Abstract
Native tandem mass spectrometry reveals marked differences in the rates at which the two polymorphic forms of the HBV capsid exchange dimeric subunits with the soluble pool.
Hepatitis B virus (HBV) is a major cause of liver disease in humans, and the leading cause of hepatic cancer.1 The development of new antiviral therapies targeting the viral capsid, and a growing interest in utilizing engineered adaptations thereof as drug delivery vehicles, is driving the need for precise biophysical characterization of these particles, including their dynamic properties.2 The structures of HBV capsids and their building-blocks have been determined by cryo-electron microscopy,3 X-ray crystallography,4,5 and NMR spectroscopy.6 Native (intra-viral) capsids are polymeric shells of a 183-residue polypeptide, whose first 149 residues suffice for capsid assembly. This assembly domain forms dimers wherein the two chains pair via formation of a central four-helix bundle. These dimers serve as building-blocks that naturally assemble into capsids of two sizes, corresponding to triangulation numbers of T = 3 (90 dimers and 32.0 nm diameter) and T = 4 (120 dimers and 35.5 nm diameter), wherein the four-helix bundles form the capsids’ characteristic protrusions.3 Recently, we have employed tandem mass spectrometry (MS/MS) to investigate the molecular composition of both HBV capsids and their stability in vacuo.7,8 These particles proved to be particularly suitable for analysis by MS, yielding mass measurements with a precision of <0.1% and thereby demonstrating for the first time that the lattices of both capsids are complete, consisting of exactly 90 and 120 dimers, respectively.
Any viral capsid must be very stable to afford protection to the enclosed genome during transit between hosts; however, it must also be capable of releasing the genome—and in some cases, completely disassembling—when the appropriate compartment of an infected cell is reached. Similarly, effective drug delivery requires cargo release and/or vehicle disassembly at the targeted site. Whereas capsid structures determined by X-ray crystallography and cryo-electron microscopy give the impression of static stable structures, there is evidence—particularly for picornaviruses, which have icosahedral capsids of similar size to those of HBV—that capsids can be quite dynamic.9–11 Thus polypeptides visualized to be internal to the closed capsid shell were found to be accessible to proteases and antibodies in the external milieu, implying that they are transiently exposed on the outer surface. This phenomenon has been termed “breathing”.
Whereas the assembly of HBV capsids has been studied extensively (see above), little is yet known about their dynamic properties and disassembly. Singh and Zlotnick12 observed hysteresis in HBV capsid assembly/disassembly and proposed that weak protein–protein interactions can account for both capsid stability and breathing. Here we have exploited the high mass discrimination achievable by MS/MS, in conjunction with isotopic labeling, to examine dimer exchange under dilute conditions in the absence of denaturants, for both capsid forms.
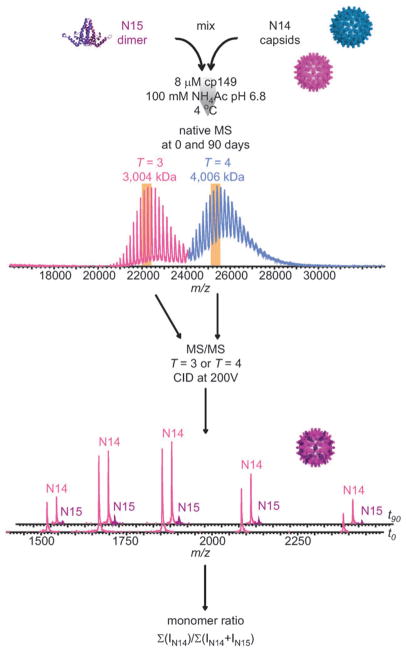
The proteins analyzed were prepared as follows. The 149-residue assembly domain (hereafter Cp149) was expressed in E. coli, purified as dimers (Cp149d), and stored in 100 mM sodium carbonate (pH 9.6), as described previously.13,14 The three cysteine residues in the native protein were substituted with alanine in the construct used. The corresponding 15N-labelled protein (15N-Cp149d) was prepared in a similar manner. Cp149d was induced to assemble into capsids (Cp149c) by adjusting solution conditions to 150 mM sodium chloride and 100 mM HEPES (pH 7.0) by gel filtration.13,14 The ratio of T = 3 : T = 4 was 43 : 57, as determined by counting 1261 capsids in cryo-electron micrographs (the wild-type Cp149 assembly domain gives a majority of T = 4 capsids).14 To observe exchange, it was necessary to co-incubate the proteins under conditions where Cp149c and Cp149d co-exist under equilibrium conditions. Cp149 remains dimeric at low ionic strength and high pH (ca. 9.6) at 4°C, but assembles into Cp149c when the ionic strength is raised and the pH is adjusted to neutral, as described above. Raising the temperature also drives the assembly reaction. Once assembled, however, Cp149c remain fully assembled at low ionic strength and high pH.12 In view of these considerations, the incubations described below were conducted under conditions of low ionic strength, near-neutral pH, and low temperature, unless otherwise indicated. Ammonium acetate was employed because previous experiments indicated that the proteins were properly folded, assembled, and soluble in this buffer and also because of its compatibility with the MS.7,8 To test for dimer exchange, Cp149c were mixed with 15N-Cp149d in equimolar ratio (in terms of dimers) at a total monomer concentration of 8 μM in 100 mM ammonium acetate (pH 6.8) at 4°C. Controls to assess the long term integrity of the Cp149c and 15N-Cp149d were prepared under identical conditions. Immediately after mixing, and after 90 days of incubation, samples were withdrawn and analyzed by MS/MS using a modified Q-ToF1 instrument (Waters, UK) as described previously.15,16 Precursor ions at m/z values of 21700 and 24800, corresponding to T = 3 and T = 4 Cp149c, respectively, were selected and subjected to collision-induced dissociation (CID) at an accelerating voltage of 200 V, whereby, as shown previously, Cp149 monomers were ejected from Cp149c.7 The relative amount of 15N-Cp149 monomer, compared to the amount of 14N-Cp149 monomer, is therefore a measure of the amount of Cp149d exchanged between the capsids and free ones in solution. In other words, monomers constitute the readout but dimers are exchanged in solution. Denaturation studies of dimers show that they are very stable, with no evidence of separation of monomers prior to global unfolding.3
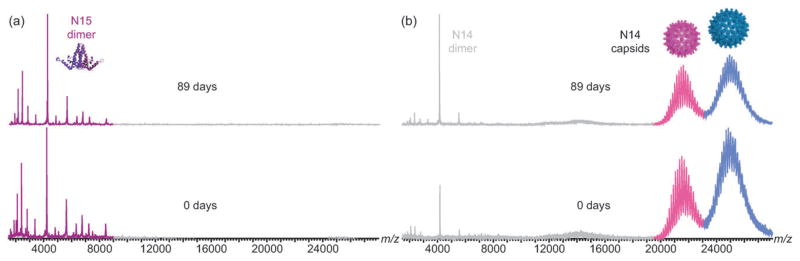
To monitor for dimer exchange, 14N-containing Cp149c were incubated with 15N-Cp149d and analyzed by MS/MS as described above. As depicted in Fig. 1, after 90 days a significant number of 15N-Cp149 monomer ions were detected, indicating that Cp149d exchange had occurred between the T = 3 Cp149c and the soluble pool. In contrast, essentially no incorporation was detected in the case of T = 4 Cp149c. The long-term integrity of the Cp149c and 15N-Cp149d controls incubated separately was assessed and confirmed as shown in Fig. 2. Both proteins were found to be completely stable under these conditions for at least 89 days; neither did Cp149d assemble to Cp149c nor did Cp149c dissociate to Cp149d. Dynamic light scattering and circular dichroism measurements of the proteins did not change over this time period (not shown).
Fig. 1.
Monitoring HBV capsid breathing by MS/MS. 15N-Cp149d was mixed with Cp149c and then analyzed at intervals for up to 90 days by MS/MS. Precursor ions of T = 3 and T = 4 Cp149c were selected (represented by vertical tan-colored bands) and individually subjected to CID. The relative amount of 15N-Cp149d incorporated into the intact Cp149c was determined from the ion intensities of the ejected monomers in the MS/MS spectrum, as shown for the T = 3 Cp149c. Monomer ratios were obtained based on the summed intensities of the different charge states.
Fig. 2.
Long-term integrity of capsids and dimers. MS of the 15N-Cp149d (a) and Cp149c (b) controls at 0 and 89 days of incubation at 4°C shows that the proteins were stable. The 15N-Cp149d did not assemble into capsids, and Cp149c did not dissociate to dimers.
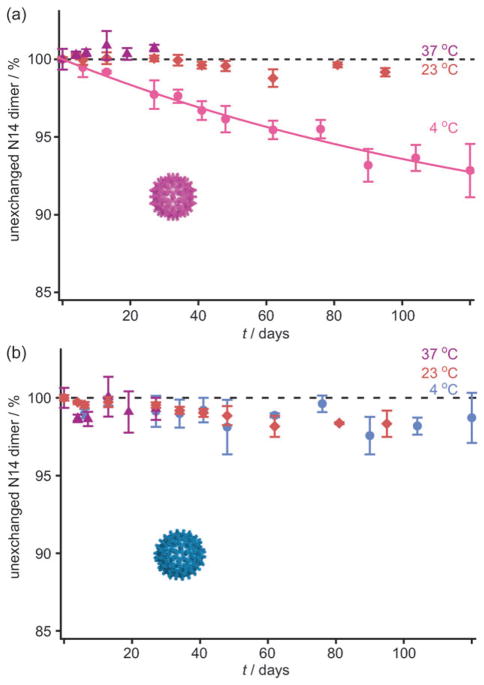
To investigate the temperature dependence of Cp149d exchange, we incubated Cp149c with 15N-Cp149d at different temperatures. Samples were withdrawn at intervals for up to 120 days and analyzed by MS/MS. The results are shown in Fig. 3. No significant exchange of Cp149d was observed in either capsid at the elevated temperatures (23°C and 37°C) but a significant portion of the Cp149d in T = 3 Cp149c was exchanged at 4°C (Fig. 3a). An exponential fit to the data suggests that only a subfraction of the dimers are subject to exchange. No significant exchange of Cp149d was observed in the case of T = 4 Cp149c at any of the tested temperatures (Fig. 3b).
Fig. 3.
Subunit exchange depends on capsid geometry and temperature. Dimer exchange was monitored by MS/MS at 4°C, 23°C, and 37°C for both T = 3 (a) and T = 4 (b) Cp149c for up to 120 days. No significant exchange was observed at 23°C (orange) and 37°C (purple) for either geometry. At 4°C, the T = 3 Cp149c exchanged Cp149d with free ones in solution (pink), an exponential fit is shown indicating an endpoint of approximately 13%. T = 4 Cp149c do not show significant levels of exchange at the same temperature (blue). Error bars correspond to the standard deviation of triplicate analysis. The dashed line represents the 100% level.
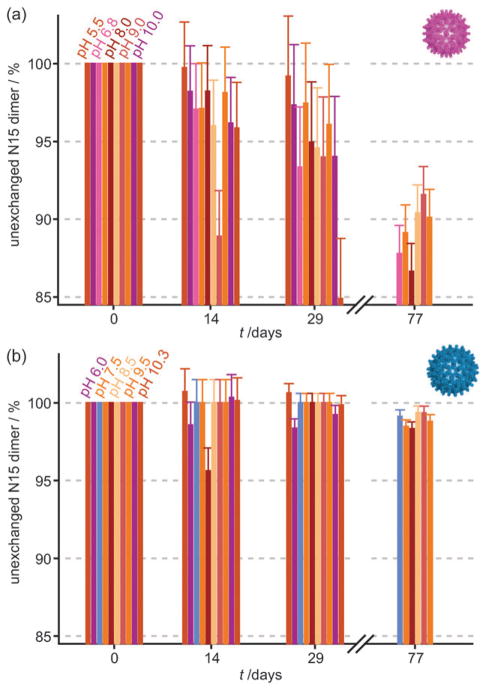
To detect a potential pH-dependence for Cp149d exchange, incubations were done at pH values ranging from 5.5 to 10.3 in 100 mM ammonium acetate at 4°C. Samples were withdrawn at intervals for up to 80 days and analyzed by MS. The results are shown in Fig. 4. For both T = 3 and T = 4 Cp149c there was no significant pH-dependence. Both capsid geometries appeared stable across this pH range as assessed by native gel electrophoresis (not shown), however, the protein was observed to aggregate at more acidic pH. Attempts to assess the ionic strength dependence of this system by additions of sodium chloride were precluded by the incompatibility of this salt with the MS.
Fig. 4.
Exchange rates are largely unaffected by change in pH. Dimer exchange was monitored in samples taken from incubations with pH values ranging from 5.5 to 10.3, for both T = 3 (a) and T = 4 Cp149c (b). Significant exchange occurred only in the T = 3 form, but there was no clear pH dependency for either geometry.
Both forms of Cp149c are stable, typically requiring > 3 M urea and pH 9.6 for dissociation. As to their relative stabilities, simple curvature or vertex–proximity considerations17 predict that the T = 3 structure should be less stable, i.e. less resistant to mechanical stress. However, atomic-force microscopy measurements of deformability have detected no difference between the two forms.7 On the other hand, results from CID during MS/MS—in which the capsids are subjected to a different kind of stress (being transferred in vacuo)—showed the T = 3 form to better maintain its integrity under these conditions.8 The results presented here demonstrate that the T = 3 capsid exchanges dimers with the soluble pool slowly but definitely over a period of months, whereas the T = 4 capsid exchanges much more slowly, if at all. This raises the questions of where and how exchange might occur.
Capsid assembly is increased in vitro by raising the temperature (our unpublished observations), and, as shown here, dimer exchange is greater in the cold. Assembly of capsids is driven by the burial of hydrophobic surface area.18 As it is well known that hydrophobic interactions decrease in strength with decreasing temperature19 this suggests that exchange occurs by the selective extraction of dimers with weaker hydrophobic contacts. In the two HBV capsids, there is a total of seven quasi-equivalent subunits with their associated environments; three (A, B, C) in the case of T = 3 and four (A, B, C, D) in the case of T = 4 (Fig. 5).3 Similarly, two kinds of dimers are distinguished by quasi-equivalence in each capsid: 60 A–B dimers and 30 C–C dimers in the T = 3 capsid, and 60 A–B dimers and 60 C–D dimers in the T = 4 capsid. The data suggest that only a small subset of the dimers in the T = 3 capsid are exchangeable. In this regard we note that the T = 3 C–C dimer is the only one within the two geometries that lies across two facets of the icosahedron (Fig. 5a). It is plausible that this extra curvature results in less stable contacts between the dimer and its neighbors.
Fig. 5.
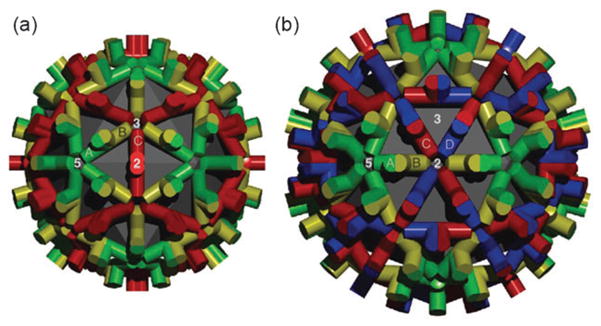
Surface lattice of T = 3 and T = 4 HBV capsids. Shown is the arrangement of dimers in the T = 3 (a) and T = 4 (b) icosahedral geometries. The quasi-equivalent sites are labelled in both cases (T = 3; A, B, C and T = 4; A, B, C, D). Note that sites with the same symbol do not have identical structures. The 5-, 3-, and 2-fold axes of symmetry are also indicated. In the T = 3 lattice, the red C dimer lies on the two-fold axis of symmetry, and across two facets of the icosahedron. No such environment exists in the T = 4 geometry. Figure reproduced from ref. 3.
In conclusion, we report the first measurements of the rates at which the two polymorphic variants of the HBV capsid exchange dimeric building-blocks with the soluble pool. Exchange is slow but real for the T = 3 capsid (~7% in 3 months) but unmeasurably slow for the T = 4 capsid. The results have implications for drug delivery vehicle stability and formulation, and for the process of uncoating during the course of natural HBV infection (also termed “breathing”),20 for instance, that the dissociation probably does not proceed spontaneously but instead involves an active mechanism.
Acknowledgments
We thank Ira Palmer and Joshua Kaufman for preparation of the proteins, and Klaus H. P. Gast and R. Seckler at the Physical Biochemistry Department of Potsdam University, Germany, for performing the dynamic light scattering and circular dichroism experiments. This work was supported by the Netherlands Proteomics Centre and by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Abbreviations
- CID
collision induced dissociation
- Cp149
capsid protein
- Cp149d
dimeric capsid protein
- Cp149c
capsid
- HBV
Hepatitis B virus
- MS
mass spectrometry
- MS/MS
tandem MS
References
- 1.Lesmana LA, Leung NW, Mahachai V, Phiet PH, Suh DJ, Yao G, Zhuang H. Liver Int. 2006;26(Suppl 2):3. [Google Scholar]
- 2.Singh P, Gonzales MJ, Manchester M. Drug Dev Res. 1996;67:23. [Google Scholar]
- 3.Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, Harris A, Stahl SJ, Wingfield PT. Adv Virus Res. 2005;64:127. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 4.Wynne SA, Crowther RA, Leslie AG. Mol Cell. 1999;3:771. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 5.Packianathan C, Katen SP, Dann CE, III, Zlotnick A. J Virol. 2010;84:1607. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freund SM, Johnson CM, Jaulent AM, Ferguson N. J Mol Biol. 2008;384:1301. doi: 10.1016/j.jmb.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJ, Wingfield PT, Steven AC, Heck AJ. Proc Natl Acad Sci U S A. 2008;105:9216. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uetrecht C, Versluis C, Watts NR, Wingfield PT, Steven AC, Heck AJ. Angew Chem, Int Ed. 2008;47:6247. doi: 10.1002/anie.200802410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis JK, Bothner B, Smith TJ, Siuzdak G. Proc Natl Acad Sci U S A. 1998;95:6774. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katpally U, Fu TM, Freed DC, Casimiro DR, Smith TJ. J Virol. 2009;83:7040. doi: 10.1128/JVI.00557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Yafal AG, Lee YM, Hogle J, Chow M. J Virol. 1994;68:3965. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh S, Zlotnick A. J Biol Chem. 2003;278:18249. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 13.Wingfield PT, Stahl SJ, Williams RW, Steven AC. Biochemistry. 1995;34:4919. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 14.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. Biochemistry. 1996;35:7412. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 15.van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Heck AJ. Anal Chem. 2006;78:7473. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 16.Heck AJ. Nat Methods. 2008;5:927. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 17.Albertazzi E, Domene C, Fowler PW, Heine T, Seifert G, Van Alsenoy C, Zerbetto F. Phys Chem Chem Phys. 1999;1:2913. [Google Scholar]
- 18.Ceres P, Zlotnick A. Biochemistry. 2002;41:11525. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 19.Scheraga HA, Nemethy G, Steinberg IZ. J Biol Chem. 1962;237:2506. [PubMed] [Google Scholar]
- 20.Rabe B, Delaleau M, Bischof A, Foss M, Sominskaya I, Pumpens P, Cazenave C, Castroviejo M, Kann M. PLoS Pathog. 2009;5:e1000563. doi: 10.1371/journal.ppat.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]