Abstract
The epigenetic mechanisms involved in establishing and maintaining genomic imprinting are steadily being unmasked. The nucleosome remodeling and histone deacetylation (NuRD) complex is implicated in regulating DNA methylation and expression of the maternally expressed H19 gene in preimplantation mouse embryos. To dissect further the function of the NuRD complex in genomic imprinting, we employed an RNA interference (RNAi) strategy to deplete the NuRD complex component Metastasis Tumor Antigen 2 (MTA2). We found that Mta2 is the only zygotically expressed Mta gene prior to the blastocyst stage, and that RNAi-mediated knockdown of Mta2 transcript leads to biallelic H19 expression and loss of DNA methylation in the differentially methylated region in blastocysts. In addition, biallelic expression of the paternally expressed Peg3 gene, but not Snrpn, is also observed in blastocysts following Mta2 knockdown. Loss of MTA2 protein does not result in a decrease in abundance of other NuRD components, including methyl-binding-CpG-binding domain protein 3 (MBD3), histone deacetylases 1 and 2 (HDACs 1 and 2), and chromodomain helicase DNA-binding protein 4 (CHD4). Taken together, our results support a role for MTA2 within the NuRD complex in genomic imprinting.
Keywords: DNA methylation, embryo, genomic imprinting, H19, MTA, NuRD complex, Peg3, RNAi
The metastasis tumor antigen 2 (MTA2) protein, a component of the NuRD chromatin remodeling complex, is involved in genomic imprinting during mouse preimplantation development.
INTRODUCTION
Dynamic alterations in chromatin structure due to ATP-dependent remodeling and covalent histone modifications play important roles in regulating gene expression [1]. The nucleosome remodeling and histone deacetylation (NuRD) complex is ideally situated to modulate gene expression because it possesses both nucleosome remodeling and histone deacetylase activities [2–7]. The NuRD complex is approximately 2 MDa in size and in mammalian cells is composed of at least seven polypeptides [7–10]. Four of the components, histone deacetylases 1 (HDAC1) and HDAC2 and two histone-binding proteins (RBBP4/RbAp46 and RBBP7/RbAp48), form a deacetylase complex that is also found in the SIN3A histone deacetylase complex [11]. The other three components, chromodomain helicase DNA-binding protein 4 (CHD4 [Mi-2β]), methyl-binding-CpG-binding domain protein 3 (MBD3), and Metastasis Tumor Antigen (MTA), however, appear to be unique to the NuRD complex [12].
CHD4 is an ATP-dependent nucleosome-remodeling enzyme [7, 13]. Although mammalian MBD3 does not bind to methylated DNA [14], when NuRD binds MBD2, the resulting NuRD complex becomes an integral component of the MeCP1 complex [13, 15] that can bind to methylated DNA, thereby leading to histone deacetylation of DNA-methylated nucleosomes [13]. MTA proteins, which are the last characterized proteins of the NuRD complex [16], are encoded by three genes [17], and it appears that only one family member is found within a given NuRD complex [18].
MTA1, the founding member of the MTA family, is associated with metastatic growth of cell lines in vitro and with invasive growth of tumors [19]. In addition to MTA1, MTA3 represses WNT4 function in mammary epithelial cells and plays important roles in invasive growth of breast cancer cells by repressing transcription of Snail [18, 20]. In contrast to the restricted expression patterns of MTA1 and MTA3, MTA2 is ubiquitously expressed. Targeted mutation of the Mta2 gene results in multiple phenotypes that include partial embryonic and perinatal lethality, female infertility, abnormal T-cell activation, and lupuslike autoimmune disease in mice [21]. Although it is likely that the three MTA family members form different NuRD complexes with distinct functions [16, 22–25], the role of MTA proteins in the function of NuRD complexes remains elusive.
Genomic imprinting is an epigenetic process that results in parent-of-origin-specific gene expression of a small number of genes [26, 27]. In mammals, many imprinted genes are located in approximately 1-Mb clusters, with regulation of the linked genes controlled by a differentially methylated region (DMR), which exhibits allele-specific epigenetic modifications, such as DNA methylation or posttranslational histone modifications. Deletion of DMRs results in the loss-of-imprinting of multiple genes in cis. Allele-specific epigenetic modifications are established in the germline or zygote and must be subsequently maintained for development to proceed normally. Maintaining these marks is especially critical during preimplantation development because the paternal genome is actively demethylated shortly after fertilization, and then both genomes are passively demethylated during preimplantation development [28]. Moreover, a dramatic reprogramming of gene expression occurs during preimplantation development, first being observed during the two-cell stage, with changes in histone acetylation serving a regulatory function [29]. Remarkably, despite these dramatic changes in DNA methylation and gene expression, the imprinting marks that are established during gametogenesis are maintained by mechanisms that remain to be elucidated fully.
Although chromatin remodeling and histone modifications play crucial roles in genomic imprinting [30–32], a direct link between the NuRD complex and genomic imprinting is lacking. Results of our previous study implicate NuRD in regulating imprinted gene expression, because knockdown of MBD3 in preimplantation mouse embryos results in both biallelic expression of H19 and loss of DNA methylation within the imprinting control region (ICR) at the blastocyst stage [33]. We also noted in that study that MBD3 depletion is accompanied by a reduction in the amount of nuclear-localized MTA2, suggesting a role for MTA2 in genomic imprinting.
To define further the function of the NuRD complex in imprinting, and in particular MTA2, we used an RNA interference (RNAi) strategy to deplete MTA2 selectively. We found that Mta2 is the only zygotically expressed Mta gene, and that depleting Mta2 transcripts leads to biallelic expression of H19 and loss of DNA methylation on the ICR in blastocysts. In addition, biallelic expression of the paternally expressed Peg3, but not Snrpn, is also observed in blastocysts following Mta2 knockdown.
MATERIALS AND METHODS
Collection and Culture of Oocytes and Preimplantation Embryos
For double-stranded RNA (dsRNA) injections, oocytes and embryos were obtained as previously described from either CF1 (Harlan, http://www.harlan.com/) females (6–8 wk of age) mated to B6D2F1/J males (Jackson Laboratory) for embryo assays [34], or from C57BL/6J (Jackson Laboratory, http://www.jax.org/) females mated to B6(P12X) males for allelic assays [33, 35]. This latter strain has Mus musculus castaneus chromosomes 7, 12, and X in a C57BL/6 background. All experiments were conducted with the approval of the Institutional Animal Care and Use Committee at the University of Pennsylvania.
Cumulus cell-free, germinal vesicle (GV)-intact oocytes were obtained from equine chorionic gonadotrophin (eCG)-primed females as previously described [36]. The collection medium for oocytes was bicarbonate-free minimal essential medium (Earle salts) containing 25 mM Hepes, pH 7.3; 3 mg/ml polyvinylpyrrolidone; and 2.5 μM milrinone to prevent GV breakdown. Oocytes were matured in vitro in Whitten medium [37] containing 0.01% polyvinyl alcohol (PVA) (Whittens/PVA).
Meiotically incompetent oocytes (i.e., oocytes that do not spontaneously resume meiosis when placed in a suitable culture medium) were obtained from 12-day-old female mice. Ovaries were placed in Ca2+- and Mg2+-free CZB medium [38] containing collagenase I (Worthington, Lakewood, NJ) and DNase I (Sigma, St. Louis, MO); prior to use, the medium was filtered using a 0.2-μm filter. The ovaries were minced into small pieces and then incubated for 5–10 min in an atmosphere of 5% CO2 in air at 37°C. Oocytes were freed from follicles by repeated pipetting with a mouth-operated pipette, and liberated oocytes were removed and transferred to CZB medium. The remaining ovarian tissue was returned to the incubator, and after another 15–30 min the procedure was repeated.
Embryos were cultured in KSOM containing amino acids [39] for up to 4 days in 5% CO2 in air at 37°C. One-cell, two-cell, four-cell, and eight-cell embryos and blastocysts that developed in vivo were flushed from either oviducts or uteri at 20–21, 41–44, 60–61, 68, and 92–96 h after eCG, respectively [29].
Microinjection of one-cell embryos was performed essentially as previously described [29]. Prior to pronucleus formation, the embryos were injected with 10 pl of dsRNA using a Picoliter Injector Microinjection System (Harvard Apparatus, Holliston, MA); the culture medium was bicarbonate-free Whitten medium containing 0.01% PVA and 25 mM Hepes, pH 7.3. Following microinjection, the embryos were cultured in KSOM containing amino acids as described above.
dsRNA Preparation
Double-stranded RNA was prepared by annealing two complementary RNAs transcribed by T7 or SP6 polymerase in vitro. The cDNA fragments were initially subcloned into the PCRII vector. Mta2 dsRNA was a 529-bp fragment prepared with 5′-CACCAATCAACAGAAACCAGC-3′ and 5′-ACACACACACACCAGGGTCAA-3′.
After in vitro transcription using T7 and SP6 RNA polymerase (Ambion), DNA templates were removed by DNase I treatment. The RNA products were extracted with phenol:chloroform and then precipitated with ethanol. To anneal sense and antisense RNAs, equimolar quantities of sense and antisense RNA were mixed in annealing buffer (10 mM Tris, pH 7.4, and 0.1 mM ethylenediaminetetraacetic acid) to a final concentration of 2 μM each, heated for 2 min at 94°C and then incubated at room temperature for at least 16 h. To remove unhybridized RNA, the mixture was treated with 2 μg/ml RNaseT1 (Calbiochem, San Diego, CA) and 1 μg/ml RNaseA (Sigma) for 30 min at 37°C. The dsRNA products were extracted with phenol:chloroform and ethanol precipitated, then dissolved in water. The quality of dsRNA was confirmed by electrophoresis in an agarose gel. Gfp dsRNA was prepared as previously described [40]. The dsRNA samples were diluted to a final concentration of 1–2 mg/ml and stored at −80°C until used.
RNA Extraction and Real-Time RT-PCR
Total RNA from 5 to 50 embryos was extracted using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA). The RT reaction, primed with random hexamers, was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Total RNA isolated was reverse transcribed in a 20-μl reaction volume. The resulting cDNA was quantified by real-time PCR (qRT-PCR). The qRT-PCR analysis was performed with the ABI Taqman Assay-on-demand probe/primer sets for Mta1(Mm00475337_m1), Mta2 (Mm00488671_m1), and Mta3 (Mm00475365_m1). One embryo equivalent of cDNA was used for each real-time PCR reaction with a minimum of three replicates as well as a minus RT and minus template controls for each gene. Unless otherwise stated, quantification was normalized to Ubtf (Mm00456972_m1) and histone H2A mRNA (Mm00501974_s1).
Allele-Specific Expression Analysis of Blastocyst-Stage Embryos
H19 and Snrpn expression assays were conducted on cDNA using the LightCycler Real Time PCR System (Roche Molecular Biochemicals) as described previously [33]. Peg3 RT-PCR expression assays were conducted on cDNA from single blastocysts by allele-specific restriction digests as previously described [41]. The digested PCR products were resolved by PAGE. The contribution of each parental allele to the total expression was determined using the Quantity One software (Bio-Rad).
Antibodies
The following antibodies were used in immunofluorescence (IF) and Western blotting (WB): anti-MTA1 goat polyclonal (sc-9445; Santa Cruz Biotechnology; WB, 1:1000; IF, 1:100), anti-MTA2 mouse monoclonal (ab50209; Abcam; WB, 1:10 000; IF, 1:200), anti-MTA3 rabbit polyclonal (sc-48799; Santa Cruz Biotechnology; WB, 1:1000; IF, 1:100), anti-CHD4/Mi2β rabbit polyclonal (A301-082A; Bethyl Laboratories; WB, 1:1000; IF, 1:150), anti-RBBP4 rabbit monoclonal (2566–1; Epitomics; WB, 1:10 000), anti-HDAC1-rabbit polyclonal (06–720; Millipore; WB, 1:5000), anti-HDAC2-mouse monoclonal (05–814; Millipore; WB, 1:10 000), anti-MBD3 rabbit polyclonal (3896; Cell Signaling Technology; WB, 1:1000), anti-POU5F1/OCT4 mouse monoclonal (sc-5279; Santa Cruz Biotechnology; IF, 1:100), anti-NANOG rabbit polyclonal (ab80892; Abcam; IF, 1:200), anti-hyperacetylate histone H4 rabbit polyclonal (06–946; Millipore; IF, 1:500), anti-YY1 rabbit polyclonal (ab12132; Abcam; WB, 1:1000; IF, 1:100), anti-β-tubulin (T8328; Sigma; WB, 1:2000).
Immunostaining of Oocytes/Eggs/Embryos and Quantification of Fluorescence Intensity
Oocytes or embryos were fixed in 2% paraformaldehyde in PBS for 20 min at room temperature and then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Immunocytochemical staining was performed by incubating the fixed samples with primary antibodies for 60 min, followed by secondary antibodies conjugated with Cy5 or fluorescein isothiocyanate for 60 min; no signal was observed when the primary antibody was omitted. Polyclonal antibodies against MTA1, MTA2, and MTA3, hyperacetylated histone H4, and monoclonal antibodies against POU5F1 and SOX2 (Upstate Biotechnology) were diluted as listed above. The DNA was stained with 1 μM SYTOX Green (Molecular Probes, Eugene, OR). The cells were then washed and mounted under a coverslip with gentle compression in VectaShield antibleaching solution (Vector Laboratories). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope.
For each MTA immunostaining, all samples—i.e., oocytes, eggs, and embryos—were processed simultaneously. For each MTA, the laser power was adjusted so that the signal intensity was below saturation for the developmental stage that displayed the highest intensity, and all images were then scanned at that laser power. The intensity of fluorescence was quantified using National Institutes of Health Image J software.
Immunoblot Analysis
Protein samples from embryos were solubilized in Laemmli sample buffer [42], resolved by SDS-PAGE (10% gel, except when MBD3 or CHD4 were analyzed, in which case the gel concentration was 15% or 8%, respectively), and transferred to a nitrocellulose membrane. The membrane was blocked by soaking in Blotto (Tris-buffered saline with 0.1% Tween-20 and 5% nonfat dried milk) for 1.5 h and incubated overnight with the primary antibody in blocking solution. The membrane was then washed three times with Tris-buffered saline with 0.1% Tween-20 (TBST), incubated with a secondary antibody conjugated with horseradish peroxidase for 45 min, and washed five times with TBST. The signal was detected with the ECL Advance Western blotting detection reagents (Amersham, Piscataway, NJ) following the manufacturer's recommendations. The primary antibodies were diluted as described above, and secondary antibodies (Amersham ECL-HRP Linked Secondary Antibody; GE Healthcare) were diluted 1:20 000.
Bisulfite Mutagenesis
The allele-specific DNA methylation patterns were examined for DMRs of H19 [43], Peg3 [44], and Snrpn [41]. DNA was isolated from 2–12 blastocysts using The QIAamp DNA Micro Kit (Qiagen, Valencia, CA). The DNA was then denatured and treated with bisulfite using the Imprint DNA Modification Kit according to the manufacturer's protocol (Sigma-Aldrich, St. Louis, MO). Partial mutagenized DNA was amplified by the Epitech Whole Bisulfitome Kit (Qiagen) according to the manufacturer's instructions. Half of a blastocyst equivalent of the mutagenized DNA or 2 μl of the amplified DNA was used for each PCR reaction, and the products were cloned and sequenced as described previously [33, 43]. Six or more clones were sequenced for each PCR reaction. The sequencing data was analyzed using the online tool BDPC (http://biochem.jacobs-university.de/BDPC/index.php) [45]. Strands from a PCR that contained an identical pattern of methylated cytosines and that could not be distinguished from other strands by polymorphisms were only counted once. DNA strands were considered hypomethylated when at least 50% of the CpGs assayed were not methylated.
Statistical Analysis
Student t-test and chi-square test were used, and differences of P < 0.05 were considered significant.
RESULTS
Temporal and Spatial Pattern of MTA Expression During Oocyte Maturation and Preimplantation Development
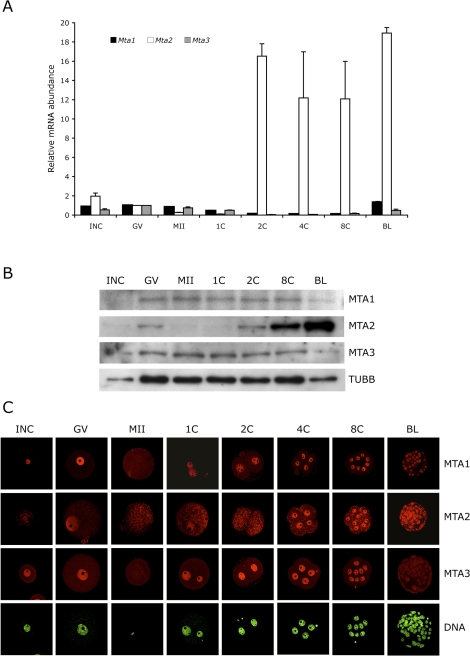
Transcripts for Mta1, Mta2, and Mta3 were assayed by qRT-PCR in oocytes and throughout preimplantation development (Fig. 1A). In contrast to Mta1 and Mta3 mRNA, which are degraded during maturation and preimplantation development and then zygotically expressed between the eight-cell and blastocyst stages, Mta2 expression is dramatically increased in the two-cell embryo, which corresponds to the time of the maternal-to-zygotic transition [46]. Consistent with these changes in transcript abundance, immunoblotting experiments demonstrated that the amounts of MTA1 and MTA3 were relatively constant during preimplantation development, whereas there was a dramatic increase in the amount of MTA2 protein starting in the two-cell embryo (Fig. 1B). This increase in Mta2 expression and protein accumulation was also observed by immunocytochemistry, in which staining was predominantly observed in the nucleus (Fig. 1C). Nuclear staining for MTA1 and MTA3 was also relatively constant up to the eight-cell stage. The amount of MTA1 assayed by immunoblotting remained essentially constant during preimplantation development, suggesting that the MTA1 decrease in nuclear signal intensity was due to the increased number of nuclei. In contrast, although the amount of MTA3 also remained essentially constant, little nuclear signal was detected in blastocysts, suggesting that the decrease is due to translocation from the nucleus to the cytoplasm.
FIG. 1.
Mta RNA and protein expression patterns in oocyte and preimplantation embryos. A) Temporal pattern of expression of Mta1, Mta2, and Mta3. The experiment was conducted twice, and the data are expressed as mean ± range and are expressed relative to the value obtained for full-grown oocytes. The amount of Mta RNA was normalized to Gfp mRNA that was added as an external control prior to RNA isolation. B) Immunoblot analysis of MTA1, MTA2, and MTA3 expression. One hundred oocytes/embryos were loaded per lane, and beta-tubulin (TUBB) was used as a loading control. The experiment was conducted three times, and similar results were obtained in each case; a representative experiment is shown. C) Immunocytochemical analysis of MTA1, MTA2, and MTA3 expression. All samples for a given MTA were processed for immunocytochemistry together, and all images were taken at the same laser power, thereby enabling direct comparison of signal intensities. The experiment was conducted three times, and at least 25 oocytes/embryos were analyzed for each sample. Shown are representative examples. INC, incompetent oocyte; GV, fully grown oocyte; MII, metaphase II; 1C, one-cell embryo; 2C, two-cell embryo; 4C, four-cell embryo; 8C, eight-cell embryo; BL, blastocyst. Original magnification x80.
RNAi-Mediated Ablation of MTA2 Reduces the Amount of MTA2 but Not Other NuRD Complex Components
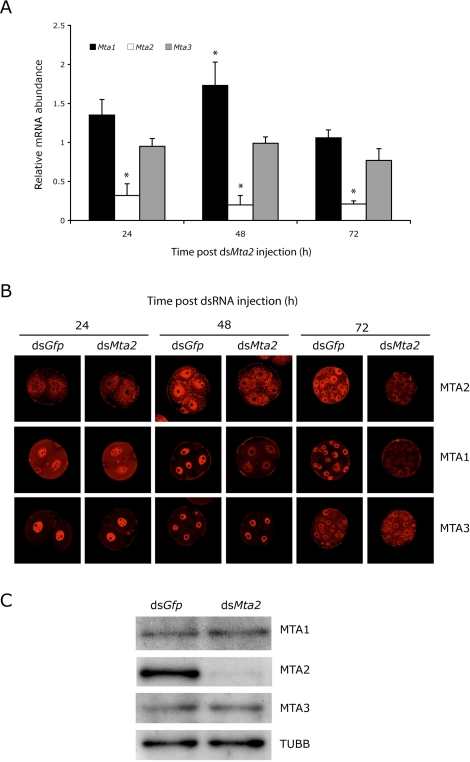
We previously demonstrated that RNAi-mediated targeting of Mbd3 resulted in reduced amounts of nuclear MTA2 protein in blastocysts and biallelic H19 expression [33]. The observation that Mta2 is upregulated during the two-cell stage made Mta2 an ideal candidate to be studied in genomic imprinting by an RNAi approach. Accordingly, one-cell embryos were injected with Mta2 dsRNA to ablate both maternal and zygotic Mta2 transcripts. Control embryos were injected with dsRNA for Gfp. The embryos were then cultured to the blastocyst stage in vitro. The RNAi strategy specifically targeted Mta2, and not Mta1 and Mta3 transcripts (Fig. 2A), although there was a transient increase in the relative abundance of Mta1 transcripts after injection.
FIG. 2.
Effects of RNAi-mediated knockdown of Mta2 on Mta1, Mta2, and Mta3 expression, and localization and amount of MTA1, MTA2, and MTA3. A) One-cell embryos were injected with either Gfp dsRNA (control) or Mta2 dsRNA and then cultured for 24, 48, or 72 h, at which time the relative abundance of Mta1, Mta2, and Mta3 transcripts was assayed by qRT-PCR and expressed relative to controls. The experiment was performed three times, and the data are expressed as mean ± SEM. B) The experiment was performed as described in A, and the samples were processed for immunocytochemical detection of MTA 1, 2, or 3 at the indicated times. The experiment was conducted three times, and at least 25 oocytes/embryos were analyzed for each sample. Original magnification x80. C) The experiment was performed as described in A, and the relative amount of MTA1 and MTA2 was determined by immunoblot analysis after 72 h of culture. The experiment was performed twice, and similar results were obtained. One hundred embryos were used for immunoblot analysis.
The amount of MTA2 protein was reduced within 48 h following injection as determined by immunocytochemistry (Fig. 2B) and remained low 72 h following injection as determined by immunocytochemistry and immunoblotting (Fig. 2, B and C). Although the amount of MTA1 protein was unchanged following depletion of Mta2 transcripts (Fig. 2C), the amount of nuclear-associated MTA1 protein was significantly reduced (Fig. 2B), suggesting that MTA1 (and any MTA1-associated proteins) was translocated to the cytoplasm, where dilution would account for the decreased immunocytochemical signal. Targeting Mta2 transcripts had little, if any, effect on MTA3 (Fig. 2, B and C).
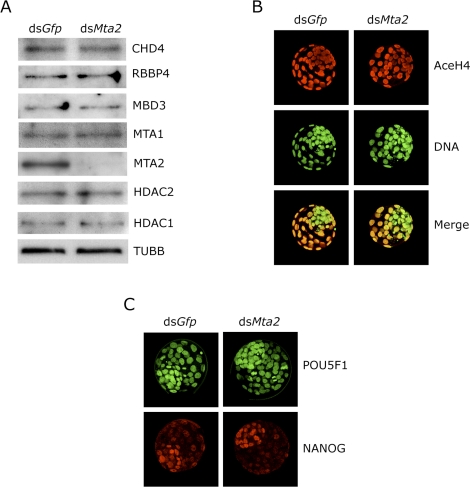
To determine whether expression of other components of the NuRD complex was perturbed in MTA2-depleted embryos, we examined expression of CHD4 (Mi-2β), RBBP4 (RbAp48), MBD3, HDAC1, and HDAC2, as well as MTA1 and MTA2, by immunoblot analysis and found no differences in these other components in blastocysts (Fig. 3A); CHD3 (Mi-2α) is not expressed in oocytes based on microarray studies [47] and therefore was not assayed. Immunocytochemical detection of these NuRD components was consistent with the immunoblotting data and revealed no change in the intensity of the nuclear signal (Supplemental Fig. S1, all supplemental data are available online at www.biolreprod.org).
FIG. 3.
Effect of Mta2 knockdown on expression of other components of the NuRD complex. A) One-cell embryos were injected with either Gfp dsRNA (control) or Mta2 dsRNA and then cultured 96 h in vitro. Equal numbers of dsGfp- and dsMta2-injected embryos were collected for immunoblot analysis; 100 embryos were loaded per lane, and TUBB was used as a loading control. The experiment was performed twice, and similar results were obtained. B) dsGfp- or dsMta2-injected embryos were processed for immunocytochemical detection of histone H4 acetylation state. At least 12 control and experimental embryos were analyzed, and the experiment was conducted four times. Shown are representative images. C) dsGfp- or dsMta2-injected embryos were processed for immunocytochemical detection of histone POU5F1 and NANOG. At least 12 control and experimental embryos were analyzed, and the experiment was conducted four times. Shown are representative images. Original magnification x80.
HDAC1 appears to be the HDAC largely responsible for the global state of histone acetylation in preimplantation mouse embryos [29]. Consistent with the observation that HDAC1 expression appeared unperturbed in MTA2-depleted blastocysts, there was no obvious effect on the global state of histone acetylation in these blastocysts (Fig. 3B). Last, the NODE complex (for NANOG- and POU5F1-associated deacetylase) identified in embryonic stem (ES) cells is composed of MTA1/MTA2, HDAC1/2, NANOG, and POU5F1 [48]. Expression of NANOG and POU5F1 in dsMta2-injected embryos was not different compared with control embryos (Fig. 3C). Results of these experiments suggest that Mta2 dsRNA effectively reduces MTA2 protein but does not affect other components of the NURD complex or another complex that contains MTA2.
Biallelic H19 Expression and Loss of DNA Methylation on the Paternal DMR in MTA2-Depleted Blastocysts
We previously reported that depletion of MBD3 is accompanied by biallelic H19 expression and reduced amount of MTA2 in blastocysts [33]. We therefore examined the effect of directly depleting MTA2 on imprinted H19 expression. Using an allele-specific real-time RT-PCR assay for H19 expression in blastocysts, we found that H19 expression was biallelic in 32% of the embryos (16/50), whereas only 10% (4/39) of the dsGfp-injected embryos exhibited biallelic H19 expression (P < 0.05, chi-square). The loss of DNA methylation in the DMR, which is located 2 kb upstream from the transcription start site, in the MTA2-depleted blastocysts provides a likely explanation for the observed biallelic H19 expression in these embryos (Fig. 4A). We observed a significant increase in the number of DNA hypomethylated paternal strands in MTA2-depleted embryos (42.5%) compared with controls (11.7%). These results strongly suggest that MTA2 is necessary for both the proper imprinted gene expression of H19 and maintenance of H19 paternal methylation in mouse early embryos.
FIG. 4.
Loss of H19 paternal DMR methylation in MTA2-depleted blastocysts. One-cell embryos were injected with either Gfp dsRNA or Mta2 dsRNA, cultured to the blastocyst stage, and collected in pools of 2–12 blastocysts that were then subjected to bisulfite mutagenesis. A) H19, paternal DNA stands are shown. B) Snrpn, maternal strands are shown. C) Peg3, maternal strands are shown. The H19 maternal and the Snprn and Peg3 paternal strands were unmethylated in all samples. Each line of circles represents a single DNA strand, with the number to the left of the line corresponding to the number of times this pattern was seen. Each circle represents a single CpG. If the CpG was methylated, the circle is filled. Those strands with less than half of the CpGs methylated are considered hypomethylated.
Snrpn and Peg3 Imprinted Gene Expression in MTA2-Depleted Embryos
Our previous study showed that reduced MBD3 had an impact on proper imprinted expression of H19 but not expression of other imprinted genes, including Snrpn and Peg3 [33]. To ascertain whether a similar situation occurs for MTA2, we examined the expression status of Snrpn and Peg3, which are paternally expressed and imprinted in blastocysts, in MTA2-depleted blastocysts. Snrpn expression remained monoallelic from the paternal allele, and there was no change in DNA methylation of the maternal allele of the Snprn DMR compared with dsGfp-injected controls (Fig. 4B). In contrast, 32% (12/38) of the MTA2-depleted embryos showed biallelic Peg3 expression compared with control embryos (2 [7.1%] of 28; P < 0.05, chi-square). Curiously, DNA methylation of the maternal allele of a potential Peg3 DMR was not significantly affected in these embryos (Fig. 4C), suggesting that this region is not responsible for controlling paternal Peg3 expression.
YY1 is implicated in proper monoallelic Peg3 expression [49–52], and YY1 can interact with MTA2 in vitro [25]. Therefore, it was formally possible that biallelic Peg3 expression was due to perturbed Yy1 expression in Mta2-depleted embryos. Such is not the case, however, because the amount of YY1 protein was unchanged in Mta2-depleted embryos (Supplemental Fig. S2). Consistent with this finding was that there was no decrease, as determined by qRT-PCR, in the relative amount of Yy1 mRNA in Mta2-depleted embryos compared with controls (data not shown).
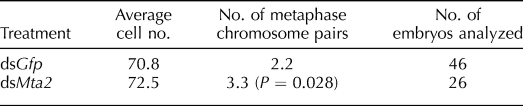
Increased Number of Metaphase Chromosome Pairs in MTA2-Depleted Blastocysts
MTA2-depleted embryos developed to the blastocyst stage at an incidence similar to controls and contained a comparable number of cells (Table 1). Nevertheless, we noted a significant increase of the number of metaphase chromosome pairs in MTA2-depleted embryos compared with dsGfp-injected embryos (Table 1), suggesting a role for MTA2 in cell division.
TABLE 1.
Cell numbers and metaphase chromosome pairs of blastocysts after dsRNA injection.
DISCUSSION
Genomic imprinting is established in the germline and early embryos. How imprinted marks are maintained during early development, and how they survive the genome-wide reprogramming after fertilization, is essentially unknown. Our previous finding [33] show that depletion of MBD3, which led to a decrease in both the amount of nuclear MTA2 and biallelic H19 expression, as well as a loss of DNA methylation in the DMR, implicated a NuRD complex in maintaining imprinted marks in preimplantation embryos. Results of experiments reported here in which the amount of MTA2 is specifically reduced further implicate a role for the NuRD complex in maintaining imprinted marks in preimplantation embryos, as well as provide new insights into NuRD function.
The composition of the NuRD complex in preimplantation embryos is not known, and the limited amounts of biological material effectively preclude undertaking such analyses. In systems in which the NuRD complex has been isolated and characterized, it appears that different MTAs are associated with different NuRD complexes [18]. Our finding of the different changes in nuclear concentration of the three MTAs between the eight-cell and blastocyst stages is consistent with each MTA residing in a distinct NuRD complex. For example, although the amount of MTA3 remains relatively constant between these two stages, there is a dramatic decrease in the nuclear MTA3 signal in blastocysts, suggesting that NuRD complexes containing MTA3 translocate to the cytoplasm. In contrast, MTA1, whose amount also remains essentially unchanged between the eight-cell and blastocyst stages, displays only a modest decrease in the nuclear signal, much of which is accounted for by the increase in the number of nuclei.
The failure of MBD3 to interact with CHD4 and the inability of antibodies against MBD3 to immunoprecipitate MBD3-containing NuRD complexes—the antibody could immunoprecipitate recombinant MBD3—suggest that MBD3 is embedded within the NuRD complex [53]. These findings, coupled with the ability of MBD3 to interact directly with MTA2 [53], provide an explanation that targeting Mbd3 mRNA results in a decrease in nuclear MTA2 [33], whereas targeting Mta2 mRNA has no apparent effect on the amount of MBD3. In this scenario, loss of MBD3 would lead to a reduced association of MTA2 with the complex and a concomitant loss from the nucleus, and possibly degradation. In contrast, a decrease in the amount of MTA2 would be predicted to have little, if any, effect on the stability of MBD3, as was observed. These observations would also account for the observation that a similar increase in the number of blastomeres arrested in metaphase is observed following depletion of either MBD3 [33] or MTA2, and that the effect is mediated by MTA2.
HDACs 1 and 2, and RBBPs 4 and 7 can form a core complex whose activity is stimulated by association with MTA2 to a level similar to that of intact NuRD; neither CHD4 nor MBD3 stimulates this core complex [53]. HDAC1 appears to be the HDAC responsible in determining the extent of global histone acetylation in preimplantation mouse embryos [29]. The finding that there is no apparent change in global histone acetylation in MTA2-depleted embryos—the amount of HDAC1 is unchanged in these embryos—suggests that either MTA2 does not stimulate HDAC activity in NuRD complexes present in preimplantation embryos or, more likely, that another HDAC1-containing complex(es), e.g., SIN3 [8, 54], is responsible.
A seminal finding reported here is that reducing the amount of MTA2 in preimplantation embryos results in biallelic expression of both H19 and Peg3, which are maternally and paternally expressed, respectively. Biallelic Peg3 expression contrasts with our previous finding that paternal monoallelic Peg3 expression is observed in MBD3-depleted preimplantation embryos [33]. A possible explanation for this difference is that MTA1- and MTA2-containing NuRD complexes control expression of Peg3 and H19, respectively. Loss of nuclear MTA1 in MTA2-depleted embryos would account for biallelic expression of both Peg3 and H19; the molecular basis for the nuclear exit of MTA1-containing NuRD complexes that occurs following MTA2 depletion is enigmatic, especially in light of finding that the amount of MTA1 protein determined by immunoblotting appears unchanged. The greater loss of MTA2, when compared with MTA1 in nuclear extracts from Mbd3−/− ES cells [55], provides an explanation for our prior finding that H19, but not Peg3, expression is biallelic following RNAi-mediated reduction of MBD3. Should a similar difference in the amounts of MTA1 and MTA2 exist in these embryos, sufficient quantities of MTA1-containing NuRD complexes may remain, thereby maintaining monoallelic paternal Peg3 expression.
In contrast to the loss of DNA methylation observed in the H19 DMR, MTA2 depletion does not affect DNA methylation in the DMR adjacent to the Peg3 promoter. This finding is not surprising, because the Peg3 DMR has not been demonstrated to function as an ICR. Szeto et al. [56] analyzed three different lines of transgenic mice that carry a bacterial artificial chromosome covering 120 kb around the Peg3 locus. Only one of three lines exhibited partial imprinted expression of Peg3 and differential methylation in the Peg3 DMR. These results suggest that the Peg3 DMR alone is not sufficient to control the imprinting in this locus, and our data indicate that MTA2 functions through an unknown element to repress the maternal Peg3.
At a molecular level, how the loss of MTA2 is mechanistically linked to biallelic expression of both H19 and Peg3, and DNA methylation of the DMR of H19 (and, presumably, a differentially methylated DNA sequence responsible for paternal Peg3 expression) remains unresolved. Attempts to conduct chromatin immunoprecipitation experiments to localize the NuRD complex to H19 and Peg3 genes were unsuccessful, largely in part because it is difficult to conduct such experiments given the small amounts of biological material that can be readily isolated. Similar experiments were also unsuccessful when model mouse cell lines were used, e.g., mouse embryonic fibroblast cells, ES cells, and trophoblast stem cells (data not shown). Nevertheless, the results presented here provide further evidence for a critical role of NuRD complexes in directing appropriate expression of imprinted genes.
Acknowledgments
The authors thank Richard A. Jimenez for his assistance with the allele-specific expression assays, Mellissa R.W. Mann and Rocio M. Rivera for their useful suggestions for DNA methylation analysis, and Gerd Blobel for his generous gifts of MTA1 and CHD4 antibodies.
Footnotes
Supported National Institutes of Health grants HD042026 to M.S.B. and R.M.S., and HD022681 to R.M.S.
REFERENCES
- Wang GG, Allis CD, Chi P.Chromatin remodeling and cancer, part II: ATP-dependent chromatin remodeling. Trends Mol Med 2007; 13: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA.The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26: 5433–5438. [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH.Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem 2001; 276: 6817–6824. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL.Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 1998; 395: 917–921. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP.Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 1999; 23: 62–66. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W.NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 1998; 2: 851–861. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D.The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 1998; 95: 279–289. [DOI] [PubMed] [Google Scholar]
- Ahringer J.NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 2000; 16: 351–356. [DOI] [PubMed] [Google Scholar]
- McDonel P, Costello I, Hendrich B.Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 2009; 41: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D.Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 1997; 89: 357–364. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN.Sin meets NuRD and other tails of repression. Cell 1999; 99: 447–450. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y.The NuRD complex: linking histone modification to nucleosome remodeling. Curr Top Microbiol Immunol 2003; 274: 269–290. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y.The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev 2001; 15: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A.Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 1998; 18: 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG.MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol 2006; 26: 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B, Kumar R.Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem 2007; 282: 1529–1533. [DOI] [PubMed] [Google Scholar]
- Kumar R, Wang RA, Bagheri-Yarmand R.Emerging roles of MTA family members in human cancers. Semin Oncol 2003; 30: 30–37. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA.MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 2003; 113: 207–219. [DOI] [PubMed] [Google Scholar]
- Toh Y, Oki E, Oda S, Tokunaga E, Ohno S, Maehara Y, Nicolson GL, Sugimachi K.Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int J Cancer 1997; 74: 459–463. [DOI] [PubMed] [Google Scholar]
- Zhang H, Singh RR, Talukder AH, Kumar R.Metastatic tumor antigen 3 is a direct corepressor of the Wnt4 pathway. Genes Dev 2006; 20: 2943–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Kovalev GI, Chang H, Kallin E, Knudsen G, Xia L, Mishra N, Ruiz P, Li E, Su L, Zhang Y.Inactivation of NuRD component Mta2 causes abnormal T cell activation and lupus-like autoimmune disease in mice. J Biol Chem 2008; 283: 13825–13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA.Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 2004; 1677: 52–57. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kajita M, Taysavang P, Wade PA.Hormonal regulation of metastasis-associated protein 3 transcription in breast cancer cells. Mol Endocrinol 2004; 18: 2937–2949. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009; 138: 660–672. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM.The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J Biol Chem 2003; 278: 42560–42568. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS.Genomic imprinting mechanisms in mammals. Mutat Res 2008; 647: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verona RI, Mann MR, Bartolomei MS.Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol 2003; 19: 237–259. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS.Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 2009; 23: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schultz RM.Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greciano PG, Goday C.Methylation of histone H3 at Lys4 differs between paternal and maternal chromosomes in Sciara ocellaris germline development. J Cell Sci 2006; 119: 4667–4677. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R.Role for DNA methylation in genomic imprinting. Nature 1993; 366: 362–365. [DOI] [PubMed] [Google Scholar]
- Yamasaki-Ishizaki Y, Kayashima T, Mapendano CK, Soejima H, Ohta T, Masuzaki H, Kinoshita A, Urano T, Yoshiura K, Matsumoto N, Ishimaru T, Mukai T, et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol Cell Biol 2007; 27: 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS.Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet 2007; 3: e137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeles GL, Ram PT, Rothstein JL, Schultz RM.Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol Reprod Dev 1994; 37: 121–129. [DOI] [PubMed] [Google Scholar]
- Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS.Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 2003; 69: 902–914. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Montgomery RR, Belanoff JR.Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol 1983; 97: 264–273. [DOI] [PubMed] [Google Scholar]
- Whitten WK.Nutrient requirements for the culture of preimplantation mouse embryo in vitro. Adv Biosci 1971; 6: 129–139. [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I.An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679–688. [DOI] [PubMed] [Google Scholar]
- Ho Y, Wigglesworth K, Eppig JE, Schultz RM.Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev 1995; 41: 232–238. [DOI] [PubMed] [Google Scholar]
- Stein P, Svoboda P, Anger M, Schultz RM.RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 2003; 9: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS.Selective loss of imprinting in the placenta following preimplantation development in culture. Development 2004; 131: 3727–3735. [DOI] [PubMed] [Google Scholar]
- Laemmli UK.Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS.A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 1997; 17: 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR.Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 2010; 19: 36–51. [DOI] [PubMed] [Google Scholar]
- Rohde C, Zhang Y, Jurkowski TP, Stamerjohanns H, Reinhardt R, Jeltsch A.Bisulfite sequencing Data Presentation and Compilation (BDPC) web server–a useful tool for DNA methylation analysis. Nucleic Acids Res 2008; 36: e34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM.Regulation of zygotic gene activation in the mouse. BioEssays 1993; 15: 531–538. [DOI] [PubMed] [Google Scholar]
- Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM.Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol 2005; 286: 493–506. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z.Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 2008; 10: 731–739. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JD.In vivo YY1 knockdown effects on genomic imprinting. Hum Mol Genet 2008; 17: 391–401. [DOI] [PubMed] [Google Scholar]
- Kim J, Kollhoff A, Bergmann A, Stubbs L.Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet 2003; 12: 233–245. [DOI] [PubMed] [Google Scholar]
- Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J.Identification of clustered YY1 binding sites in imprinting control regions. Genome Res 2006; 16: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Yu S, Choo JH, Kim J.Two evolutionarily conserved sequence elements for Peg3/Usp29 transcription. BMC Mol Biol 2008; 9: 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D.Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 1999; 13: 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C.Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol 2009; 53: 275–289. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B.The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 2006; 8: 285–292. [DOI] [PubMed] [Google Scholar]
- Szeto IY, Barton SC, Keverne EB, Surani AM.Analysis of imprinted murine Peg3 locus in transgenic mice. Mamm Genome 2004; 15: 284–295. [DOI] [PubMed] [Google Scholar]