Abstract
Background
With the exception of the oyster herpesvirus OsHV-1, all herpesviruses characterized thus far infect only vertebrates. Some cause neurological disease in their hosts, while others replicate or become latent in neurological tissues. Recently a new herpesvirus causing ganglioneuritis in abalone, a gastropod, was discovered. Molecular analysis of new herpesviruses, such as this one and others, still to be discovered in invertebrates, will provide insight into the evolution of herpesviruses.
Results
We sequenced the genome of a neurotropic virus linked to a fatal ganglioneuritis devastating parts of a valuable wild abalone fishery in Australia. We show that the newly identified virus forms part of an ancient clade with its nearest relatives being a herpesvirus infecting bivalves (oyster) and, unexpectedly, one we identified, from published data, apparently integrated within the genome of amphioxus, an invertebrate chordate. Predicted protein sequences from the abalone virus genome have significant similarity to several herpesvirus proteins including the DNA packaging ATPase subunit of (putative) terminase and DNA polymerase. Conservation of amino acid sequences in the terminase across all herpesviruses and phylogenetic analysis using the DNA polymerase and terminase proteins demonstrate that the herpesviruses infecting the molluscs, oyster and abalone, are distantly related. The terminase and polymerase protein sequences from the putative amphioxus herpesvirus share more sequence similarity with those of the mollusc viruses than with sequences from any of the vertebrate herpesviruses analysed.
Conclusions
A family of mollusc herpesviruses, Malacoherpesviridae, that was based on a single virus infecting oyster can now be further established by including a distantly related herpesvirus infecting abalone, which, like many vertebrate viruses is neurotropic. The genome of Branchiostoma floridae (amphioxus) provides evidence for the existence of a herpesvirus associated with this invertebrate chordate. The virus which likely infected amphioxus is, by molecular phylogenetic analysis, more closely related to the other 2 invertebrate viruses than to herpesviruses infecting vertebrates (ie chordates).
Findings
In 2005 there was an outbreak of acute ganglioneuritis in an Australian population of the edible gastropod mollusc, abalone or Haliotis spp[1]. Using transmission electron microscopy, herpes-like particles were observed in ganglia of affected abalone[2] and purified virions from moribund abalone nervous tissues were identified as resembling those of herpesviruses, having an icosohedral capsid approximately 100 nm in diameter surrounded by a 150 nm diameter spiked envelope[3]. Potential herpesvirus particles were also identified previously in Taiwan following mortalities in Haliotis diversicolor [4]. Recently a diagnostic PCR test has been developed to detect the abalone virus [5]. The test has detected viral DNA sequences in diseased abalone from separate geographical locations in Australia and in DNA isolated from a herpes-like virus found some time ago in Taiwan[4].
Three herpesvirus families have been described in the order Herpesvirales - the Herpesviridae which infect Mammalia, Aves and Reptilia, the Alloherpesviridae infecting Amphibia and Osteichthyes (bony fish), and the mollusc-infecting Malacoherpesviridae containing a single virus that infects an invertebrate class, Bivalvia [6-8]. The phylogenetic relationships of these herpesviruses have been well studied and their evolution over epochs is largely synchronous with host lineages [7,8]. Gastropods separated early in the Cambrian period from all other known herpesvirus hosts. This unique evolutionary positioning[6] combined with our discovery of a related herpesvirus genome apparently integrated into the genome of another invertebrate, amphioxus, expands the Herpesvirales order and provides two key links to understanding the nature of the ancient ancestors of mollusc and vertebrate herpesviruses. To understand the structural and evolutionary relationships of the abalone virus to other herpesviruses, we purified abalone virus particles and isolated and sequenced genomic DNA using methods previously described[3,9]. The DNA was subjected to multiple displacement amplification[10] and sequenced using the Roche 454 GS-FLX system followed by partial genome assembly using the Newbler algorithm (Roche).
Based on the assembled DNA sequences of the abalone virus, several protein coding sequences predicted using Artemis[11] showed varying distant homology to herpesvirus proteins, most notably those of Ostreid herpesvirus 1 (oyster herpesvirus 1, OsHV-1), a virus infecting bivalve mollusc species[12,13]. BLAST analysis[14] of assembled sequence contigs based on predicted proteins identified 39 full length homologues of OsHV-1 genes (Table 1). These coding sequences, within partial genome scaffold sequences, or as individual coding sequences, have been submitted to Genbank. None of the coding sequences identified appear to be split by introns. Full-length sequences encoding homologues of DNA polymerase and the DNA packaging ATPase subunit of the (putative) terminase (henceforth referred to as the polymerase and terminase respectively), were identified and chosen for use in sequence alignments and phylogenetic analysis (Figures 1 &2). Hereafter, we will refer to the new abalone virus as abalone herpesvirus or AbHV-1.
Table 1.
OsHV-1 homologues of AbHV-1 coding sequences
| AbHV-1 | OsHV-1 | BLASTP result | ||||
|---|---|---|---|---|---|---|
| Gene | Genbank | Genbank | Description | Ident. | Score | E value |
| AbHVp002c | ADJ95315.1 | YP_024647.1 | ORF109 terminase | 42% | 620 | 3e-175 |
| AbHVp003 | ADL16651.1 | YP_024565.1 | ORF20 RNR2 | 37% | 252 | 1e-64 |
| AbHVp005c | ADL16652.1 | YP_024602.1 | ORF59 | 24% | 90 | 1e-15 |
| AbHVp006 | ADL16653.1 | YP_024573.1 | ORF28 | 26% | 239 | 2e-60 |
| AbHVp013c | ADL16656.1 | YP_024591.1 | ORF47 | 27% | 336 | 1e-89 |
| AbHVp018c | ADL16657.1 | YP_024590.1 | ORF46 | 31% | 48 | 6e-04 |
| AbHVp019c | ADL16658.1 |
YP_024552.1 YP_024552.1 |
ORF49, ORF7 primase/helicase |
24%, 24% | 94, 74 | 5e-17, 1e-10 |
| AbHVp024 | ADL16662.1 | YP_024567.1 | ORF22 | 23% | 234 | 7e-59 |
| AbHVp031c | ADL16665.1 | YP_024606.1 | ORF66 | 27% | 375 | 2e-101 |
| AbHVp032 | ADL16666.1 | YP_024607.1 | ORF67 | 32% | 247 | 3e-63 |
| AbHVp034 | ADL16667.1 | YP_024575.1 | ORF30 | 27% | 53 | 3e-05 |
| AbHVp037c | ADL16668.1 | YP_024616.1 | ORF77 | 23% | 170 | 1e-39 |
| AbHVp038c | ADL16669.1 | YP_024587.1 | ORF43 | 27% | 70 | 1e-10 |
| AbHVp039c | ADL16670.1 | YP_024634.1 | ORF95 | 27% | 94 | 2e-17 |
| AbHVp043c | ADL16671.1 | YP_024611.1 | ORF71 | 23% | 108 | 1e-21 |
| AbHVp045c | ADL16672.1 | YP_024604.1 | ORF61 | 29% | 185 | 1e-44 |
| # AbHVp050 | ADL16674.1 |
YP_024593.1, YP_024552.1 |
ORF49, ORF7 primase/helicase |
21% 20% | 125, 90 | 4e-26, 1e-15 |
| AbHVp057c | ADJ95314.1 | YP_024639.1 | ORF100 DNA polymerase |
31% | 673 | 0.0 |
| AbHVp064 | HQ400676 | YP_024619.1 | ORF80 | 38.5 | 0.29 | |
| AbHVp070c | HQ400677 | YP_024651.1 | ORF113 | 25% | 105 | 8e-21 |
| AbHVp073c | HQ400678 | YP_024650.1 | ORF112 | 26% | 119 | 7e-25 |
| AbHVp075 | HQ400679 | YP_024649.1 | ORF111 | 32% | 198 | 6e-49 |
| AbHVp086 | HQ400681 | YP_024645.1 | ORF107 | 26% | 134 | 4e-29 |
| AbHVp093 | HQ400682 | YP_024622.1 | ORF83 | 19% | 49 | 3e-04 |
| AbHVp102 | HQ400683 | YP_024630.1 | ORF91 | 30% | 151 | 1e-34 |
| AbHVp104c | HQ400684 | YP_024584.1 | ORF40 | 30% | 238 | 2e-60 |
| AbHVp110 | HQ400685 | YP_024595.1 | ORF52 | 34% | 68 | 4e-10 |
| AbHVp111 | HQ400686 | YP_024596.1 | ORF53 | 23% | 50 | 9e-04 |
| AbHVp112 | HQ400687 |
YP_024597.1, YP_024608.1 |
ORF54, ORF68 | 43% | 643 | 0.0 |
| AbHVp113c | HQ400688 | YP_024657.1 | ORF115 | 32% | 80 | 3e-13 |
| AbHVp117c | HQ400689 | YP_024635.1 | ORF96 | 23% | 53 | 4e-05 |
| AbHVp121 | HQ400690 | YP_024633.1 | ORF94 | 28% | 114 | 2e-23 |
| AbHVp130c | HQ400691 | YP_024605.1 | ORF64 | 36% | 212 | 6e-53 |
| AbHVp131 | HQ400692 | YP_024615.1 | ORF76 | 29% | 202 | 1e-59 |
| AbHVp133 | HQ400693 | YP_024569.1 | ORF24 | 23% | 67 | 3e-09 |
| AbHVp134c | HQ400694 |
YP_024608.1, YP_024597.1 |
ORF68, ORF54 | 53% | 784 | 0.0 |
| AbHVp135c | HQ400695 | YP_024624.1 | ORF85 | 26% | 225 | 1e-56 |
| AbHVp136c | HQ400696 | YP_024588.1 | ORF44 | 32% | 134 | 1e-29 |
| AbHVp137 | HQ400697 | YP_024609.1 | ORF69 | 29% | 172 | 7e-41 |
Figure 1.
Alignment of ATP hydrolase domains from terminase protein sequences. ClustalW alignment of one of the conserved regions of the putative terminase gene - the ATP hydrolase (ATPase) domain from various herpesviruses taken from Table 3, identified using Interproscan. Grey background = >90% conserved amino acids.
Figure 2.
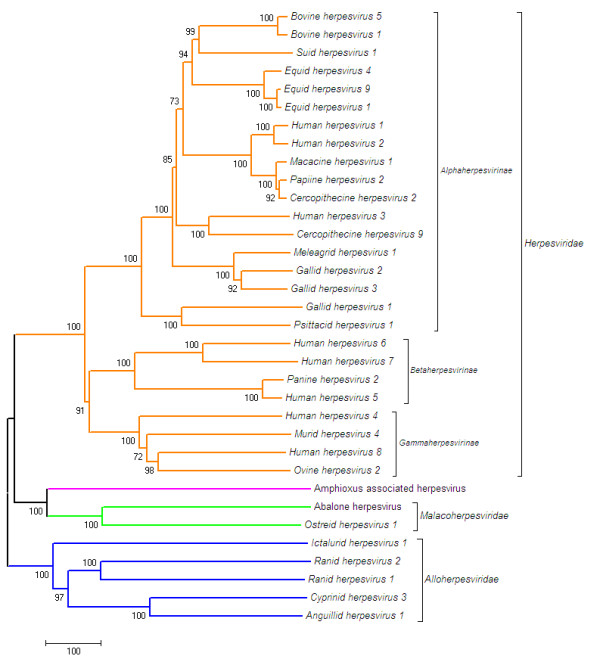
Dendrogram of concatenated DNA polymerase and terminase protein sequences from 34 herpesviruses. Dendrogram illustrating the evolutionary relationship of abalone and amphioxus herpesviruses to 32 other herpesviruses based on the concatenated full length protein sequences of DNA polymerase and the ATPase subunit of the putative terminase for each virus. The tree was inferred with MEGA4[32] using the Minimum Evolution (ME) method and a model based on the number of amino acid differences detected after an alignment using ClustalW[19]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2000 replicates) are shown next to the branches. The scale bar for the branch lengths = 100 amino acid sequence differences.
During the search for homologues of predicted AbHV-1 proteins using BLAST we identified, in the non-redundant (nr) Genbank protein sequence database, Branchiostoma floridae (amphioxus) coding sequences with significant homology to some of those in AbHV-1. The genome of amphioxus has been recently sequenced[15] although final assembly of chromosomes is not yet complete. On further analysis of amphioxus coding sequences using BLASTP with the predicted protein sequences of the oyster herpesvirus OsHV-1 genome (Genbank NC_005881), we identified 19 herpesvirus gene homologues. Consistent with this being an integrated virus, we found that 18 of these genes are clustered within a 150 kb region of a single amphioxus scaffold BRAFLscaffold_217, including the herpesvirus specific terminase gene[16] and all but 4 of these genes do not contain introns. These virus coding sequences appear to be legitimately assembled within published genome sequence scaffolds and are therefore probably integrated within the amphioxus genome. Further experiments such as fluorescence in situ hybridisation of chromosomes would confirm this. The 19 coding sequences identified are listed in Table 2 along with their OsHV-1 homologues and BLAST scores. We utilised the amphioxus virus terminase and polymerase protein sequence homologues in our analyses.
Table 2.
Branchiostoma floridae (amphioxus) homologues of OsHV-1 coding sequences
| OsHV-1 | Branchiostoma floridae | BLASTP result | |||
|---|---|---|---|---|---|
| Accession/ORF | Accession | Location | Ident. | Score | E value |
|
YP_024639.1 ORF100 DNA polymerase |
XP_002591163.1 DNA polymerase |
55013..60373 no introns |
28% | 379 | e-102 |
| YP_024567.1 ORF22 | XP_002591166.1 | 67372..72375 no introns |
24% | 128 | 8e-27 |
|
YP_024552.1 YP_024593.1 ORF7, ORF49 family primase-helicase |
XP_002591168.1 | 76325..79696 no introns |
24% | 122 | 3e-25 |
| YP_024630.1 ORF91 | XP_002591169.1 | 80230..85225 introns predicted |
29% | 114 | 2e-23 |
| YP_024606.1 ORF66 AE_Prim_S_like primase | XP_002591170.1 | 86185..88995 no introns |
24% | 239 | 2e-60 |
| YP_024573.1 ORF28 | XP_002591172.1 | 94244..96667 no introns |
22% | 107 | 4e-21 |
| YP_024641.1 ORF102 | XP_002591174.1 | 99529..101919 no introns |
20% | 78 | 4e-12 |
| YP_024645.1 ORF107 | XP_002591175.1 contains PAT1 domain pfam09770 | 103007..105292 no introns |
24% | 65 | 2e-08 |
| YP_024584.1 ORF40 | XP_002591176.1 | 105441..107045 no introns |
29% | 180 | 4e-43 |
| YP_024643.1 ORF104 | XP_002591178.1 | 108452..110641 no introns |
19% | 101 | 4e-19 |
| YP_024615.1 ORF76 | XP_002591179.1 | 112401..114281 no introns |
26% | 71 | 4e-10 |
| YP_024624.1 ORF85 | XP_002591189.1 | 137878..148379 introns predicted |
22% | 70 | 1e-09 |
|
YP_024597.1 YP_024608.1 ORF54, ORF68 family membrane glycoprotein |
XP_002591190.1 XP_002591197.1 (possible gene family) |
148508..150751 174789..176912 no introns |
30% | 332 | 1e-88 |
| YP_024591.1 ORF47 | XP_002591194.1 | 163504..167571 no introns |
23% | 275 | 4e-71 |
| YP_024647.1 ORF109 terminase | XP_002591195.1 terminase | 168081..170354 no introns |
31% | 308 | 2e-81 |
| YP_024650.1 ORF112 | XP_002591198.1 | 177489..179961 introns predicted |
23% | 68 | 2e-09 |
| YP_024609.1 ORF69 | XP_002591200.1 | 187709..188944 no introns |
25% | 80 | 3e-13 |
| YP_024600.1 ORF57 | XP_002610653.1 chloride channel | BRAFLscaffold_25 2304811..2311488 introns predicted |
30% | 86 | 4e-15 |
Note: B. floridae OsHV homologue locations are all on scaffold BRAFLscaffold_217, except for OsHV ORF57. All OsHV and B. floridae predicted proteins listed are of unknown function unless stated otherwise. Four B. floridae genes are predicted to contain introns. Also 4 other B. floridae genes in the scaffold BRAFLscaffold_217 between 60 kb and 150 kb encode proteins similar to apoptosis regulators like IAP-3 often present in herpesvirus genomes (not listed and not detected using OsHV sequences).
The putative terminase, or DNA packaging ATPase, appears specific to herpesviruses and some bacteriophages, such as T4[16] and is thought to be an enzyme motor involved in packaging viral DNA into preformed capsids[17]. We used the ATPase motif from this protein to investigate the phylogeny of the herpesviruses.
The ATP hydrolase (ATPase) motif sequences from 20 of the 34 terminase proteins listed in Table 3, plus their T4 bacteriophage homologue and the amphioxus terminase homologue (XP_002591195.1, listed in Table 2), were identified using Interproscan[18] and aligned using ClustalW[19]. Figure 1 shows that 12 amino acids are conserved across all herpesvirus ATPase domain sequences, including those from the abalone, oyster and amphioxus virus genomes, indicating the placement of the abalone virus and putative amphioxus virus within the Herpesvirales order. A common ancestral origin for the mollusc and amphioxus viruses is confirmed by the absence of introns in the terminase gene and the presence of additional amino acid loops (Figure 1). Although being in the same clade (Figure 2), at a protein sequence level the mollusc viruses are only moderately related with 40% amino acid identity in this conserved viral protein, across their full length.
Table 3.
Genbank Accessions of Herpesvirus Polymerase and Terminase protein sequences used for phylogenetic analysis
| Virus | Polymerase | Terminase |
|---|---|---|
| Abalone_herpesvirus | ADJ95314.1 | ADJ95315.1 |
| Amphioxus_associated_virus | XP_002591163.1 | XP_002591195.1 |
| Anguillid_herpesvirus_1 | YP_003358194.1 | YP_003358149.1 |
| Bovine_herpesvirus_1 | NP_045328.1 | NP_045342.1 |
| Bovine_herpesvirus_5 | NP_954917.1 | NP_954931.1 |
| Cercopithecine_herpesvirus_2 | YP_164473.1 | YP_164457.1 |
| Cercopithecine_herpesvirus_9 | NP_077443.1 | NP_077457.1 |
| Cyprinid_herpesvirus_3 | YP_001096114.1 | YP_001096069.1 |
| Equid_herpesvirus_1 | YP_053075.1 | YP_053090.1 |
| Equid_herpesvirus_4 | NP_045247.1 | NP_045262.1 |
| Equid_herpesvirus_9 | YP_002333511.1 | YP_002333526.2 |
| Gallid_herpesvirus_1 | YP_182359.1 | YP_182378.2 |
| Gallid_herpesvirus_2 | AAF66765.1 | YP_001033943.1 |
| Gallid_herpesvirus_3 | NP_066862.1 | NP_066845.1 |
| Human_herpesvirus_1 | NP_044632.1 | NP_044616.1 |
| Human_herpesvirus_2 | P07918.1 | NP_044484.1 |
| Human_herpesvirus_3 | NP_040151.1 | NP_040165.1 |
| Human_herpesvirus_4 | YP_401712.1 | YP_401690.1 |
| Human_herpesvirus_5 | P08546.2 | P16732.1 |
| Human_herpesvirus_6 | NP_042931.1 | NP_042953.2 |
| Human_herpesvirus_7 | P52342.1 | YP_073802.1 |
| Human_herpesvirus_8 | AAC57086.1 | YP_001129382.1 |
| Ictalurid_herpesvirus_1 | NP_041148.2 | NP_041153.2 |
| Macacine_herpesvirus_1 | NP_851890.1 | NP_851874.1 |
| Meleagrid_herpesvirus_1 | NP_073324.1 | NP_073308.1 |
| Murid_herpesvirus_4 | NP_044849.1 | NP_044866.2 |
| Ostreid_herpesvirus_1 | YP_024639.1 | YP_024647.1 |
| Ovine_herpesvirus_2 | YP_438136.1 | YP_438152.1 |
| Panine_herpesvirus_2 | NP_612698.1 | NP_612722.1 |
| Papiine_herpesvirus_2 | YP_443877.1 | YP_443861.1 |
| Psittacid_herpesvirus_1 | NP_944403.1 | NP_944422.2 |
| Ranid_herpesvirus_1 | YP_656727.1 | YP_656697.1 |
| Ranid_herpesvirus_2 | YP_656618.1 | YP_656576.1 |
| Suid_herpesvirus_1 | YP_068333.1 | YP_068358.1 |
The phylogenetic analysis comparing concatenated polymerase and terminase full-length proteins (Figure 2, Table 3), illustrates the evolutionary relationships within the Herpesvirales order. The five Alloherpesviridae viruses are clustered together, with separate clades for frog and fish viruses as found previously [8], and the Herpesviridae are clustered into separate major clades reflecting their taxonomic groupings of alpha-, beta- and gammaherpesvirinae sub-families. The phylogenetic analysis confirms a relationship between the amphioxus virus and the abalone and oyster viruses in a deep invertebrate clade. The level of divergence makes estimation of the relative divergence times of the 3 herpesvirus families difficult. Interestingly, the amphioxus virus is in the clade with mollusc viruses, which may not have been expected given the amphioxus chordate host lineage is more aligned with vertebrates than molluscs.
The invertebrate herpesvirus clade provides a unique branching point to inform the metazoan diversification of the herpesviruses. It is thought that during the Cambrian era, the Bilaterial species diverged to generate the Protostomes (evolving into such animals as flatworms, molluscs and arthropods) and the Deuterostomes (from which the chordates and then the vertebrates evolved)[20,21]. Molluscs emerged more than 100 My before vertebrates with a bony skeleton (the current known range of herpesviruses in vertebrates). One hypothesis to explain the diversity of viruses within vertebrates and the positioning of the mollusc viruses among them, rather than as an ancestral outgroup, is the existence of diverse herpesviruses in Cambrian metazoans. Consistent with this hypothesis, previous estimates for the divergence of just the Herpesviridae in vertebrates indicate a divergence of alpha-, beta- and gammaherpesviruses to over 400 Mya, and longer times are predicted for divergence of Alloherpesviridae and Malacoherpesviridae[7]. An alternate hypothesis to explain the branching of the 3 herpesvirus families is that molluscs acquired herpesviruses by transmission in the aquatic environment, for example through association such as mollusc predation of early chordates. It appears that modern Malacoherpesviridae may have the ability to infect across species, a feature not typically observed in vertebrate herpesviruses, although the infection observed is restricted to related mollusc species[22].
As more sequence data and gene structure for Alloherpesviridae, Malacoherpesviridae and other invertebrate herpesviruses become available it will allow a more informative analysis of their evolution. Of particular interest will be new herpesviruses yet to be discovered in species which share bilateral symmetry such as amphioxus, sea squirts, flatworms or squid. Our discovery of clustered intact herpesvirus genes in amphioxus suggests an opportunistic integration has occurred in the amphioxus genome. This may not be a normal feature of infection and latency, but herpesviruses can occasionally integrate into the genome of their host[23]. Surprisingly, the nearest relatives of this chordate virus seem to be the viruses infecting molluscs rather than those of fish or frogs. Although herpesvirus particles have not been seen in the more primitive metazoan species, their existence is suspected; short herpes-like DNA sequences having been found in a metagenomic study of Hawaian coral[24]. Further metagenomic approaches similar to those described previously[25] and PCR-directed approaches[26] based on new sequences described here will enable these evolutionary questions to be addressed. The sequence information is also crucial for the development of molecular diagnostic tools to monitor and manage disease outbreaks.
The neurotropism of certain herpesviruses is well documented but this behaviour is not known outside the families of herpesviruses infecting terrestrial vertebrates[27,28]. The neurotropic tissue infection profile of the new gastropod virus analysed here is shared with some viruses within the Herpesviridae family. Convergent evolution may have given rise to the neurotropism seen in some members of the Herpesviridae and now the Malacoherpesviridae families. The rooting of a neurotropic invertebrate virus near or before the divergence of alpha-, beta-, and gammaherpesviruses, may also suggest that early mammalian herpesvirus precursors were neurotropic and that some have retained this feature over time. It is interesting to speculate as to the earliest functional interactions between sensory cells and viruses, as the first sign of neurons appeared over 600 million years ago in "cnidarians," (eg: hydra), but organisms basal to them like sponges do not have neurons or synapses[29]. Recent evidence indicates sponges have gene networks in cells which were precursors to nerve cells including proteins related to virus nerve entry receptors[30]. Others[24] have speculated on a link between herpesvirus neurotropism and the evolution of modern herpesviruses from ancestors infecting invertebrates such as Cnidaria (for example, coral or sea anemones), thought to be related to the first species with sensory receptors[31]. Further, the discovery reported here of a putative herpesvirus integrated into the genome of amphioxus hints at a wide diversity of herpesviruses within the invertebrate community, perhaps dating back to before the divergence of arthropods, molluscs and chordates. It will be exciting to discover such invertebrate herpesviruses and explore their links to ancient herpesvirus ancestors.
To accommodate the new abalone virus, which we have suggested naming abalone herpesvirus or AbHV-1, within the Herpesvirales order, we suggest the creation of a new genus called Haliotivirus within the Malacoherpesviridae family and assignment of AbHV-1 as a species under Haliotivirus (as Haliotid herpesvirus 1).
We have referred to the putative virus genome integrated into the Branchiostomid species chromosome as amphioxus-associated virus, AaHV-1. We suggest the species name Branchiostomid herpesvirus 1. Given the unique nature of the virus revealed by phylogenetic analysis and the unique evolutionary positioning of amphioxus as an invertebrate chordate, we suggest this virus, if classified, could be a member of a new family, Aspondyloherpesviridae (from the Greek for "no spine").
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KWS, FW, BGC, SW conceived and designed the experiments; FW, NC performed the experiments; KWS, TS, DS analyzed the data; FW, SW, TS, DS, NC contributed reagents, materials, analysis tools; KWS, BGC wrote the paper. All authors have contributed to the editing or revision of the manuscript and approve its publication.
Contributor Information
Keith W Savin, Email: keith.savin@dpi.vic.gov.au.
Benjamin G Cocks, Email: ben.cocks@dpi.vic.gov.au.
Frank Wong, Email: frank.wong@csiro.au.
Tim Sawbridge, Email: tim.sawbridge@dpi.vic.gov.au.
Noel Cogan, Email: noel.cogan@dpi.vic.gov.au.
David Savage, Email: david.savage@mac.com.
Simone Warner, Email: simone.warner@dpi.vic.gov.au.
Acknowledgements
The authors wish to thank Fisheries Victoria for supplying infected abalone, German Spangenberg for facilitating the genome sequencing and Megan Vardy for technical assistance during generation of DNA sequence data. Funding was provided by the Department of Primary Industries, Victoria, Australia, The Commonwealth Scientific & Industrial Organisation, Australia and the Fisheries Research & Development Corp., Australia. The funding bodies had no role in the study design, data collection, analysis or interpretation, manuscript preparation or submission other than contributing to author salaries and experiment costs.
References
- Hooper C, Hardy-Smith P, Handlinger J. Ganglioneuritis causing high mortalities in farmed Australian abalone (Haliotis laevigata and Haliotis rubra) Australian Veterinary Journal. 2007;85:188–193. doi: 10.1111/j.1751-0813.2007.00155.x. [DOI] [PubMed] [Google Scholar]
- NACA. Quarterly Aquatic Animal Disease Report (Asia and Pacific Region) 2006/1. Book Quarterly Aquatic Animal Disease Report (Asia and Pacific Region) 2006/1 (Editor ed.^eds.). City. 2006. p. 5.
- Tan J, Lancaster M, Hyatt A, van Driel R, Wong F, Warner S. Purification of a herpes-like virus from abalone (Haliotis spp.) with ganglioneuritis and detection by transmission electron microscopy. Journal of Virological Methods. 2008;149:338–341. doi: 10.1016/j.jviromet.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Chang PH, Kuo ST, Lai SH, Yang HS, Ting YY, Hsu CL, Chen HC. Herpes-like virus infection causing mortality of cultured abalone Haliotis diversicolor supertexta in Taiwan. Dis Aquat Organ. 2005;65:23–27. doi: 10.3354/dao065023. [DOI] [PubMed] [Google Scholar]
- Corbeil S, Colling A, Williams LM, Wong FYK, Savin K, Warner S, Murdoch B, Cogan NOI, Sawbridge TI, Fegan M. et al. Development and validation of a TaqMan PCR assay for the Australian abalone herpes-like virus. Dis Aquat Organ. 2010;91:1–10. doi: 10.3354/dao02277. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Research. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Waltzek TB, Kelley GO, Alfaro ME, Kurobe T, Davison AJ, Hedrick RP. Phylogenetic relationships in the family Alloherpesviridae. Dis Aquat Organ. 2009;84:179–194. doi: 10.3354/dao02023. [DOI] [PubMed] [Google Scholar]
- Le Deuff RM, Renault T. Purification and partial genome characterization of a herpes-like virus infecting the Japanese oyster, Crassostrea gigas. J Gen Virol. 1999;80:1317–1322. doi: 10.1099/0022-1317-80-5-1317. [DOI] [PubMed] [Google Scholar]
- Silander K, Saarela J. Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol Biol. 2008;439:1–18. doi: 10.1007/978-1-59745-188-8_1. full_text. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Farley CA, Banfield WG, Kasnic G Jr, Foster WS. Oyster herpes-type virus. Science. 1972;178:759–760. doi: 10.1126/science.178.4062.759. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Trus BL, Cheng N, Steven AC, Watson MS, Cunningham C, Le Deuff RM, Renault T. A novel class of herpesvirus with bivalve hosts. J Gen Virol. 2005;86:41–53. doi: 10.1099/vir.0.80382-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Davison AJ. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-U. [DOI] [PubMed] [Google Scholar]
- Yang K, Homa F, Baines JD. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J Virol. 2007;81:6419–6433. doi: 10.1128/JVI.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. Six major steps in animal evolution: are we derived sponge larvae? Evol Dev. 2008;10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- Arzul I, Renault T, Lipart C, Davison AJ. Evidence for interspecies transmission of oyster herpesvirus in marine bivalves. J Gen Virol. 2001;82:865–870. doi: 10.1099/0022-1317-82-4-865. [DOI] [PubMed] [Google Scholar]
- Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG. et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, Edwards RA, Haynes M, Angly FE, Wegley L, Rohwer FL. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Ehlers B, Dural G, Yasmum N, Lembo T, de Thoisy B, Ryser-Degiorgis MP, Ulrich RG, McGeoch DJ. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J Virol. 2008;82:3509–3516. doi: 10.1128/JVI.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW, Husak PJ, Banfield BW, Smith GA. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. full_text. [DOI] [PubMed] [Google Scholar]
- Terry LA, Stewart JP, Nash AA, Fazakerley JK. Murine gammaherpesvirus-68 infection of and persistence in the central nervous system. J Gen Virol. 2000;81:2635–2643. doi: 10.1099/0022-1317-81-11-2635. [DOI] [PubMed] [Google Scholar]
- Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS. A post-synaptic scaffold at the origin of the animal kingdom. PLoS One. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV. et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]



