Abstract
Maintenance of cellular function relies on the expression of genetic information with high fidelity, a process in which RNA molecules form an important link. mRNAs are intermediates that define the proteome, rRNAs and tRNAs are effector molecules that act together to decode mRNA sequence information, and small noncoding RNAs can regulate mRNA half-life and translatability. The steady-state levels of these RNAs occur through transcriptional and post-transcriptional regulatory mechanisms, of which RNA decay pathways are integral components. RNA decay can initiate from the ends of a transcript or through endonucleolytic cleavage, and numerous factors that catalyze or promote these reactions have been identified and characterized. The rate at which decay occurs depends on RNA sequence or structural elements and usually requires the RNA to be modified in a way that allows recruitment of the decay machinery to the transcript through the binding of accessory factors or small RNAs. The major RNA decay pathways also play important roles in the quality control of gene expression. Acting in both the nucleus and cytoplasm, multiple quality control factors monitor newly synthesized transcripts, or mRNAs undergoing translation, for properties essential to function, including structural integrity or the presence of complete open reading frames. Transcripts targeted by these surveillance mechanisms are rapidly shunted into conventional decay pathways where they are degraded rapidly to ensure that they do not interfere with the normal course of gene expression. Collectively, degradative mechanisms are important determinants of the extent of gene expression and play key roles in maintaining its accuracy.
Keywords: RNA quality control, RNA half-life, stability and instability elements, nuclear RNA decay, cytoplasmic RNA decay, exosome, decapping, exonucleolytic digestion, NMD, P bodies, closed-loop mRNP
The respective levels of individual cellular mRNAs are an important determinant of gene expression. The advent of accurate multiplexed quantitative methodologies, such as microarray analyses, have allowed genome-wide views of changes in mRNA levels under different conditions [1], leading to the use of transcript abundance as a hallmark with which to predict changes in pathway activities. In addition to mRNA, expression of the genome at the protein level also relies on rRNA and tRNA because both are also essential for enabling polypeptide synthesis. Although transcriptional activation has been a predominant focus of studies seeking to elucidate the mechanisms by which steady-state levels of all RNAs are controlled, it is critical to recognize that these levels are reflective of the sum of both transcript synthesis as well as degradation. Accordingly, the role of RNA decay pathways is becoming increasingly appreciated [2].
Broadly speaking, RNA decay mechanisms are involved in the maintenance of normal decay rates, the regulation of the stability of certain RNAs, and the elimination of nonfunctional or otherwise defective transcripts. Since every step in the life cycle of an RNA molecule is subject to error, the latter quality control (QC) mechanisms have evolved to dispose of nuclear or cytoplasmic molecules deemed defective. In general, degradation of such spurious transcripts is initiated by specialized factors capable of first recognizing them and, subsequently, funneling them to the common RNA decay machinery. Similarly, the regulated expression of certain transcripts usually relies on the binding of protein factors and/or small antisense RNAs that alter the translational status and/or stabilities of the mRNAs by changing accessibility of the mRNAs to the decay machinery.
In the sections that follow, we provide an overview of RNA decay pathways, consider the diversity of their substrates, and assess their overall impact on the extent and fidelity of gene expression.
General cytoplasmic mRNA turnover
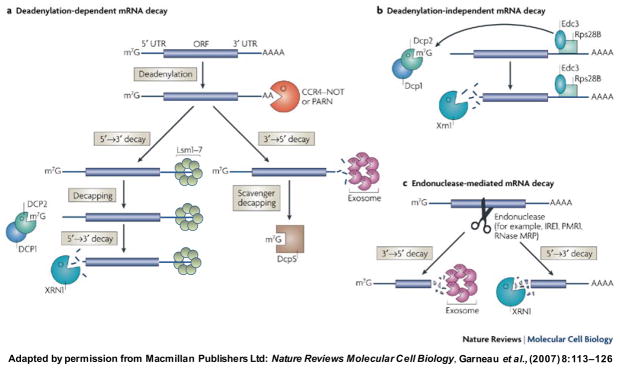
RNA decay mechanisms have been most extensively studied to understand the fate of mRNAs in the cytoplasm. The generation of most mRNAs involves nuclear processing events that result in the modification of the 5′ end with a 7-methyl guanosine cap structure and, with the exception of metazoan histone mRNAs, a 3′ poly(A) tail. Proper mRNP packaging allows the export of the mRNA to the cytoplasm where it interacts with the protein synthesis machinery and undergoes translation. The mRNP is configured in a manner that promotes its “circularization,” a state in which the mRNA 5′ and 3′ ends communicate via interactions of the poly(A)-binding protein (Pab1p in yeast; PABP in higher eukaryotes) at the 3′ end of the mRNA and the cap-binding protein (eIF4E) at the 5′ end that are bridged by the initiation factor, eIF4G [3, 4]. This “closed-loop” conformation serves to both protect the mRNA from exonucleolytic digestion at either end and to enhance the overall translatability of the mRNA. During the life of an mRNA molecule in the cytoplasm, progressive deadenylation of the 3′-poly(A) tail (by the Ccr4-NOT deadenylase complex in yeast, or PARN in human cells) leads to the gradual loss of bound Pab1p and presumably disruption of the closed-loop structure of the mRNP. This leads to the association of additional factors with the mRNA, accessibility of the 5′ cap to the Dcp1p/Dcp2p decapping complex and subsequent 5′→3′ exonucleolytic digestion of the decapped mRNA by the Xrn1p exoribonuclease [5]. Thus, one mechanism for turnover of many cytoplasmic mRNAs is a deadenylation-dependent, 5′→3′ process that is regulated by the rate of decapping (Figure 1). Given its importance to the decay pathway, it is not surprising that efficient decapping requires the activity of several accessory factors. Following deadenylation, the activities of the heptameric Lsm1-7p complex, the Dhh1p RNA helicase (also known as RCK/p54 or Me31B in higher eukaryotes), and Pat1p are proposed to enable recruitment of the decapping complex, since loss of any of these factors leads to the accumulation of capped, deadenylated mRNAs. In addition to their roles as stimulators of Dcp1p/Dcp2p activity, Dhh1p and Pat1p also behave as translational repressors. Other factors that act as enhancers of mRNA decapping (EDC) activity are the Edc1, Edc2, and Edc3 (Lsm16) proteins. Edc1p and Edc2p were shown to be RNA-binding proteins that genetically interact with the Dcp1p/Dcp2p complex [6]. Edc3p is unique in seemingly being required for the regulated degradation of only two transcripts in yeast – the YRA1 pre-mRNA and RPS28B mRNA [7, 8].
Figure 1.
a. Deadenylation-dependent mRNA decay pathways. The 3′-poly(A) tail is removed by the Ccr4–NOT or PARN deadenylases. Following deadenylation, two mechanisms can degrade the mRNA further: either decapping-dependent 5′→3′ decay or 3′→5′ exosome-mediated mRNA decay. In the 5′→3′ decay pathway, the Lsm1–7p complex associates with the 3′ end of the mRNA transcript and induces decapping by the Dcp1p/Dcp2p complex. The mRNA is then degraded by the 5′–3′ exoribonuclease, Xrn1p. Alternatively, the exosome can mediate 3′–5′ digestion of the deadenylated transcript. b. Deadenylation-independent mRNA decay pathways require recruitment of the decapping machinery. For example, in yeast, Rps28B protein interacts with Edc3p to recruit the Dcp1p/Dcp2p decapping enzyme. Following decapping, the mRNA is degraded by Xrn1p. c. Endonuclease-mediated mRNA decay involves an internal cleavage event in an mRNA, generating two fragments with unprotected ends. These fragments subsequently undergo digestion by Xrn1p or the exosome. (Adapted from reference 5).
Following deadenylation, an alternative pathway can degrade mRNA (Figure 1). The exosome, a large multisubunit complex that acts in the nucleus and the cytoplasm can mediate 3′→5′ mRNA decay. A nine subunit exosome core comprised of catalytically inactive 3′→5′ exonuclease homologues is present in both the nucleus and the cytoplasm, with the complex in each compartment possessing at least one additional defining catalytic factor. Structural and functional studies have determined that the yeast exosome is composed of 11 subunits, nine of which (Rrp4p, Rrp40p, Csl4p, Ski6p/Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p, Mtr3p) comprise the nuclease-free scaffold. The tenth subunit, Dis3p/Rrp44p, is a nuclease component, and the eleventh subunit is either the 3′→5′ exonuclease Rrp6p (found in the nuclear exosome) or Ski7p (in the cytoplasmic exosome). The exosome structure appears to be conserved in humans, except that: i) the association of Dis3p/Rrp44p is not as stable with the 9-subunit scaffold as in yeast; ii) Rrp6p in humans may be present in both the nucleus and cytoplasm; and iii) humans may have an additional nuclear subunit, MPP6, not present in yeast. Modeling of the exosome suggests a channel-containing ring-like structure formed by the 9-subunit scaffold, with the location of Dis3p/Rrp44p remaining ill-defined. Until recently, it was thought that the exosome possessed only 3′→5′ exonuclease activity. However, recent work has demonstrated that Dis3p/Rrp44p has an endoribonuclease activity mediated through a highly conserved PIN domain at its N-terminus [9]. Exosome activity in the cytoplasm requires the Ski2p/Ski3p/Ski8p complex which is thought to be recruited to the exosome via interaction with Ski7p.
In the yeast Schizosaccharomyces pombe, an additional pathway has been identified in which some mRNAs do not undergo deadenylation prior to decapping. Instead, Lsm1p-dependent decapping is preceded by the addition of one to two uridyl residues to the 3′ end of the polyadenylated mRNA by Cid1p, a putative poly(U) polymerase (PUP) and/or terminal uridyl transferase (TUTase) [10]. Additional transcripts that undergo poly(U)-dependent degradation are the metazoan histone RNAs, which are unique in not possessing a poly(A) tail [11]. Instead, the 3′-end of the transcript ends in a conserved stem-loop that is the binding site for a stem-loop binding protein, SLBP1. Bridging of histone mRNA 5′ and 3′ ends is effected by SLBP-interacting protein 1 (SLIP1), which interacts with both SLBP and eIF4G. Accelerated degradation of histone mRNAs occurs upon termination of DNA synthesis at the end of S-phase in cells. SLBP, in the presence of the ATR kinase and the NMD factor Upf1 (see below), acts to recruit factors involved in poly(U) addition, followed by binding of the heptameric Lsm1-7 complex, and the triggering of exosome-mediated 3′→5′ decay. This pathway is not the exclusive mode of histone mRNA decay since these RNAs can also serve as substrates for decapping by the Dcp1/Dcp2 complex and 5′→3′ decay by Xrn1 [12].
It has been proposed that degradation of mRNAs in the cytoplasm may take place in P bodies, discrete foci highly enriched in components of the decay machinery (Dcp1p/Dcp2p, Pat1p, Dhh1p, Lsm1-7p, and Xrn1p, among others) into which translationally repressed mRNAs are thought to localize to undergo degradation [13]. P bodies have been detected in all eukaryotes in which they have been sought, although their composition varies depending on the organism [13]. P body assembly and size appear to depend on the pool of nontranslating, ribosome-free mRNAs. Overexpression of a nontranslating mRNA fragment leads to larger P bodies, as does inhibition of translation initiation under conditions of stress, or through the use of conditional alleles of initiation factors. However, inhibition of translation elongation by treatment of cells with cycloheximide, which traps mRNAs associated with ribosomes, leads to a loss of P bodies, suggesting an inverse relationship between ribosome-associated mRNAs and P bodies. Translational repression is thought to be an upstream step in the compartmentalization of mRNPs to P bodies, since loss of Pat1p and/or Dhh1p, which can act as translational repressors, leads to a loss in observable P bodies and increased accumulation of the mRNAs in polysomes [14]. Conversely, overexpression of Dhh1p or Pat1p leads to increased translation repression and increased P body formation. However, the significance of P bodies has recently been questioned. Evidence showing that decapped decay intermediates can be polyribosome-associated suggests that assembly of these structures is not essential for mRNA decay in yeast [15, 16], consistent with earlier observations that some factors required for mRNA decay can co-fractionate with polyribosomes [17]. Moreover, in both yeast and metazoan cells, disruption of P bodies by depletion of central P body components failed to manifest decay phenotypes for several different mRNAs [18–20].
Conditional mRNA decay through protein binding
The presence of a cap and poly(A) tail at either end of an mRNA and the factors that associate with them to confer stability from cellular nucleases highlight the importance of both cis-acting sequence elements and sequence-specific trans-acting factors that function individually or in combination [21] to regulate mRNA decay. mRNA stability determinants are usually present in the 3′-untranslated regions (UTRs) of the mRNAs they regulate, although coding region and 5′-UTR elements have also been described [22].
One of the most well-characterized 3′-UTR sequence determinants is the mammalian adenylate-uridylate rich element (ARE) present in many metazoan transcripts that exhibit rapid response to cellular cues such as those encoding cytokines, proto-oncogenes, and interferons [23]. AREs were originally defined as having the pentameric sequence AUUUA, but have since been further subclassified according to the number and context of the AUUUA pentamers [24]. These sequences can be bound by ARE-binding proteins (ARE-BPs) to stabilize or destabilize ARE-containing transcripts. The Hu family of ARE-BPs is comprised of four members - HuR (HuA), HuB (Hel-N1), HuC, and HuD – and these proteins are mammalian homologs of Drosophila ELAV proteins that possess mRNA stabilizing effects. HuR, the best studied member, is ubiquitous, contains three RNA recognition motifs (RRMs), and targets transcripts such as those derived from the TNF, IL-3, and VEGF genes. Other classes of ARE-BPs target mRNAs for degradation. These include the RRM-containing AU-rich binding factor 1 (AUF1) and the KH splicing regulatory protein, KSRP. Originally identified as a promoter of c-myc degradation in vitro, AUF1 has four functionally distinct isoforms (p37, p40, p42, and p45) [25]. A different structural class of destabilizing ARE-BPs, typified by CCCH-type zinc fingers, includes tristetraprolin (TTP), BRF1, and BRF2. TTP has been found to bind and destabilize the TNF-α, IL-3, GM-CSF, and COX-2 transcripts. T-cell internal antigen 1 (TIA-1) and TIA-related protein (TIAR) constitute a set of ARE-BPs that do not lead to mRNA decay upon binding, but instead induce translational silencing and aggregation of TNF-α mRNA into stress granules. RHAU is an RNA helicase that was isolated as an ARE-BP of the urokinase plasminogen activator mRNA [26]. Destabilizing AREs appear to function by enhancing the recruitment of decay factors to target mRNAs. The ARE itself has been seen to interact directly with the exosome [27] and ARE-BPs can directly/indirectly interact with mRNA decay factors. AUF1(p37), KSRP, RHAU, and TTP have also all been found to interact with the exosome. KSRP and RHAU also bind to the PARN deadenylase, while TTP has been shown to interact with the decapping enzyme and the CCR4 deadenylase to modulate PARN activity in vitro. The mechanism by which the Hu proteins stabilize their targets is not fully understood. It is thought that they might either compete with destabilizing factors for binding to AREs or somehow enhance PABP:poly(A) interaction to prevent deadenylation. This is thought to be the mechanism for other mRNA stabilizing elements such as the U-rich ARE sequence recognized by Pub1p in yeast or poly(C)-rich elements recognized by KH-domain RNA binding proteins.
Another 3′-UTR sequence-specific modulator of mRNA stability is the evolutionarily conserved PUF family of proteins that have been proposed to support the mitotic proliferation of stem cells [28]. PUF proteins usually have eight consecutive Puf repeats, each about 40 amino acids long, that form a three-helix domain that stacks into a crescent-shaped structure thought to bind RNA targets on one surface and interacting proteins on the other. PUF target sequences are sometimes referred to as Nanos Response Elements (NREs), and have a “UGUR” core recognition sequence [28]. Individual PUFs can target multiple mRNAs, and PUF target specificity is mediated in part by interactions with other 3′-UTR-binding proteins such as Nanos and cytoplasmic polyadenylation element-binding proteins (CPEBs). PUFs function to enhance degradation of target mRNAs by accelerating deadenylation. Puf3p in yeast can also promote decapping after deadenylation, while Pumilio may effect deadenylation-independent repression of hunchback mRNAs in Drosophila.
The iron-response element (IRE), a cis-acting stem-loop sequence that is present in the 5′ or 3′ UTRs of certain mammalian mRNAs involved in iron metabolism, is modulated through the binding of the iron response proteins, IRP1 and IRP2. Whereas 5′-UTR localized IREs act to repress translation of mRNA (e.g., ferritin mRNA), those in the 3′-UTR appear to modulate mRNA stability. Transferrin receptor 1 (TfR1) mRNA has a 3′-UTR IRE, consisting of five stem-loops, that functions as an instability element. The TfR1 mRNA also contains an ARE, which confers a short half-life to the mRNA through a deadenylation-independent endonucleolytic cleavage event in the absence of IRPs. IRP binding to the IREs in iron-deficient cells blocks this cleavage event and increases the half-life of the TfR1 mRNA, leading to production of the receptor and to increased iron uptake into cells [29].
As previously mentioned, metazoan histone RNA decay is regulated through the binding of SLBP to the stem-loop at the 3′ end of the histone RNAs [11]. This degradation pathway also requires Upf1, an RNA helicase involved in the elimination of mRNAs containing premature translation termination codons (see below), which is thought to bind SLBP and stimulate poly(U) addition. Another mammalian pathway that utilizes Upf1 in the absence of an abnormally positioned stop codon is the Staufen 1-mediated decay (SMD) pathway, in which the double-stranded RNA-binding protein Staufen 1 (STAU1) mediates degradation of certain mRNA substrates by recruiting Upf1 to their 3′-UTRs [30]. SMD is thought to compete with nonsense-mediated mRNA decay (NMD; see below) by competing for Upf1 binding to Upf2, another factor involved in NMD [31].
Conditional mRNA decay through RNA interference
Small non-coding RNAs (ncRNAs) – which include antisense transcripts, small interfering RNAs (siRNAs), and microRNAs (miRNAs) – play critical roles in maintaining gene expression in eukaryotes by binding to complementary/near-complementary sites on target RNAs and affecting their degradation or translation. Although originally thought to be restricted to multicellular organisms, recent evidence suggests that similar mechanisms may operate in some yeasts [32]. RNA interference (RNAi) has mainly been characterized through the effects of siRNAs and miRNAs, each of which follows a distinct pathway for biogenesis [33]. siRNAs are 21–23 nucleotide RNA molecules that are usually perfectly complementary to their target sequences. They constitute a part of the Argonaute (AGO) RNA-induced silencing complex (RISC), a multiprotein complex that uses the guide RNA to recognize complementary target sites on mRNAs and induce their degradation. miRNAs are 20–25 nucleotides long, may have mismatches to their target sequences and are also present in multiprotein miRNP/RISC complexes containing an AGO family member. mRNA degradation by RISCs/miRNPs is thought to occur by inducing cleavages at sites of the RNA duplexes, although recent evidence suggests that miRNAs can trigger deadenylation by the CAF1-CCR4 deadenylase [34]. These mechanisms are thought to function in innate immune responses against viruses and transposable elements [35]. Many endogenous genes also contain miRNA binding sites in their 3′ UTRs, which serve to regulate target mRNA expression during different cellular processes [36].
Antisense transcripts to specific mRNAs can also act to stabilize their target transcripts [32], e.g. an antisense transcript to the induced nitric oxide synthase (iNOS) mRNA stabilizes the mRNA by binding to the HuR ARE-binding protein and the target, presumably inhibiting deadenylation and decay [37].
Nuclear processing and exosome-mediated decay
The synthesis of all functional RNAs in the nucleus of a cell requires not only transcription of precursor molecules, but subsequent processing events that involve RNA cleavage and trimming, leading to the generation of processed transcripts and waste products [2]. Transcriptomes also contain a large number of rapidly degraded transcripts that originate from bidirectional promoters or intergenic regions, and are collectively called cryptic unstable transcripts (CUTs). These include “transcriptional noise” and antisense transcripts that can induce gene silencing if left unchecked, or serve to regulate expression of nearby genes. Additionally, precursors to many noncoding RNA transcripts, including small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and microRNAs (miRNAs), undergo endo- and exonucleolytic processing steps to yield mature transcripts [38].
Trimming of many RNA precursors as well as elimination of most of the RNA waste in the nucleus is carried out by the nuclear Rrp6p-containing exosome. Exosome-mediated degradation in the nucleus is greatly enhanced by TRAMP, a polyadenylation complex consisting of a non-canonical poly(A) polymerase (Trf4p or Trf5p), a zinc knuckle protein (Air1p or Air2p), and the Mtr4p helicase, which is thought to connect the TRAMP and exosome complexes [39]. It has been suggested that TRAMP mediates substrate recognition by the exosome through recruitment of RNA targets via Air1p/Air2p followed by the addition of short poly(A) tails to their 3′ ends. In turn, these poly(A) tails are thought to make the substrates more accessible to the exosome for subsequent digestion by providing them with an unstructured 3′ end. Exosome mutants have been found to accumulate polyadenylated degradation intermediates as well as transcripts with extended 3′ ends, highlighting the importance of this degradative machine in processing and degradation of RNA intermediates.
RNA decay and RNAP II transcription termination
Termination of RNA polymerase (RNAP) II transcriptional activity releases the polymerase for further rounds of RNA synthesis as well as prevents the synthesis of abnormally long transcripts that may be detrimental to the cell. Transcription termination is coupled to 3′-end processing events that result in the endonucleolytic cleavage of the nascent transcript, followed by polyadenylation of the 5′ cleavage product and its subsequent binding by the nuclear poly(A)-binding protein, which protects the transcript from 3′→5′ exonucleolytic digestion [40]. The 3′ cleavage fragment from the initial endonucleolytic event (with an unprotected 5′ end) remains attached to the RNAPII, which must be released to conduct further rounds of transcription. The Rat1p (in yeast; Xrn2 in human) nuclear 5′→3′ exoribonuclease is thought to play a role in this event since mutants of Rat1p or Rai1p (enhancer of Rap1p activity) show readthrough of normal termination sites, a phenomenon seen not only in yeast, but in human cells as well. The “torpedo model” proposes that the Rat1p 5′→3′ exoribonuclease acts on the unprotected 5′ end of the 3′ cleavage fragment and degrades it in a manner that results in the release of the RNAPII through a poorly understood mechanism. Recent in vitro studies have found that recombinant Rat1p/Rai1p complex by itself or with Rtt103p (a 3′-end processing factor that associates with Rat1p/Rai1p in vivo) is insufficient to terminate RNAPII activity, suggesting the requirement of other factors and/or post-translational modifications to the factors involved in this process [41].
Depending on the phosphorylation status of the Rpb1p C-terminal heptad repeat domain (CTD) of RNAPII, an alternate method for transcriptional termination appears to take place for nonpolyadenylated short stable transcripts, such as snRNAs and snoRNAs, as well as CUTs, which relies on the cotranscriptional binding of the Nrd1 complex (Nrd1p, Nab3p and Sen1p) to specific binding sites on these transcripts. Association of the Nrd1 complex with their target transcripts is thought to lead to transcriptional termination and recruitment of the exosome/TRAMP complex, which either trims the 3′-end of snRNAs and snoRNAs, or degrades CUTs [40].
RNA decay and telomeric maintenance
Maintenance of telomeres, the physical ends of chromosomes, is required for genome stability and shortening of telomeres correlates with increased aging [42, 43]. Telomere size is maintained by protecting the telomeric regions in a heterochromatic state through binding of several protein factors, including the abundant shelterin complex, and a large noncoding telomeric repeat-containing RNA (TERRA or TelRNAs). TERRA is transcribed by RNA polymerase (RNAP) II from subtelomeric regions, is heterogeneous, contains a poly(A) tail at its 3′-end, is thought to play a role in organizing and/or maintaining the telomeric structure, and inhibits telomerase activity. TERRA has a short half-life, and degradation appears to be mediated by the nuclear 5′→3′ exoribonuclease, Rat1p, since loss of Rat1p in Saccharomyces cerevisiae leads to accumulation of TERRA and shortened telomeres [44]. Decreased expression of TERRA is seen in human cancer cell lines and it is thought that this downregulation leads to increased telomerase activity and carcinogenesis.
Quality control (QC) mechanisms in the nucleus
Apart from functioning in the processing and regulation of normal transcripts, several RNA decay pathways in both the nucleus and the cytoplasm ensure the removal of structurally and/or functionally faulty transcripts. Nuclear surveillance occurs for RNAPI, II, and III transcripts and extensive studies from yeast have contributed to the understanding of these mechanisms. Defective rRNAs in the nucleus are eliminated through the TRAMP/exosome complex [45, 46]. Hypomodified tRNAiMet can be recognized by the Rrp44p subunit of the exosome and requires the activity of the TRAMP complex [47]. Recent evidence suggests that Rat1p and Xrn1p-mediated degradation may be the predominant pathway for degradation of several mature hypomodified tRNAs [48]. Aberrant RNAPII transcripts can undergo surveillance early on during synthesis. Transcription elongation and mRNP assembly require the activity of the THO (Tho2p, Hpr1p, Mft1p, and Thp2p) complex and factors involved in mRNA export (Sub2p helicase, Yra1p, and Tex1p). Together, these factors associate to form the transcription-export (TREX) complex. Yeast mutants in the THO complex or the associated Sub2p show a rapid decrease in steady-state levels of several RNAs in an exosome-dependent manner [46, 49]. Defects in THO/Sub2p also result in inefficient polyadenylation, leading to subsequent TRAMP/exosome-dependent transcript degradation. Pre-mRNAs that reach the nuclear pore are subject to perinuclear mRNP surveillance, mediated in part by the pre-mRNA retention and splicing (RES) complex, consisting of Snu17p, Bud13p, and Pml1p. The nuclear pore complex (NPC) factor Mlp1p also functions in this pathway, along with Nup60p, Pml39p, and Esc1p in a pathway regulated by Ulp1p desumoylating enzyme [49]. Degradation of perinuclearly retained mRNPs is thought to be initiated by the Swt1p endonuclease and may involve subsequent digestion by the exosome [50].
Cytoplasmic RNA quality control
RNPs that successfully exit the nucleus are still subject to further checkpoints. Studies from yeast have shown that rRNAs in mature ribosomes with deleterious point mutations in the peptidyl transferase center (25S rRNA) or the decoding site (18S rRNA) undergo more rapid decay than their wild-type counterparts [51, 52]. This process, termed nonfunctional rRNA decay (NRD), is a late stage ribosome quality control pathway that takes place in the cytoplasm. Degradation of the mutant 18S rRNA, but not 25S rRNA, is dependent on translation elongation, since addition of elongation inhibitors such as cycloheximide or hygromycin B stabilized these transcripts. Whereas both mutant transcripts were partly subject to exosome-mediated decay, only the aberrant 18S transcript was sensitive to Ski7p and Xrn1p and appeared to localize in P bodies. Analysis of the half-lives of mutant 18S transcripts in strains containing deletions of factors involved in various cytoplasmic decay pathways provided evidence that 18S NRD was also dependent on Dom34p and Hbs1p, factors that participate in the “no-go decay” (NGD) pathway (discussed below) used to eliminate transcripts containing stalled ribosomes.
mRNAs that exhibit translational defects are also subject to accelerated degradation. The most well studied pathway in this category is the evolutionarily conserved nonsense-mediated mRNA decay (NMD) pathway [53, 54]. This pathway targets mRNAs that contain premature translation termination codons (PTCs), thus minimizing the accumulation of potentially toxic truncated polypeptides. However, the mere presence of a premature nonsense codon in the mRNA is insufficient to lead to NMD. Translation of the mRNA and recognition of the early stop are necessary first steps and mutations or drugs that impair translation initiation, elongation, or termination stabilize nonsense-containing mRNAs. All eukaryotes have a conserved set of core NMD factors encoded by the UPF1, UPF2/NMD2, and UPF3 genes. Upf1p has been shown to interact with Upf2p/Nmd2p, which in turn can also interact with Upf3p, and it has been proposed that all three factors function as a complex. Higher eukaryotes additionally require the products encoded by the SMG1 and SMG5-9 genes [54, 55]. The central component of the NMD pathway is the ATPase/helicase phosphoprotein Upf1p, with the other factors serving to regulate its function. In higher eukaryotes, cycles of phosphorylation/dephosphorylation mediate Upf1 function. Smg1, Upf2, and Upf3 promote phospho-Upf1, whereas Smgs 5, 6, and 7 serve to dephosphorylate Upf1.
Recognition of the PTC in most eukaryotes, which occurs in a UPF-dependent manner, triggers decapping of the substrate mRNA, usually without prior deadenylation, and the decapped transcript is then degraded primarily by the 5′→3′ Xrn1p exonuclease, with some degradation occurring 3′→5′ by the cytoplasmic exosome. Decay activity in Drosophila NMD is instituted by endonucleolytic cleavage of the nonsense mRNA by the PIN-containing Smg6 protein followed by exosome-mediated digestion of the 5′ cleavage fragment and Xrn1-mediated degradation of the unprotected 3′ cleavage fragment (Figure 1) [56].
Despite the conservation of the decay pathway, the detailed mechanism by which PTCs are recognized has yet to be resolved. In mammalian cells, NMD is activated when a PTC is 50–55 nucleotides upstream of an exon-exon junction, the site of deposition of the exon junction complex (EJC). The latter contains the Upf2 and Upf3X proteins and is thought to mediate metazoan NMD. The “pioneer round model” proposes that translation by the first, or pioneer, ribosome is able to displace EJCs from each spliced junction. Hence, the presence of a PTC upstream of an EJC would allow Upf1 that is part of the SURF (Smg1, Upf1, eRF1, and eRF3) complex on the terminating ribosome to interact with the EJC through Upf2, leading to Upf1 phosphorylation by the Smg1 kinase, and subsequent decapping and decay. Amongst other observations, tethering EJC factors downstream of a normal termination codon leads to mRNA destabilization, providing support for this model [57]. Other observations suggest that excessive distance between the premature termination codon and the poly(A) tail of metazoan mRNAs may also trigger NMD [58].
In lower eukaryotes, NMD is predicated on the idea that PTC recognition occurs due to an absence of factors 3′ to the nonsense codon that are associated with a normal 3′-UTR. According to this “faux 3′-UTR” model, normal termination codons are located in the vicinity of bound Pab1p. Interaction between 3′-bound Pab1p (or a factor whose UTR-association may be enhanced by Pab1p) and the terminating ribosome precludes the stable interaction of the Upf1p/Upf2p/Upf3p (Upf) complex to the ribosome and mediates efficient peptide hydrolysis and ribosome release. At a PTC, the absence of these factors leads to inefficient termination and stable binding of the Upf proteins, a prerequisite for decapping and subsequent Xrn1p-mediated degradation. In support of this model tethering Pab1p downstream of a nonsense codon leads to mRNA stabilization in yeast, fly, and human cells [59–61]. The recent observation that neither Pab1p nor a poly(A) tail are required to preclude NMD in yeast suggests that, under some circumstances, other factors may compensate for the Pab1p function in normal termination [62].
In contrast to mRNAs with abbreviated ORFs, transcripts lacking in-frame stop codons undergo non-stop decay (NSD), which serves to not only downregulate the production of extended polypeptides, but also to release ribosomes that are stalled at the 3′ end of an mRNA with no codon at the A-site. NSD requires eRF3 and the exosome co-factor, Ski7p. The latter protein shows similarity to translation elongation factor eEF1A and may serve to recruit the exosome to the mRNA. In addition, translation through the poly(A) tail likely displaces Pab1p, leading to the promotion of decapping and Xrn1p-mediated decay [2, 57].
Transcripts in yeast on which translation elongation is blocked, e.g. by the introduction of a stem-loop structure, are recognized and degraded by the “no-go” mRNA decay (NGD) pathway. Unlike NMD or NSD which initiate exonucleolytic digestion of their transcripts, NGD initiates an endonucleolytic cleavage event close to the stalled ribosome by an as yet unidentified factor. Similar to other decay pathways initiated by endonucleases, the cleaved fragments are further degraded by the combined actions of the 3′→5′ exosome and the 5′→3′ Xrn1 proteins. NGD involves the activity of the eRF1-related factor, Dom34p, and Hbs1p, a GTPase family member related to eEF1A, and may mimic the roles of eRF1 and eRF3 (also a GTPase) in ribosome dissociation from the mRNA [2, 57].
Conclusion
The bottom line...
Degradative mechanisms are important determinants of the steady-state levels of RNAs in a cell and constitute an important means of gene expression regulation. RNA decay pathways influence the fidelity of gene expression by maintaining error-free rRNA, mRNA, and tRNA at every step during their synthesis and utilization. Moreover, the regulation of mRNA stability constitutes a rapid means of modulating protein expression to ensure adaptability of an organism to changing environments. For example, virus survival upon infection in many cases requires the virus to co-opt the host translation apparatus for viral protein synthesis. In order to minimize competition for the host protein synthetic machinery, some viruses express factors that target host cell mRNAs for degradation. Herpes simplex virus 2 (HSV-2) viral host shutoff protein, Vhs, is a nuclease that degrades host cell mRNA [63], and KSHV encodes the shutoff and exonuclease (SOX) protein which, despite having no apparent nuclease activity, induces bulk mRNA degradation through hyperadenylation of host cell mRNAs [64]. In some instances, functional decay pathways can lead to, or aggravate, disease states, such as in the case of genetic disorders caused by the downregulation of PTC-containing mRNAs by the NMD pathway [54, 65]. The importance of understanding RNA decay mechanisms thus not only provides insight into added layers of gene regulation that can be exploited for therapeutic use [66], but has also identified mechanisms such as RNA interference that have proved invaluable in the understanding of numerous biological phenomena.
Acknowledgments
We thank members of the Jacobson laboratory for editorial comments and discussions. This work was supported by grants to A.J. from the National Institutes of Health (R37 GM27757) and the Human Frontier Science Program (RGP0018).
References
- 1.Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci. 2005;30:506–14. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–8. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 3.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223.1–223.14. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 6.Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Molec Cell. 2004 doi: 10.1016/j.molcel.2004.06.028. (in press) [DOI] [PubMed] [Google Scholar]
- 8.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 10.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangus DA, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stalder L, Muhlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009;15:1265–1273. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008;14:246–262. doi: 10.1261/rna.847408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–95. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 23.Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- 24.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 25.Inoue A, Arao Y, Omori A, Ichinose S, Nishio K, Yamamoto N, Kinoshita Y, Mita S. Identification of S1 proteins B2, C1 and D1 as AUF1 isoforms and their major role as heterogeneous nuclear ribonucleoprotein proteins. Biochem J. 2003;372:775–785. doi: 10.1042/BJ20021719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 27.Anderson JR, Mukherjee D, Muthukumaraswamy K, Moraes KC, Wilusz CJ, Wilusz J. Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. Rna. 2006;12:1810–6. doi: 10.1261/rna.144606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 29.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 30.Maquat LE, Gong C. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans. 2009;37:1287–1292. doi: 10.1042/BST0371287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–9. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 34.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, 3rd, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gitlin L, Andino R. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2009 doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 37.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–97. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 38.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends in Biochemical Sciences. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 40.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Dengl S, Cramer P. Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro. J Biol Chem. 2009;284:21270–21279. doi: 10.1074/jbc.M109.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoeftner S, Blasco MA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. Embo J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. Embo J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Lebreton A, Seraphin B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim Biophys Acta. 2008;1779:558–565. doi: 10.1016/j.bbagrm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Andersen KR, Jensen TH, Brodersen DE. Take the “A” tail--quality control of ribosomal and transfer RNA. Biochim Biophys Acta. 2008;1779:532–537. doi: 10.1016/j.bbagrm.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 50.Skruzny M, Schneider C, Racz A, Weng J, Tollervey D, Hurt E. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 2009;7:0164–0174. doi: 10.1371/journal.pbio.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell. 2006;24:619–626. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 54.Jacobson A, Izaurralde E. Nonsense-mediated mRNA decay: from yeast to metazoans. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 655–687. [Google Scholar]
- 55.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 57.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–56. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 58.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 62.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–140. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korom M, Wylie KM, Morrison LA. Selective ablation of virion host shutoff protein RNase activity attenuates herpes simplex virus 2 in mice. J Virol. 2008;82:3642–3653. doi: 10.1128/JVI.02409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YJ, Glaunsinger BA. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009;7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neu-Yilik G, Kulozik AE. NMD: multitasking between mRNA surveillance and modulation of gene expression. Adv Genet. 2008;62:185–243. doi: 10.1016/S0065-2660(08)00604-4. [DOI] [PubMed] [Google Scholar]
- 66.Peltz SW, Welch EM, Trotta CR, Davis T, Jacobson A. Targeting post-transcriptional control for drug discovery. RNA Biol. 2009;6:329–334. doi: 10.4161/rna.6.3.8953. [DOI] [PubMed] [Google Scholar]