol Chem. 1965 Apr;240:1758–1765. [PubMed] [Google Scholar]
- RUTTER W. J. EVOLUTION OF ALDOLASE. Fed Proc. 1964 Nov-Dec;23:1248–1257. [PubMed] [Google Scholar]
- Richardson J. S. Describing patterns of protein tertiary structure. Methods Enzymol. 1985;115:341–358. doi: 10.1016/0076-6879(85)15025-1. [DOI] [PubMed] [Google Scholar]
- Rose I. A., O'Connell E. L. Studies on the interaction of aldolase with substrate analogues. J Biol Chem. 1969 Jan 10;244(1):126–134. [PubMed] [Google Scholar]
- Sajgó M., Hajós G. The amino acid sequence of rabbit muscle aldolase. Acta Biochim Biophys Acad Sci Hung. 1974;9(3):239–241. [PubMed] [Google Scholar]
- Sawyer L. A fourth crystal form of rabbit muscle aldolase. J Mol Biol. 1972 Nov 14;71(2):503–505. doi: 10.1016/0022-2836(72)90365-8. [DOI] [PubMed] [Google Scholar]
- Schneider G., Lindqvist Y., Brändén C. I., Lorimer G. Three-dimensional structure of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum at 2.9 A resolution. EMBO J. 1986 Dec 20;5(13):3409–3415. doi: 10.1002/j.1460-2075.1986.tb04662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamala N., Lim L. W., Mathews F. S., McIntire W., Singer T. P., Hopper D. J. Structure of an intermolecular electron-transfer complex: p-cresol methylhydroxylase at 6.0-A resolution. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4626–4630. doi: 10.1073/pnas.83.13.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro S., Enser M., Pugh E., Horecker B. L. The effect of pyridoxal phosphate on rabbit muscle aldolase. Arch Biochem Biophys. 1968 Nov;128(2):554–562. doi: 10.1016/0003-9861(68)90062-3. [DOI] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Sygusch J., Beaudry D. Catalytic activity of rabbit skeletal muscle aldolase in the crystalline state. J Biol Chem. 1984 Aug 25;259(16):10222–10227. [PubMed] [Google Scholar]
- Sygusch J., Beaudry D. Preliminary crystallographic investigation of rabbit liver aldolase. J Mol Biol. 1985 Nov 5;186(1):215–217. doi: 10.1016/0022-2836(85)90274-8. [DOI] [PubMed] [Google Scholar]
- Sygusch J., Boulet H., Beaudry D. Structure of rabbit muscle aldolase at low resolution. J Biol Chem. 1985 Dec 5;260(28):15286–15290. [PubMed] [Google Scholar]
- Tolan D. R., Amsden A. B., Putney S. D., Urdea M. S., Penhoet E. E. The complete nucleotide sequence for rabbit muscle aldolase A messenger RNA. J Biol Chem. 1984 Jan 25;259(2):1127–1131. [PubMed] [Google Scholar]
- Xia Z. X., Shamala N., Bethge P. H., Lim L. W., Bellamy H. D., Xuong N. H., Lederer F., Mathews F. S. Three-dimensional structure of flavocytochrome b2 from baker's yeast at 3.0-A resolution. Proc Natl Acad Sci U S A. 1987 May;84(9):2629–2633. doi: 10.1073/pnas.84.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]



