Abstract
Background
H9N2 avian influenza A viruses have become panzootic in Eurasia over the last decade and have caused several human infections in Asia since 1998. To study their evolution and zoonotic potential, we conducted an in silico analysis of H9N2 viruses that have infected humans between 1997 and 2009 and identified potential novel reassortments.
Results
A total of 22 hemagglutinin (HA) and neuraminidase (NA) nucleotide and deduced amino acid sequences were retrieved from the NCBI flu database. It was identified that mature peptide sequences of HA genes isolated from humans in 2009 had glutamine at position 226 (H3) of the receptor binding site, indicating a preference to bind to the human α (2-6) sialic acid receptors, which is different from previously isolated viruses and studies where the presence of leucine at the same position contributes to preference for human receptors and presence of glutamine towards avian receptors. Similarly, strains isolated in 2009 possessed new motif R-S-N-R in spite of typical R-S-S-R at the cleavage site of HA, which isn't reported before for H9N2 cases in humans. Other changes involved loss, addition, and variations in potential glycosylation sites as well as in predicted epitopes. The results of phylogenetic analysis indicated that HA and NA gene segments of H9N2 including those from current and proposed vaccine strains belong to two different Eurasian phylogenetic lineages confirming possible genetic reassortments.
Conclusions
These findings support the continuous evolution of avian H9N2 viruses towards human as host and are in favor of effective surveillance and better characterization studies to address this issue.
Background
The H9N2 influenza A viruses have been known to cause infection in the poultry population around the globe including Ireland, Iran, Germany, Italy, Pakistan, Saudi Arabia, South Africa and USA since mid-1990 s [1]. In 1998, domestic pigs from Hong Kong were also observed to be infected with H9N2 influenza Y280-like viruses [2]. Several human cases of H9N2 infection have been recorded since 1997 from Hong Kong and China in children and adults exhibiting influenza like symptoms and mild upper respiratory tract infections [2-6]. Genetic analysis of H9N2 viruses from Hong Kong live bird markets showed the preferential binding of viruses to 2, 6-linked sialic acid, human-like receptors [6,7]. All these findings pointed towards the possibility of interspecies transmission of H9N2 viruses and its persistent threat to the human population.
Influenza viruses belonging to the Orthomyxoviradae family of viruses are divided into eight single stranded RNA segments encoding ten proteins. These include two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), along with nucleoproteins (NP), three polymerase proteins (PA, PB1, PB2) two matrix proteins (M1, M2) and non-structural proteins (NS1, NS2) [8-11]. Of these ten proteins HA and NA are primarily responsible for facilitating influenza virus infection. There are 16 HA and nine NA subtypes. HA is involved in the early stages of infection, causing the binding of the sialic acid receptor present on the host cell surface, and leading to fusion of the viral and endosomal membrane and subsequent entry into the host cell [11]. Virus aggregation is prevented by the NA glycoprotein and by the cleavage of the α-ketosodic linkage between sialic acid and an adjacent sugar residue. This facilitates the movement of the virus to and from the site of infection by destruction of receptors recognized by HA [12]. Previous studies have defined two distinct lineages of H9N2 influenza viruses: North American and Eurasian. The Eurasian lineage can be further divided into three major sublineages; the G1 lineage, represented by A/Quail/Hong Kong/G1/97 (G1-like); the Y280 lineage, represented by three prototype viruses A/duck/Hong Kong/Y280/97 (Y280-like), A/Chicken/Beijing/1/94 (BJ94-like), and A/Chicken/Hong Kong/G9/97 (G9-like) and the Korean lineage, represented by A/chicken/Korea/38349-p96323/96 (Korean-like) and A/duck/Hong Kong/Y439/97 (Y439-like) [7,13,14].
It is important to study the evolution of H9N2 viruses because of their constant prevalence in poultry flocks and repeated emergence in the human population. The present study involved computational molecular analysis and phylogenetic characterization of 11 influenza A (H9N2) viruses which have been isolated between 1997 and 2009. The aim of this study was to aid in understanding the evolution of pandemic H9N2 strains, which have circulated various animal populations in the indicated period.
Methods
Viruses
To perform this study, a computational search of all reported cases of influenza A H9N2 human infections from 1997 to 2009 was conducted. A total of eleven nucleotide and their respective deduced amino acid sequences for each of hemagglutinin (HA) and neuraminidase (NA) segments were retrieved from the NCBI flu database [15] accessed on April, 19, 2010. The viruses used in this study are listed in Table 1. Sequencing data was obtained together with information of the host, subtype, isolation year, and isolation place. The selected sequences of H9N2 human cases were then aligned and compared by using multiple sequence alignment software ClustalW2 [16].
Table 1.
GenBank accession numbers genes and proteins of avian influenza A (H9N2) viruses isolated from 1997 to 2009 from humans.
| Accession Numbers | |||||
|---|---|---|---|---|---|
| Year | Strains | Hemagglutinin | Neuraminidase | ||
| Genes | Proteins | Genes | Proteins | ||
| 1997 | A/Hong Kong/1074/97 | GU053179.1 | ACZ48627 | GU053180.1 | ACZ45242 |
| 1998 | A/Shantou/239/98 | AY043015.1 | AAL32476 | AY043021.1 | AAL32481 |
| A/Shaoguan/408/98 | AY043017.1 | AAL32477 | AY043022.1 | AAL32482 | |
| A/Shaoguan/447/98 | AY043018.1 | AAL32478 | AY043023.1 | AAL32483 | |
| 1999 | A/Hong Kong/1073/99a | AJ404626.1 | CAB95856 | AJ404629.1 | CAB95859 |
| A/Hong Kong/1074/99 | AJ404627.1 | CAB95857 | AJ404628.1 | CAB95858 | |
| A/Guangzhou/333/99 | AY043019.1 | AAL32479 | AY043024.1 | AAL32484 | |
| 2003 | A/Hong Kong/2108/03 | DQ226106.1 | ABB58945 | DQ226128.1 | ABB58956 |
| 2008 | A/Hong Kong/3239/08 | CY055156.1 | ADC41863 | CY055158.1 | ADC41865 |
| 2009 | A/Hong Kong/33982009b | CY055140.1 | ADC41843 | CY055142.1 | ADC41845 |
| A/Hong Kong/35820/09 | CY055148.1 | ADC41853 | CY055150.1 | ADC41855 | |
a A(H9N2) Available vaccine virus (G1 Clade; WHO report, 2010)
b A(H9N2) Proposed vaccine virus (G1 Clade; WHO report, 2010)
Potential Glycosylation sites and antigenic variations
Identification and comparison of N-glycosylation sites into reported protein sequences of HA and NA was performed by an online server ScanProsite [17] and the extent of antigenic variations between viruses was checked by the CTL epitope prediction method [18]. Each amino acid sequence of HA and NA was evaluated separately using consensus approach.
Phylogenetic characterization and tree construction
Phylogenetic patterns of NA and HA nucleotide sequences of H9N2 influenza viruses isolated between 1997 and 2009 from humans were observed using MEGA4.0.2 [19]. The selected nucleotide sequences based on local alignment and homology searches using BLAST were aligned by using CLUSTALW. Unrooted phylogenetic trees were constructed by using minimum evolution analysis with maximum composite likelihood and the Tamura-Nei model. Internal branching probabilities were determined by bootstrap analysis of 1000 replicates and are indicated by percentage value on each branch.
Results and Discussion
Molecular Analysis
Influenza A viruses of subtype H9N2 are now considered to be widespread in poultry and have demonstrated the ability to infect humans [20]. The recurring presence of H9N2 infections in humans has raised concerns about the possibility of H9N2 viruses evolving into pandemic strains. Therefore, it is crucial to evaluate the potential pandemic threat posed by H9N2 viruses using experimental and computational approaches. During this study, bioinformatics analysis of HA and NA from H9N2 viruses was performed and the key residues in receptor binding sites (RBS), the cleavage motifs of HA and NA hemadsorbing sites (HB), stalk length and enzyme active sites were studied in detail.
Hemagglutinin
It has been well documented that the receptor binding site motif of HA is critical for cellular receptor specificity and determining virus host range [21,22]. Out of five conserved amino acids in the pocket of the HA glycoprotein, two positions showed the maximum number of mutations (198, 234) and three remained 99% conserved (191, 235, and 236). The pattern of observed mutations at position 198 was E198T, E198A, E198V and E198D, whereas for position 234 was L234Q, Q234L and Q234M as summarized in Table 2. Mutations in these regions are considered strong factors for change in sugar specificity thus leading to change in host specificity. The presence of glutamine (Q) at position 234 (H3 numbering: 226) is a typical avian virus signature, and it has been reported that presence of this amino acid results in a preference for binding to 2,3-linked sialic acid (avian receptors) whereas, in the case of leucine (L) at the same position, there is a preference for 2,6-linked sialic acid (human receptors) and potential cause of reported human infections [5,6,23-25]. However, upon analysis of amino acids at the receptor binding site of HA glycoproteins, we have identified that H9N2 viruses isolated in 2009 from infected patients in Hong Kong possessed glutamine at position 234 (H3 numbering: 226) instead of leucine as has been found in previous isolates, yet still somehow managed to bind with human sialic receptors. As shown in Table 2, this residue variation in avian H9N2 viruses has not been reported before in human cases. This observation can be correlated with a number of phenomena such as genetic evolution of influenza viruses in order to evade host defense mechanisms, and transfer of this virus from swine to humans instead of avian to human transmission. It is also known that pigs act as a "mixing vessel" because viruses isolated from pigs recognize both types of sialic receptors [26] and most importantly represent the balance that exists between the activities of HA and NA. Both the genes PB2 and HA are known to be critical for the pathogenicity of the virus. Therefore, the role of other proteins such as the PB2 segment [4,27] must be viewed as potential causes of zoonotic H9N2 possibly resulting in future human to human transmission.
Table 2.
Comparison of critical amino acid residues in hemagglutinin and neuraminidase proteins.
| Hemagglutinin | Neuraminidase | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viruses | Receptor Binding Site | cleavage Site | Stalk deletions | Hemadsorbing Site | drug binding pocket | ||||||||||||||||||||||||||||
| 191 | 198 | 234 | 235 | 236 | 335 - 338 | 38-39 | 46-50 | 62-64 | 366 - 373 | 399 - 404 | 431 - 433 | 118 | 119 | 151 | 152 | 277 | 292 | 371 | |||||||||||||||
| H | E | L | Q | G | RSSR | I | K | K | D | S | R | S | G | D | S | D | I | R | S | P | Q | E | R | E | D | R | E | R | R | ||||
| A/Quail/Hong Kong/G1/971 | - | - | - | - | - | - | Yes | No | No | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A/Duck/Hong Kong/Y280/972 | - | T | - | - | - | - | No | No | Yes | - | - | E | - | - | - | - | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Chicken/Beijing/1/942 | - | V | Q | - | - | - | |||||||||||||||||||||||||||
| A/Chicken/Hong Kong/G9/972 | - | A | - | - | - | - | |||||||||||||||||||||||||||
| A/Duck/Hong Kong/Y439/973 | - | - | Q | - | - | A-N- | |||||||||||||||||||||||||||
| A/Ck/Korea/38349-p96323/963 | - | - | Q | - | - | A-Y- | No | No | No | - | S | - | - | - | - | - | - | - | N | N | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Hong Kong/1074/97 | - | - | - | - | - | - | Yes | No | No | - | - | - | - | - | - | A | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Shantou/239/98 | N | A | - | - | - | - | No | No | No | - | - | - | - | - | - | - | - | - | - | - | S | S | - | - | K | - | - | - | - | - | - | - | - |
| A/Shaoguan/408/98 | N | A | - | - | - | - | No | No | Yes | - | - | E | - | - | - | - | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Shaoguan/447/98 | N | A | - | - | - | - | No | No | Yes | - | - | E | - | - | - | - | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Hong Kong/1073/99a | - | - | - | - | - | - | Yes | No | No | - | - | - | - | - | - | - | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Hong Kong/1074/99 | - | - | - | - | - | - | Yes | No | No | - | - | - | - | - | - | - | - | - | - | - | N | W | - | - | - | - | - | - | - | - | - | - | - |
| A/Guangzhou/333/99 | N | V | M | - | - | - | No | No | No | - | E | - | - | - | - | - | - | - | - | - | N | S | - | - | K | - | - | - | - | - | - | - | - |
| A/Hong Kong/2108/03 | N | V | - | - | - | - | No | No | No | - | E | - | - | - | - | - | - | - | R | - | N | S | - | - | K | - | - | - | - | - | - | - | - |
| A/Hong Kong/3239/08 | N | A | - | - | - | - | No | No | No | - | E | - | - | - | - | - | - | - | - | - | N | S | - | - | K | - | - | - | - | - | - | - | - |
| A/Hong Kong/33982/09b | - | D | Q | - | - | --N- | No | No | No | - | E | - | - | - | - | - | - | - | R | - | N | S | - | - | K | - | - | - | - | - | - | - | - |
| A/Hong Kong/35820/09 | - | D | Q | - | - | --N- | No | No | No | - | E | - | - | - | - | - | - | - | R | - | N | S | - | - | K | - | - | - | - | - | - | - | - |
Amino acid residues were compared in the hemagglutinin receptor binding site and cleavage sites, as well as in the neuraminidase stalk region, HB site and antiviral drug binding sites of influenza A H9N2 viruses isolated during 1997-2009 from humans. Residues differences and similarities are compared with reference strains of lineages (G1-like1, Y280-like2 and Korean-like3) and vaccine strains
a, b. Dashes indicate identical/conserved residues.
a A(H9N2) Available vaccine virus (G1 Clade; WHO report, 2010) b A(H9N2) Proposed vaccine virus (G1 Clade; WHO report, 2010)
Analysis of the HA cleavage site showed that strains isolated in 2009 have a different cleavage site compare to those previously reported. From 1997 to 2008, all H9N2 viruses retained a conserved amino acid pattern at the cleavage site; 335RSSR338GLF341 except for the sublineage prototype strain (A/chicken/Kr/38349-p96323/96) that had the cleavage site 335ASYR338GLF341. Presence of the R-S-S-R motif is suggestive of H9N2 viruses with low pathogenicity that have adapted to chicken host [28-30]. However, it has been observed that viruses isolated in 2009 have the different pattern (335RSNR338GLF341) due to the substitution mutation S337N (Table 2). To our knowledge, this is the first report to show presence of the R-S-N-R motif in avian H9N2 influenza viruses isolated from human patients although the R-S-N-R motif had been reported before in different studies on avian H9N2 viruses from around the globe [31-33]. The significance of this mutation on viral stability or increased pathogenicity is not fully understood as yet and requires further studies, as the cleavage site is considered an indicator of pathogenicity [34,35]. Comparison with vaccines strains also revealed variations at positions 198 and 234 in the HA glycoprotein. The currently available vaccine virus to the virus has an E and an L at positions 198 and 234, respectively, whereas strains associated with human infections reported in 2009 contain D and Q residues at these respective positions.
Neuraminidase
The major factors known to influence the functional activities of the NA glycoprotein are enzyme active sites, the stalk length, HB site and potential glycosylation sites. The HB site is located on the surface of the NA molecule, away from the neuraminidase enzyme active site [36]. Analysis of the HB site showed substitutions similar to those detected in the Y280 lineage prototype strain (A/Duck/Hong Kong/Y280/97) and in H9N2 human isolates from Hong Kong in 1999. These mutations are typical of human pandemic H2N2 and H3N2 viruses [7,14]. The most prominent mutation is at position 402 from isoleucine to asparagine/serine and at position 403 from arginine to tryptophan/serine. Strains from 1999 - 2009 also showed mutations at position 367 from lysine to glutamic acid and at position 432 from glutamine to lysine as described in Table 2. However, the biological significance of any of these substitutions in the HB site is not yet known.
The NA protein consists of a box-like head with an enzymatic active site that is connected to a fibrous stalk region of variable length. The NA stalk is important for balancing the complementary activity of HA and NA and has been correlated with efficiency of virus replication and pathogenesis. From various studies previously performed it has been concluded that longer stalk length of the virus results in better replication [37-40]. Analysis of stalk length revealed that the two prototype viruses, Qa/Hong Kong/G1/97 and A/Hong Kong/1073/97, of the G1-lineage contained a two amino acid deletion in the NA stalk region at positions 38 and 39, a deletion at position 62 - 64 was observed in A/Duck/Hong Kong/Y280/97, A/Shaoguan/408/98 and A/Shaoguan/447/98. The particular 46-50 amino acid deletion, which is important for poultry adaptation of the virus [41], is not found in any of the analyzed sequences even though these viruses are thought to be transmitted from avian species to humans. Furthermore, as shown in Table 2, from 1999 - 2009 none of the reported strains contained a stalk deletion at any position, which may also be an indication of evolution in the viral structure leading to a NA protein with better replication rate in humans.
Sequence analysis of binding pocket residues in NA for drugs such as zanamivir (Relenza®) and oseltamivir (Tamiflu®) was also performed. These analyses showed that for each virus, amino acids in the enzyme active site were conserved and no substitution mutations were seen, which may lead to an evolution of sialidase inhibitor resistant viruses (Table 2). It is known that mutations at positions other than the active sites still tend to alter the activity of active site amino acids; therefore this possibility cannot be excluded. Such mutations are not always captured or extensively studied in homology models. Therefore, wet lab studies are required to check the level of effectiveness possessed by NA inhibitors against recently reported strains.
Phylogenetic characterization
Evolutionary relationships of HA and NA nucleotide sequences were determined by comparing H9N2 human isolates from 1997 - 2009 with the established Eurasian H9N2 lineages: namely, the G1, Y280 and Korean-lineages represented by their respective prototype viruses (Table 1). The strains of current and proposed vaccines were also included in this analysis.
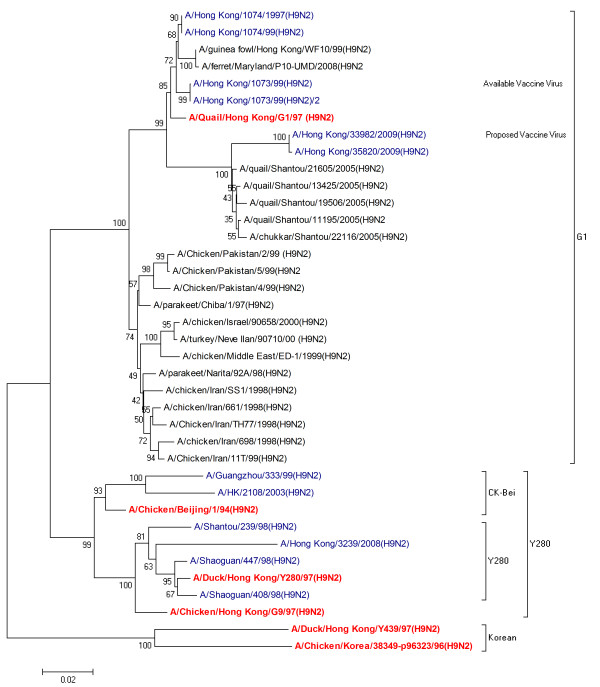
Two distinct groups were observed in the unrooted phylogenetic tree for HA (Figure 1). Viruses isolated from Hong Kong in 1997, 1999, and in 2009 showed a more intimate relationship with the G1 lineage strain and clustered together in one group. Sequences from Hong Kong isolated in 2003 and 2008, along with Chinese strains from 1998 and 1999 showed the close association with the Y280 lineage and clustered together in second group. None of the reported sequences clustered within the Korean-like lineage.
Figure 1.
Phylogenetic relationships of HA genes in H9N2 influenza viruses isolated from humans between 1997 and 2009. A phylogenetic tree was generated using minimum evolution analysis with maximum composite likelihood using the Tamura-Nei model with MEGA software version 4.0.2. Numbers below branches indicate bootstrap value percentages from 1000 replicates. The scale bar represents the distance unit between sequence pairs. Representative prototype viruses for different Eurasian lineages are indicated as red. The sequences of H9N2 influenza viruses isolated from the human population are indicated as blue.
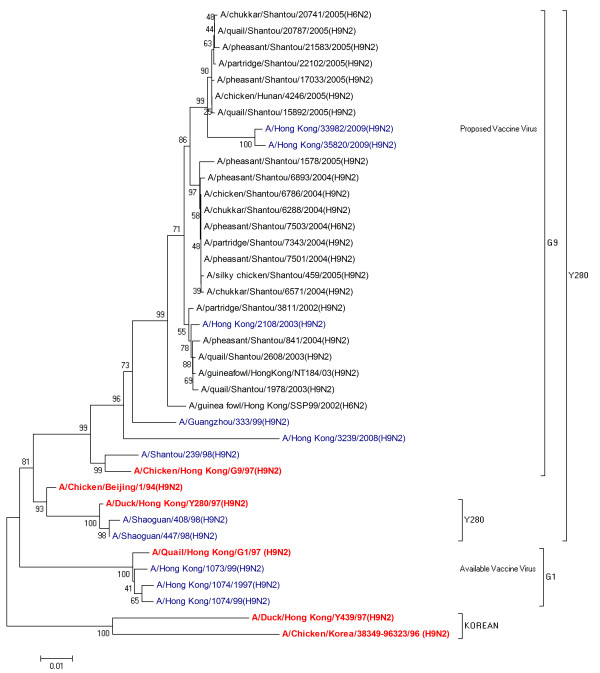
The phylogenetic tree for NA (Figure 2) showed clustering pattern different from that observed in the HA phylogenetic results. Similar to HA, none of the sequences grouped within the Korean-like lineage. However, sequences isolated from Hong Kong in 2003, 2008 and 2009 clustered within G9-like lineage which is one of the prototype strains from the Y280 lineage. It was observed that the proposed vaccine strain and the 2009 strain from Hong Kong appear in the same sub group with 100% similarity. Sequences isolated from Shaoguan, China in 1998 clustered together with the Y280 lineage strain in one subgroup, while another contained the G1 lineage strain along with available vaccine strain and strains from Hong Kong isolated in 1997 and 1999.
Figure 2.
Phylogenetic relationships of NA genes in H9N2 influenza viruses isolated from humans between 1997 and 2009. The phylogenetic methods and abbreviations were as described for figure 1.
Collectively, we observed that phylogenetic relationship of strains at different times and geographical regions displayed complexity and diversity. It was identified that both HA and NA glycoproteins do not belong to a single lineage but originate from different Eurasian sublineages which relates to genetic heterogeneity of H9N2 viruses. These results were also in consensus with the sequence analysis results of strains exhibiting high sequence similarity in the same lineages and sublineages.
N-Glycosylation sites of HA and NA
The N-linked glycosidic moieties have been found to play a vital role in mediating viral infectivity, receptor binding capacity and cell associated host immune responses, as well as protecting critical viral protein epitopes from immune attack [42,43]. Potential glycosylation sites with the N-X-T/S motif (in which X may be any amino acid except proline) were identified, which revealed the region based similarities and yearly variations. For H9N2 HA protein sequences, the N-glycosylation sites of viruses isolated from Hong Kong in 1997 were similar to those isolated in 1999 (current vaccine strain) from Hong Kong. The strains isolated from China in 1998 exhibited a different pattern of glycosylation sites in terms of position and sequence pattern. A similar case was observed for another virus isolated from China (A/Guangzhou/333/99) in 1999, which showed a totally different pattern of glycosylation sites as shown in Table 3. It may be because viruses from China fall into the Y280 lineage while other cases are closest to the G1 sublineage as shown in our phylogenetic analysis. Sequence analysis results also showed that Chinese isolates contained mutations in the HA receptor binding site at positions 191N and 198A, as compared to other viruses having 191 H and 198E or 198 D (Table 2). Viruses from Hong Kong isolated during 2003 contained altogether different N- glycosylation sites at different positions, which were not found in the analysis of previous entries. Glycosylation sites of viral strains from 2008 and 2009 again had high similarity with each other but were different from all the previous strains except at one site with a single mutation 305 - 308 (NISK - NCSK) in 2008 strains, which was also present in Hong Kong viruses from 1997 - 1999.
Table 3.
Comparison of predicted N-glycosylation sites of the virus hemagglutinin proteins.
| Hemagglutinin (HA) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1997 | 1998 | 1999 | 2003 | 2008 | 2009 | ||||||||
| A/Hong Kong/1074/97 |
A/Shantou/239/98 A/Shaoguan/408/98 A/Shaoguan/447/98 |
A/Hong Kong/1073/99 A/Hong Kong/1074/99 |
A/Guangzhou/333/99 | A/Hong Kong/2108/03 | A/Hong Kong/3239/08 |
A/Hong Kong/33982/09 A/Hong Kong/35820/09 |
|||||||
| Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence |
| 29 - 32 | NSTE | 12 - 15 | NSTE | 29 - 32 | NSTE | 124 - 127 | NVTY | 19 - 22 | NSTE | 21 - 24 | NSTE | 21 - 24 | NSTE |
| 105 - 108 | NGTC | 124 - 127 | NVSY | 105 - 108 | NGTC | 281 - 284 | NTTL | 131 - 134 | NVTY | 133 - 136 | NVSY | 97 - 100 | NGTC |
| 141 - 144 | NVTY | 201 - 204 | NRTF | 141 - 144 | NVTY | 288 - 291 | NVSK | 288 - 291 | NTTL | 210 - 213 | NRTF | 133 - 136 | NVTY |
| 206 - 209 | NDTT | 281 - 284 | NTTL | 206 - 209 | NDTT | 475 - 478 | NGTY | 295 - 298 | NVSK | 290 - 293 | NTTL | 198 - 201 | NDTT |
| 218 - 221 | NRTF | 288 - 291 | NVSK | 218 - 221 | NRTF | 482 - 485 | NGTY | 297 - 300 | NVSK | 290 - 293 | NSTL | ||
| 298 - 301 | NSTL | 475 - 478 | NGTY | 298 - 301 | NSTL | 305 - 308 | NCSK | 297 - 300 | NISK | ||||
| 305 - 308 | NISK | 305 - 308 | NISK | 484 - 487 | NGTY | 484 - 487 | NGTY | ||||||
| 492 - 495 | NGTY | 492 - 495 | NGTY | ||||||||||
| 551 - 554 | NGSC | 551 - 554 | NGSC | ||||||||||
Similar to HA, NA sequences isolated from Hong Kong in 1997 and 1999 showed similar glycosylation sites, as did the sequences isolated from China in 1999 and from Hong Kong in 2003. Sequences isolated from Hong Kong in 2008 and 2009 also had similar glycosylation sites. An interesting observation was that in some cases of HA and NA, the glycosylation sites remained the same as those seen in previous or later strains. However, there was a change of position for these sites, such as 198 - 201 (NATA) in Hong Kong/1074/1997 and 183 - 186 (NATA) in Shantou/239/98 (Table 4).
Table 4.
Comparison of predicted N-glycosylation sites of the virus neuraminidase proteins.
| 1997 | 1998 | 1999 | 2003 | 2008 | 2009 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/Hong Kong/1074/97 | A/Shantou/239/98 | A/Shaoguan/408/98 | A/Shaoguan/447/98 |
A/Hong Kong/1073/99 A/Hong Kong/1074/99 |
A/Guangzhou/333/99 | A/Hong Kong/2108/03 | A/Hong Kong/3239/08 |
A/Hong Kong/33982/2009 A/Hong Kong/35820/2009 |
|||||||||
| Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence |
| 59 - 62 | NITE | 27 - 30 | NSSN | 66 - 69 | NSTT | 47 - 50 | NSTT | 59 - 62 | NITE | 44 - 47 | NSSN | 44 - 47 | NSSD | 36 - 39 | NSSD | 36 - 39 | NSSD |
| 67 - 70 | NNTT | 44 - 47 | NITE | 83 - 86 | NWSK | 64 - 67 | NWSK | 67 - 70 | NNTT | 61 - 64 | NITE | 61 - 64 | NITE | 53 - 56 | NITE | 53 - 56 | NITE |
| 68 - 71 | NTTI | 52 - 55 | NSTT | 143 - 146 | NGTT | 124 - 127 | NGTT | 68 - 71 | NTTI | 69 - 72 | NSTT | 69 - 72 | NSTT | 61 - 64 | NSTT | 61 - 64 | NSTT |
| 84 - 87 | NWSK | 69 - 72 | NWSK | 197 - 200 | NATA | 178 - 181 | NATA | 84 - 87 | NWSK | 86 - 89 | NWSK | 86 - 89 | NWSK | 138 - 141 | NGTS | 78 - 81 | NWSK |
| 144 - 147 | NGTI | 129 - 132 | NGTS | 231 - 234 | NGTC | 212 - 215 | NGTC | 144 - 147 | NGTI | 146 - 149 | NGTA | 146 - 149 | NGTA | 192 - 195 | NATA | 138 - 141 | NGTT |
| 198 - 201 | NATA | 183 - 186 | NATA | 399 - 402 | NWSG | 380 - 383 | NWSG | 198 - 201 | NATA | 200 - 203 | NATA | 200 - 203 | NATA | 226 - 229 | NGTC | 192 - 195 | NATA |
| 232 - 235 | NGTC | 217 - 220 | NGTC | 232 - 235 | NGTC | 234 - 237 | NGTC | 234 - 237 | NGTC | 394 - 397 | NSSG | 226 - 229 | NGTC | ||||
| 400 - 403 | NWSG | 400 - 403 | NWSG | 402 - 405 | NSSG | 402 - 405 | NSSG | 394 - 397 | NSSG | ||||||||
Antigenic sites of HA and NA
Analysis of antigenic sites of HA and NA also showed region based similarities and yearly variations as observed for glycosylation sites distribution. Viruses isolated from Hong Kong in 1997 and 1999 had the same antigenic sites as strains isolated from China in 1998 and 1999, except for A/Shaoguan/447/98 which had only one antigenic site (219 - GRIDYYWSV) similar to other Chinese strains. Strains isolated from Hong Kong in 2003, 2008 and 2009 had similar antigens as the earlier strains from the region but at different positions compared to the strains from 2008 and 2009, but again presenting similar antigenic site positions (Table 5).
Table 5.
Comparison of antigenic sites in amino acid sequences of hemagglutinin proteins (H9N2).
| 1997 | 1998 | 1999 | 2003 | 2008 | 2009 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/Hong Kong/1074/97 | A/Shantou/239/98 | A/Shaoguan/408/98 | A/Shaoguan/447/98 |
A/Hong Kong/1073/99 A/Hong Kong/1074/99 |
A/Guangzhou/333/99 | A/Hong Kong/2108/03 | A/Hong Kong/3239/08 |
A/Hong Kong/33982/2009 A/Hong Kong/35820/2009 |
|||||||||
| Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence |
| 431 | AYNAELLVL | 414 | AYNAELLVL | 414 | AYNAELLVL | 253 | GESHGRILK | 431 | AYNAELLVL | 414 | AYNAELLVL | 421 | AYNAELLVL | 423 | AYNAELLVL | 423 | AYNAELLVL |
| 108 | CYPGNVENL | 91 | CYPGNVENL | 91 | CYPGNVENL | 473 | IRNGTYNRR | 108 | CYPGNVENL | 91 | CYPGNVENL | 98 | CYPGNVENL | 100 | CYPGNVENL | 100 | CYPGNVENL |
| 236 | GRIDYYWSV | 219 | GRIDYYWSV | 428 | TLDDHDANV | 219 | GRIDYYWSV | 236 | GRIDYYWSV | 219 | GRIDYYWSV | 226 | GRIDYYWSV | 228 | GRIDYYWSV | 228 | GRIDYYWSV |
| 312 | GTCPKYVRV | 117 | IFPDTIWNV | 219 | GRIDYYWSV | 360 | DSTQKAIDK | 312 | GTCPKYVRV | 82 | ERPSAVNEM | 124 | IFPDTIWNV | 126 | IFPDTIWNV | 304 | GTCPKYVKV |
| 134 | IFPDTTWNV | 392 | SEVENRLNM | 117 | IFPDTIWNV | 396 | TRLNMINNK | 134 | IFPDTTWNV | 117 | IFPDTIWNV | 314 | KLAVGMRNV | 316 | KLAVGLRNV | 316 | KLAIGLRNV |
For NA, the sequences isolated from Hong Kong in 1997 and 1999 had the same antigenic sites. Sequences from China isolated in 1998 had similar antigenic sites but at different positions with variation at one site. Chinese sequences from 1999 had three sites that were the same as those in the Hong Kong viruses isolated during 2003. Sequences isolated from Hong Kong in 2008 and 2009 also had similar antigenic sites as previous strains from the region but at different positions. Furthermore, in 2008, one new antigenic site was observed in a sequence (102 - ASGDIWVTR) and another two in 2009 sequences at positions 19 (ALFATTMTL) and 240 (GRADTRILF) as shown in Table 6. Another observation was that both the available and proposed vaccine strains are not fully coordinated with the antigenic epitope regions of the HA and NA genes. Therefore, in case of reappearance of incompatible viral strains, vaccine products may become less effective.
Table 6.
Comparison of antigenic sites in amino acid sequences of neuraminidase proteins (H9N2).
| 1997 | 1998 | 1999 | 2003 | 2008 | 2009 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/Hong Kong/1074/97 | A/Shantou/239/98 | A/Shaoguan/408/98 | A/Shaoguan/447/98 |
A/Hong Kong/1073/99 A/Hong Kong/1074/99 |
A/Guangzhou/333/99 | A/Hong Kong/2108/03 | A/Hong Kong/3239/08 |
A/Hong Kong/33982/2009 A/Hong Kong/35820/2009 |
|||||||||
| Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | Sequence |
| 98 | FSKDNSIRL | 83 | FSKDNSIRL | 97 | FSKDNSIRL | 78 | FSKDNSIRL | 98 | FSKDNSIRL | 100 | FSKDNSIRL | 100 | FSKDNSIRL | 92 | FSKDNSIRL | 92 | FSKDNSIRL |
| 295 | GSNRPVLYI | 280 | GSNRPVLYI | 294 | GSNRPVLYI | 275 | GSNRPVLYI | 295 | GSNRPVLYI | 248 | GRADTRILF | 297 | GSNRPVLYI | 289 | GSNRPVLYI | 19 | ALFATTMTL |
| 459 | GANINFMSI | 231 | GRADTRILF | 152 | HRTLLMNEL | 133 | HRTLLMNEL | 459 | GANINFMSI | 155 | HRTLLMNEL | 248 | GRADTRILF | 102 | ASGDIWVTR | 240 | GRADTRILF |
| 153 | HRTLLMNEL | 138 | HRTLLMNEL | 145 | TTHDRIPHR | 126 | TTHDRIPHR | 153 | HRTLLMNEL | 378 | RVIGGWITA | 155 | HRTLLMNEL | 147 | HRTLLMNEL | 289 | GSNRPVLFI |
| 277 | SCYPRYPEV | 282 | NRPVLYINM | 296 | NRPVLYINM | 277 | NRPVLYINM | 277 | SCYPRYPEV | 279 | SCYPRYPEV | 458 | WPDGANINL | 291 | NRPVLYINM | 102 | AAGDIWVTR |
The potential for differences in antigenic variations is high, especially when compared across sublineages of H9N2 viruses. Xu et al [44] demonstrated antigenic diversity in H9N2 viruses using monoclonal antibodies, which corresponded with phylogenetic relationships. Findings of the study by Xu et al also correspond with our results where H9N2 viruses showed sequence variations and antigenic diversity based on evolution. Furthermore, differences in glycosylation of the virion surface proteins may contribute to antigenic variations, however this requires further evaluation.
Conclusions
In summary, we have reported an in silico molecular analysis of HA and NA genes and respective deduced amino acid sequences from H9N2 avian influenza viruses that were isolated from humans between 1997 and 2009. We found that H9N2 surface genes belonged to two distinct lineages - G1 and Y280, indicating that they have different sources of origin. Sequence analysis revealed unique variations in antigenic and N - linked glycosylation sites. Drug binding pockets remained highly conserved in all reported strains and hence, the activity of NA inhibitors should remain unaffected. However, matching with vaccine strains showed variations, requiring further investigations in animal models. The RBS modification of leucine to glutamine (Leu226Glu) instead of glutamine to leucine and a new cleavage site motif (R-S-N-R) for HA is related to the balancing activity of NA. Findings from the study support the genetic instability of influenza A (H9N2) viruses and highlight the necessity for more comprehensive surveillance and further evaluation of H9N2 viruses with proper in vitro and in vivo models.
Abbreviations
HA: Hemagglutinin; NA: Neuraminidase; PGS: Potential Glycosylation Sites; RBS: Receptor Binding Site; HB: Hemadsorbing Site.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AMB designed the study, performed molecular analysis and drafted the manuscript. SS performed phylogenetic and antigenic analysis. MI and YT analyzed the data. YT finalized the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Azeem M Butt, Email: azeem@cemb.edu.pk.
Samerene Siddique, Email: crankycockroach@hotmail.com.
Muhammad Idrees, Email: idreeskhan@cemb.edu.pk.
Yigang Tong, Email: tong.yigang@gmail.com.
Acknowledgements
We are thankful to the Director of the National Centre of Excellence in Molecular Biology (CEMB) for encouragement and providing facilities to carry out this work.
References
- Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. doi: 10.1016/S0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- Guo Y, Li J, Cheng X. [Discovery of men infected by avian influenza A (H9N2) virus] Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999;13:105–108. [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary "human" H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97(17):9654–8. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. J Virol. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza a virus hemagglutinin subtype (h16) obtained from black-headed gulls. J Virol. 2005;79(5):2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal RP, Tosh C, Pattnaik B, Behera P, Nagarajan S, Gounalan S, Shrivastava N, Shankar BP, Pradhan HK. Analysis of the pb2 gene reveals that indian h5n1 influenza virus belongs to a mixed-migratory bird sub-lineage possessing the amino acid lysine at position 627 of the pb2 protein. Arch Virol. 2007;152(9):1637–1644. doi: 10.1007/s00705-007-1002-5. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000;355:827–835. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the "internal" genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology. 2007;361:45–55. doi: 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The Influenza Virus Resource at the National Center for Biotechnology Information. J Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin M, Raghava GPS. Prediction of ctl epitopes using qm, svm and ann techniques. Vaccine. 2004;22(23-24):3195–3204. doi: 10.1016/j.vaccine.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- Gambaryan A, Webster R, Matrosovich M. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol. 2002;147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol. 2007;81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Renshaw M, Tumpey TM, Kelly GD, Hu-Primmer J, Katz JM. Immunity to influenza A H9N2 viruses induced by infection and vaccination. J Virol. 2001;75:4896–4901. doi: 10.1128/JVI.75.10.4896-4901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golender N, Panshin A, Banet-Noach C, Nagar S, Pokamunski S, Pirak M, Tendler Y, Davidson I, Garcia M, Perk S. Genetic characterization of avian influenza viruses isolated in Israel during 2000-2006. Virus Genes. 2008;37:289–297. doi: 10.1007/s11262-008-0272-7. [DOI] [PubMed] [Google Scholar]
- Monne I, Cattoli G, Mazzacan E, Amarin NM, Al Maaitah HM, Al-Natour MQ, Capua I. Genetic comparison of H9N2 AI viruses isolated in Jordan in 2003. Avian Dis. 2007;51:451–454. doi: 10.1637/7563-033106R.1. [DOI] [PubMed] [Google Scholar]
- Banks J, Speidel EC, Harris PA, Alexander DJ. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol. 2000;29:353–359. doi: 10.1080/03079450050118485. [DOI] [PubMed] [Google Scholar]
- Perk S, Golender N, Banet-Noach C, Shihmanter E, Pokamunsky S, Pirak M, Tendler Y, Lipkind M, Panshin A. Phylogenetic analysis of hemagglutinin, neuraminidase, and nucleoprotein genes of H9N2 avian influenza viruses isolated in Israel during the 2000-2005 epizootic. Comp Immunol Microbiol Infect Dis. 2009;32:221–238. doi: 10.1016/j.cimid.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Perk S, Banet-Noach C, Shihmanter E, Pokamunski S, Pirak M, Lipkind M, Panshin A. Genetic characterization of the H9N2 influenza viruses circulated in the poultry population in Israel. Comp Immunol Microbiol Infect Dis. 2006;29:207–223. doi: 10.1016/j.cimid.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Tosh C, Nagarajan S, Behera P, Rajukumar K, Purohit K, Kamal RP, Murugkar HV, Gounalan S, Pattnaik B, Vanamayya PR, Pradhan HK, Dubey SC. Genetic analysis of H9N2 avian influenza viruses isolated from India. Arch Virol. 2008;153:1433–1439. doi: 10.1007/s00705-008-0131-9. [DOI] [PubMed] [Google Scholar]
- Klenk HD, Rott R. The molecular biology of influenza virus pathogenicity. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. full_text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, Fukutomi A, Katunuma N. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem. 1992;267:13573–9. [PubMed] [Google Scholar]
- Kobasa D, Wells K, Kawaoka Y. Amino acids responsible for the absolute sialidase activity of the influenza A virus neuraminidase: relationship to growth in the duck intestine. J Virol. 2001;75:11773–11780. doi: 10.1128/JVI.75.23.11773-11780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A° resolution. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. Bioessays. 2003;25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000;74:6015–6020. doi: 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M, Yaqub T, Reddy K, McCauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One. 2009;4:e5788. doi: 10.1371/journal.pone.0005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 1997;71:8377–8384. doi: 10.1128/jvi.71.11.8377-8384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Xu KM, Li KS, Smith GJD, Li JW, Tai H, Zhang JX, Webster RG, Peiris JSM, Chen H, Guan Y. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol. 2007;81:2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]