Abstract
Objective
Chemokines coordinate leukocyte trafficking in homeostasis and during immune responses. Prior studies of their role in arthritis have employed animal models with both an initial adaptive immune response and an inflammatory effector phase. This study focused on chemokines and their receptors in the effector phase of arthritis using the K/BxN serum-transfer model.
Methods
A time-course microarray analysis of serum-transferred arthritis was performed, examining ankle, synovial fluid and peripheral blood. Upregulation of chemokines was confirmed by quantitative RT-PCR. The functional relevance of chemokine induction was assessed by transferring serum into mice deficient in CCR1–CCR7, CCR9, CXCR2, CXCR3, CXCR5, CX3CR1, CCL2 or CCL3. Further mechanistic analysis of CXCR2 involved treatment of arthritic mice with a CXCR2 antagonist, bone-marrow transfers with CXCR2+/− and CXCR2−/− donors and recipients, flow cytometry of synovial cells, and competition experiments measuring enrichment of CXCR2-expressing neutrophils in arthritic joints of mice with mixed CXCR2+/+ and CXCR2−/− bone-marrow.
Results
Gene-expression profiling revealed upregulation of the CXCR2 ligands CXCL1, CXCL2 and CXCL5 in the joint in parallel with disease activity. CXCR2−/− mice had attenuated disease relative to CXCR2+/− littermates, as did mice receiving the CXCR2 inhibitor, while deficiency of other chemokine receptors did not affect arthritis severity. CXCR2 was required only on hematopoietic cells and was widely expressed on synovial neutrophils. CXCR2-expressing neutrophils were preferentially recruited to arthritic joints in the presence of CXCR2-deficient neutrophils.
Conclusion
CXCR2 (but not other chemokine receptors) is critical for the development of autoantibody-mediated arthritis, exhibiting a cell-autonomous role in neutrophil recruitment to inflamed joints.
Introduction
Chemokines are a family of over 50 small chemotactic proteins, divided into four structural subfamilies (CC, CXC, C, and CX3C). They have wide-ranging roles in immunity, including the organization of lymphoid organ architecture; homeostatic trafficking of naïve lymphocytes; migration of T and B cells to functional niches within lymphoid organs (such as germinal centers) during adaptive immune responses; constitutive trafficking of mast cells, monocytes/macrophages and memory T cells to non-inflamed tissues; and leukocyte recruitment to sites of inflammation (1). Chemokines act by increasing integrin-mediated leukocyte adhesion to endothelium, thereby facilitating leukocyte extravasion, and by establishing ordered gradients that direct leukocyte movement within tissues. Certain of them have additional functions such as promoting angiogenesis, inducing leukocyte degranulation and activating cells to secrete inflammatory mediators. Chemokines exert their biological activity via binding to G-protein-coupled receptors; over 20 are known, classed according to their ligand subfamily (CCR, CXCR, XCR, and CX3CR). Considerable redundancy exists in chemokine-chemokine receptor pairings, with most chemokine receptors binding multiple chemokines and many chemokines binding to more than one receptor.
There has been extensive interest in characterizing the function of chemokines and their receptors in rheumatoid arthritis (RA). Numerous chemokines have been identified in the synovium and synovial fluid (SF) of RA patients, including, but not limited to, CCL2–5, CCL8, CCL19–21, CXCL1, CXCL5–10, CXCL12–13, CXCL16, CX3CL1, and XCL1 (2;3). Synovial macrophages and fibroblast-like synoviocytes are thought to be the primary sources of chemokines in RA tissue, although other cells like neutrophils, mast cells, lymphocytes and endothelial cells also contribute to the chemokine milieu. Most chemokine receptors are expressed in RA synovium, generally on multiple cell types; these include CCR1–7, CXCR1–6, CX3CR1 and XCR1 (2;3). There are only limited data on chemokine and chemokine receptor inhibition in RA: small clinical trials of CCR1, CCR2 and CCL2 inhibitors, with only a non-significant trend toward improvement in the trial of the CCR1 antagonist (3–5).
Multiple chemokines and chemokine receptors have been implicated in animal models of arthritis on the basis of experiments employing inhibitory antibodies, pharmacological antagonists, and/or gene-targeted mice, although we are unaware of any model that has been comprehensively examined. For collagen-induced arthritis (CIA), blockade of CCL2, CCL3, CXCL2, CXCL13, CXCL16, CCR1 or CXCR4 inhibited disease, whereas genetic deficiency of CCR2 exacerbated it (2;6–10). Adjuvant-induced arthritis was ameliorated by blockade of CCL2, CCL5, CXCL1, CXCL5, CXCL10, CCR2, CXCR2 or CXCR3 (2;11–14). Antigen-induced arthritis was dampened by blockade of CXCR2 or genetic deficiency of CCR7 or CXCR5 (15;16). These models are dependent on both an initial adaptive immune response and a downstream inflammatory cascade involving innate immune cells and cytokines such as IL-1 and TNFα (17). As chemokines could be involved in both phases of disease, it remains unclear to what extent the role of chemokines is to orchestrate T- and B-cell responses or to mediate inflammatory cell recruitment to arthritic joints.
In the K/BxN arthritis model, the upstream adaptive immune response required to generate arthritogenic autoantibodies can be conveniently separated from the downstream autoantibody-mediated effector phase. The initial transgenic model was discovered after nonobese diabetic (NOD) mice were fortuitously crossed with C57Bl/6 (B6) mice carrying the KRN T-cell receptor transgenes (18). KRN recognizes a peptide from a ubiquitous glycolytic enzyme, glucose-6-phosphate isomerase (GPI), presented by the NOD-derived Ag7 major histocompatibility complex molecule (17). In K/BxN mice, an autoimmune response develops, with production of pathogenic anti-GPI antibodies that induce arthritis when transferred into normal recipients. These antibodies deposit on joint surfaces and mediate arthritis by activating the alternative complement pathway and triggering Fcγ receptors (19). Neutrophils and mast cells are both required for serum-transferred arthritis, and recruitment of neutrophils is critically dependent on the leukotriene B4 and its receptor, BLT1, such that disease does not develop in their absence (20–22).
Here we present a comprehensive analysis of chemokines and their receptors in K/BxN serum-transferred arthritis, coupling gene-expression profiling and knockout mice. Our results highlight the contribution of chemokines that trigger CXCR2, in particular in the recruitment of neutrophils.
Materials and Methods
Mice
B6, Balb/c, CXCR2−/− (N8), CCR2−/− (N10), CCR5−/− (N6), CCL2−/− (N10) and CCL3−/− (N7) mice were obtained from the Jackson Laboratory; CXCR3−/− (N11), CXCR5−/− (N2, a gift from Dr. Martin Lipp), CCR1−/− (N6), CCR3−/− (N9), CCR4−/− (N10), CCR6−/− (N20, a gift from Dr. Sergio Lira), CCR7−/− (N8, a gift from Dr. Martin Lipp), CCR9−/− (N10), CX3CR1−/− (N12, a gift from Dr. Philip Murphy) and KRN-transgenic mice have been previously described (16;18;23–29). N indicates the backcross generation, including additional crosses in our facility using mice from the Jackson Laboratory. The lines used were on the B6 genetic background, except CXCR2 and CCR3, which were of the Balb/c background. K/BxN mice were generated by crossing KRN-transgenic B6 mice with NOD mice.
Serum-transfer experiments used matched littermate controls generated from heterozygote or heterozygote/homozygote crosses, with the exception of the CCR4 and CCR9 lines, in which B6 mice from Charles River Laboratories (the source of mice used to backcross these lines) were taken as controls. Genotypes were assessed by genomic polymerase chain reaction (PCR). These experiments were reviewed by the Harvard Medical School Institutional Animal Care and Use Committee, protocol numbers 02956 and 03024.
Serum-transferred arthritis
K/BxN serum was collected from 8-week-old K/BxN mice and pooled for each experiment. Arthritis was induced by intraperitoneal injection of 200 µl (for microarray experiments), 150 µl (B6 strains), 75 µl (Balb/c strains) or 2.5 µl/g (CXCR2 line, due to its variable size) of K/BxN serum on days 0 and 2. Ankle thickness was measured with a J15 Blet micrometer. Inhibition experiments utilized the CXCR2 antagonist, SB-332235 (a gift from GlaxoSmithKline), administered at 50 mg/kg by oral gavage daily; controls received water by oral gavage daily (15).
RNA isolation
Ankles were dissected to expose the articular cavity and repeatedly flushed with 1 mM EDTA in phosphate buffered saline (PBS) to collect SF. RNA was extracted from SF cells and peripheral blood leukocytes (PBLs) using Trizol (Ambion) per the manufacturer’s instructions. RNA was isolated from dissected ankles using a modified LiCl/Urea protocol. Briefly, dissected ankle joints were digested in 6 M Urea/2% SDS to liberate RNA into solution. Genomic DNA was sheared by homogenization, and RNA was selectively precipitated with LiCl. The resulting RNA-containing sample was treated with proteinase K, extracted with chloroform:phenol, mixed with Glycoblue (Ambion), and precipitated with ethanol. All RNA samples were reprecipitated with NaOAc/ethanol and treated with DNase (Invitrogen).
Microarray Analysis
RNA samples were amplified and biotinylated using the MessageAmp kit (Ambion), fragmented, hybridized to U74Av2 gene chips (Affymetrix), conjugated with streptavidin-phycoerythrin and read by the Affymetrix GeneArray Scanner. Probe-level hybridization intensities were processed into expression values using the Robust Multi-chip Analysis (RMA) method (30). RMA gene expression values were log2 transformed and averaged for each time-point.
The magnitude of gene upregulation or downregulation was measured by averaging the fold-change for consecutive time-points and using the greatest value as a metric (two-time-point fold-change). The false-discovery rate of two-time-point fold-change cutoffs was estimated using random datasets generated from Gaussian distributions. Hierarchical clustering was performed using Cluster 3.0 (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/).
Quantitative RT-PCR
Single-stranded cDNA was generated from RNA samples using SuperscriptII reverse-transcriptase (InVitrogen) per the manufacturer’s instructions. The cDNA was amplified by PCR using Sybr Green Master Mix (Applied Biosystems) and the Mx3000p thermal cycler (Stratagene). Primers were designed using PerlPrimer (http://perlprimer.sourceforge.net), meeting the following criteria: spanned introns, aligned only with the intended mRNA target on BLAST search, and produced amplification products of <300 base-pairs. Reaction curves were scored by the cycle number (Ct) at which they met a preset fluorescence threshold. These values were converted into quantitative expression values using linear regression of a 1:4 dilution series of reference cDNA. Gene-expression values were normalized to that of the hprt gene.
Bone-marrow cell transfers
Bone marrow (BM) was collected from donor mice by flushing dissected tibia and femurs with PBS. Red blood cells were lysed with ACK buffer and a single-cell suspension was prepared by passing the BM flush over a nylon mesh filter. The BM single-cell suspension was stained with biotin-labeled antibodies against CD3, CD4 and CD8α; treated with magnetic streptavidin-linked beads (Miltenyi Biotech); and passed through magnetic separation columns. Recipient Balb/c mice were lethally irradiated with 2 doses of 600 Rads, separated by 4 hours, then reconstituted with 3×106 BM cells in DMEM transferred by tail-vein injection.
Flow cytometry
SF cells and PBLs were stained with fluorophore-labeled antibodies, washed and analyzed on a Beckman Coulter. The following antibodies were used: FITC anti-GR1 (eBioscience), PE anti-CXCR2 (R&D Systems), and PE-Cy5 anti-CD11b (eBioscience). Analysis was performed using Expo32 (Beckman).
Statistical analysis
Disease severity and quantitative RT-PCR data are represented as mean ± SEM. P-values were calculated from microarray gene-expression data using one-way ANOVA. P-values were calculated from ankle-thickening data by taking the area under the curve over 20 days for each mouse and performing two-tailed Students’ t-tests.
Results
Gene-expression profiling of K/BxN serum-transferred arthritis
A microarray analysis was performed to characterize the molecular events underlying pathology in K/BxN serum-transferred arthritis. Ankle tissue, PBLs, and SF samples were collected at six time-points (or four for SF): day 0 (baseline), day 1 (pre-clinical), day 3 (disease onset), day 7 (early disease), day 12 (disease plateau) and day 18 (late disease) (Fig. 1A). Affymetrix oligonucleotide microarrays were used to generate three datasets, one for each sample type, that were analyzed independently. The significance of RNA expression changes was assessed using “two-time-point fold-change” (TTFC), an approach that utilizes temporal relationships in the time-series data to discriminate true gene-expression changes from random fluctuations. The ratio of the number of genes meeting a given TTFC cutoff in each of the datasets to the number in randomly generated datasets was used to estimate false-discovery rates (FDR) (Fig. 1B). Differentially expressed genes were defined as those with an FDR less than 0.1, yielding 486 genes in ankle tissue and 492 in SF out of the 12488 probesets on the Affymetrix chip. In peripheral blood, gene expression changes were indistinguishable from those seen with randomized data (Fig. 1B). The PBL dataset, then, served in effect as a negative-control comparison for the gene-expression changes seen in ankle tissue and SF.
Figure 1. Microarray analysis of ankle tissue, SF and PBLs during serum-transferred arthritis.
(A) Kinetics of arthritis in B6 mice used for the microarray analysis, represented as mean ankle thickening (in mm) over time. The time-points at which samples were collected are indicated by dashed lines. n=3–4 samples per time-point per sample type (except n=2 for ankle day 1 and PBL day 18); each sample represents pooled RNA from 2–3 mice. (B) Two-time-point fold-change (TTFC) – in which fold-changes from day 0 for consecutive time-points (e.g. days 1 and 3, days 3 and 7, etc.) were averaged and the greatest such value used as a metric – was calculated for each gene on the Affymetrix microarray. The distribution of TTFCs for each of the three sample types is shown, with the vertical axis showing the number of genes with a TTFC equal to or greater than a given TTFC plotted on the horizontal axis. Comparison of the collected data (“Real Data”) is made to an aggregate of 100 randomly generated datasets (“Random Data”). Dashed lines indicate cut-off TTFC values for an estimated false-discovery rate (FDR) of 0.1 in the ankle and synovial fluid datasets.
The differentially expressed genes in ankle and SF were further analyzed by hierarchical clustering (Fig. 2A–B). Four kinetic patterns were identified in the ankle dataset: early-response genes (peaking on day 3, 25 genes), genes upregulated in parallel with disease activity (peaking on day 7, 201 genes), late-upregulated genes (peaking on day 12, 23 genes) and downregulated genes (bottoming on day 7, 208 genes). Genes differentially expressed in the SF dataset were predominantly in two clusters, with upregulation (291 genes) or downregulation (138 genes) occurring between days 3 and 7; a third, smaller, cluster contained genes upregulated between days 7 and 12 (37 genes). The ankle and synovial fluid datasets overlapped significantly: 96 of the upregulated genes in the ankle dataset were also upregulated in SF (Fig. 2C). The differentially expressed genes in both datasets represent diverse functional categories including cytokines, chemokines, proteases, extracellular matrix components and adhesion molecules. Similar categories of differentially expressed genes have been reported in a microarray analysis comparing synovial tissue from rheumatoid arthritis patients to synovial tissue from osteoarthritis patients (31).
Figure 2. Hierarchical clustering of differentially expressed genes in ankle tissue and SF during serum-transferred arthritis.
Genes with TTFC greater than the FDR=0.1 cut-offs in the ankle (A) and SF (B) datasets were grouped by hierarchical clustering. Clusters are identified by numbered shaded bars adjacent to the hierarchically clustered data. Kinetic patterns in each cluster are represented by plotting relative gene-expression values on a minimum to maximum (min-max) scale, with a blue trendline showing average min-max values for the cluster. (C) Gene- expression patterns of the 486 differentially expressed ankle genes are compared across the ankle (ANK), synovial fluid (SF), and peripheral blood leukocyte (PBL) datasets. Note the lack of upregulation or downregulation of these genes in PBL as compared with the visibly increased expression in SF of many upregulated ankle genes.
CXCR2 ligands strongly upregulated during serum-transferred arthritis
Chemokines were prominent among the genes upregulated in ankle tissue and SF. As both sample types shared similar differentially expressed chemokines and ankle tissue could be sampled from day 0, we focused our analysis of chemokine gene expression on the ankle dataset. Chemokines showed distinct expression patterns according to whether they were C- C chemokines (CCL2, 6, 7, 8, and 9), which were upregulated early, or C-X-C chemokines bearing the ELR motif (CXCL1, 2, and 5), which were upregulated in parallel with disease (Fig. 3A). These observed patterns of chemokine gene expression were validated by quantitative RT-PCR (qRT-PCR) of independently collected ankle RNA samples from days 0, 1, 3 and 7. The qRT-PCR data were largely concordant with the microarray results although the magnitude of fold-changes was greater (Fig. 3B). The most highly upregulated chemokines by day 7 were the CXCR2 ligands: CXCL1 (11-fold), CXCL2 (36-fold) and CXCL5 (97-fold). Upregulation of CCL8 and CCL9 could not be confirmed by qRT-PCR, and qRT-PCR results for CXCL12 were discordant with the microarray results.
Figure 3. Chemokine gene expression in ankle tissue during serum-transferred arthritis.
(A) Microarray data for upregulated chemokines in ankle tissue is shown in a min-max projection, with CXC chemokines in the upper panel and CC chemokines in the lower panel. The CXC chemokines are divided into CXCR2 ligands (black) and the CXCR4 ligand CXCL12 (gray). The CC chemokines are divided into CCR2 ligands (black) and CCR1 ligands (gray). (B) Gene expression of chemokines upregulated in the microarray analysis was assessed by quantitative real-time PCR (qRT-PCR) of independent ankle RNA samples collected on days 0, 1, 3 and 7 (n=3). qRT-PCR data (black solid line) was compared with microarray data (gray dashed line) for each chemokine. Gene expression is represented as fold-change (FC) from day 0.
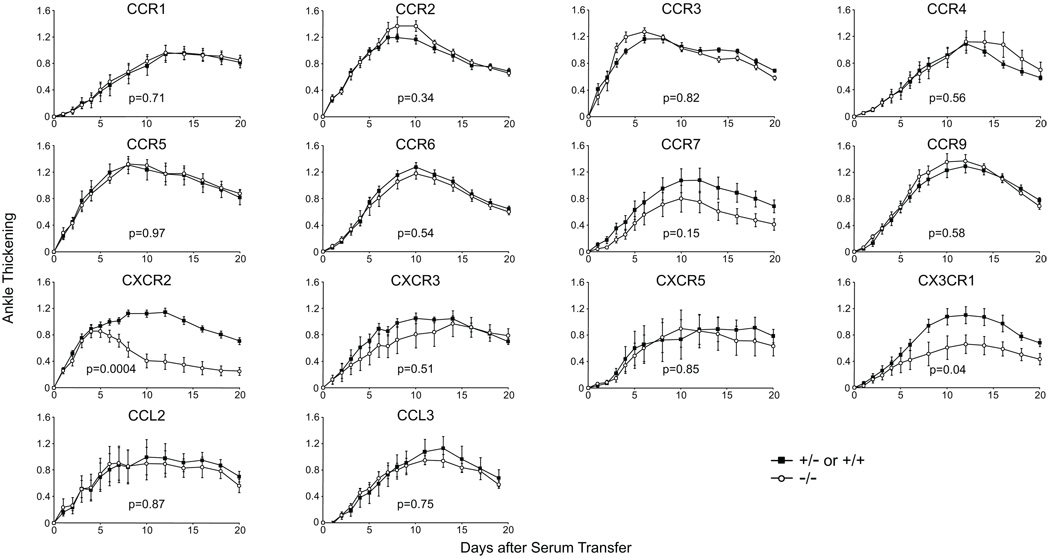
Attenuated serum-transferred arthritis in CXCR2−/− mice but not other chemokine receptor knockout mice
The role of chemokines in arthritis was further explored by studying serum-transferred arthritis in a large panel of knockout mice. We focused on the chemokine receptors as only a few chemokine-deficient mice exist whereas knockout mice have been generated for most chemokine receptors. Serum-transferred arthritis was induced in mice deficient in the following receptors: CCR1 through 7, CCR9, CXCR2, CXCR3, CXCR5 and CX3CR1. Only CXCR2 deficiency resulted in a statistically significant difference in arthritis compared with disease in heterozygote littermate controls when using criteria corrected for multiple hypothesis testing (Fig. 4). CXCR2−/− mice showed a distinct pattern in which early arthritis was comparable with that of controls but then rapidly began to resolve. CX3CR1−/− mice trended toward less severe disease than that of their littermate controls, but this divergence did not reach the threshold for statistical significance when accounting for the multiple receptors tested.
Figure 4. Attenuated course of serum-transferred arthritis in CXCR2−/− mice but not in other chemokine-receptor-deficient animals.
Serum-transferred arthritis in chemokine receptor and chemokine knockout mice was compared with that of heterozygote littermate controls (or to wild-type mice for CCR4 and CCR9 or hemizygous wild-type males for CXCR3). N=10 for CXCR2, CCR1, CCR6, CCR7, and CX3CR1; n=5 for other lines. P-values were calculated for each knockout line using area under the curve (AUC) analysis of ankle thickening over the first 20 days. The p=0.05 significance level adjusted using the Bonferroni correction was p=0.0036.
Surprisingly, neither CCR1−/− nor CCR2−/− mice had reduced arthritis, although their ligands were upregulated during serum-transferred arthritis. As receptor redundancy could have accounted for the lack of effect, we investigated candidate chemokines for which knockout mice existed. CCL2, in particular, was greatly upregulated (7–8 fold by qRT-PCR) during serum-transferred arthritis and has been implicated in other arthritis models (6;11). CCL3, although not among the differentially expressed chemokines in the microarray analysis, was also of interest as it is reported to be critical for anti-collagen-antibody-induced arthritis, an effector phase model mechanistically similar to K/BxN serum-transferred arthritis (32). CCL2−/− and CCL3−/− mice showed disease severity equivalent to that of heterozygote littermates (Fig. 4).
We then sought to confirm the contribution of CXCR2 to serum-transferred arthritis using a pharmacological approach. Treatment with the CXCR2 inhibitor, SB-332235, greatly reduced the severity of serum-transferred arthritis compared to controls (Fig. 5A). The effect of CXCR2 inhibition was apparent from the onset of disease, with a reduced slope of disease progression compared to controls throughout the 10 day course of treatment. This earlier attenuation of disease compared to the CXCR2−/− phenotype may be due to genetic variation between mouse strains. CXCR2−/− mice were on the Balb/c background, whereas B6 mice were used for the inhibition experiments to facilitate comparison with the gene expression data (which derived from B6 mice) and to confirm that the CXCR2 phenotype was not specific to the Balb/c background.
Figure 5. CXCR2 promotes neutrophil recruitment to inflamed joints in a cell autonomous manner.
(A) B6 mice with serum-transferred arthritis received either the CXCR2 antagonist SB-332235 or water daily from day 0 to 9 (n=9). P=0.0001 using AUC of ankle thickening over the first 10 days. (B) BM transfers were performed with all four combinations of +/− and −/− marrow cell donors and recipients. Serum-transferred arthritis values are shown for each group (n=6–9). P-values calculated for +/− → +/− vs. −/− → +/−, +/− → −/− vs. −/− → −/−, and +/− → +/− vs. +/− → −/− were 0.00053, 0.0052, and 0.047, respectively. (C) CXCR2 expression on neutrophils (N, GR1highCD11b+) and monocytes/macrophages (M, GR1lowCD11b+) from SF and peripheral blood was assessed by flow cytometry. Equivalent results were obtained for days 4 and 10 (representative samples shown). (D) Lethally irradiated wild-type mice were reconstituted with CXCR2−/− and CXCR2+/+ BM in a 16:1 ratio. Serum-transferred arthritis was induced after 6 weeks. SF and peripheral blood neutrophils were tested for CXCR2 expression on days 4 and 10 (representative samples shown). The percentage of SF neutrophils expressing CXCR2 was plotted against the percentage of peripheral blood neutrophils expressing CXCR2 (n=8).
CXCR2 deficiency on hematopoietic cells responsible for disease protection
CXCR2 is displayed and operates on diverse cell types, including neutrophils, monocytes/macrophages, mast cells, endothelial cells and epithelial cells (33–35). A criss-cross BM-cell transfer experiment was performed to determine whether the CXCR2−/− effect in this context was due its absence on radio-sensitive cells, radio-resistant cells or both. The experimental design involved BM cell transfers in all four combinations of CXCR2+/− and CXCR2−/− hosts and donors (Fig. 5B). CXCR2 deficiency in hematopoietic cells (−/− donor → +/− recipient) reproduced the CXCR2−/− effect (−/− → −/−), demonstrating that expression of this receptor on hematopoietic cells was required. Conversely, CXCR2 display by hematopoietic cells only (+/− → −/−) was sufficient to restore normal disease kinetics, indicating that it was largely dispensable in non-hematopoietic cells. However, there was a mild decrease in arthritis severity in these mice compared with control mice (+/− → +/−), which barely achieved statistical significance (p=0.047).
CXCR2-expressing neutrophils preferentially recruited to inflamed joints in the presence of CXCR2-deficient neutrophils
CXCR2 expression was assessed on SF cells and PBLs from mice with serum-transferred arthritis. Neutrophils and monocytes/macrophages are the predominant cells of the hematopoeitic lineage found in SF, together comprising >90% of cells (data not shown) (18). Nearly all circulating neutrophils, but few, if any, monocytes/macrophages, expressed CXCR2 (Fig. 5C), consistent with published reports (33). In synovial fluid, CXCR2 was found on 80% of neutrophils, suggesting some receptor downregulation, and on only 1–2% of monocytes/macrophages, as in peripheral blood.
Hence, we inferred that neutrophils were a likely candidate cell-type to be affected by loss of CXCR2. We performed competition experiments in which CXCR2-expressing hosts were reconstituted with a 16:1 mix of CXCR2−/− and CXCR2+/+ BM cells, such that the resulting chimeras would have a neutrophil population consisting predominantly of KO cells with a small number of WT cells (Fig. 5D). Arthritis was induced and the WT cells were tracked by staining for CXCR2 in PBLs and SF after 4 or 10 days of arthritis. CXCR2-expressing neutrophils were enriched in SF compared with PBLs at both time-points, suggesting preferential recruitment. The mean odds ratio of CXCR2 expression by neutrophils in synovial fluid as compared to peripheral blood was 2.1 (95% confidence interval: 1.1–3.9) on day 4 and 3.2 (2.2–4.7) on day 10. This technique almost certainly underestimates the enrichment as about 20% of wild-type neutrophils lose detectable expression of CXCR2 after migrating to the SF (Fig. 5C).
Discussion
Focusing on the roles of chemokines and their receptors in the effector phase of antibody-induced arthritis, we demonstrated that CXCR2 is necessary to sustain joint inflammation. CXCR2 recognizes C-X-C chemokines containing the ELR motif; in mice, such chemokines include CXCL1, CXCL2, CXCL3, CXCL5 and CXCL7 (2). Of these, CXCL1, CXCL2, and CXCL5 were upregulated in inflamed joints during K/BxN serum-transferred arthritis. In addition, it was recently reported that CXCR2 also recognizes extracellular matrix degradation products produced by matrix metalloproteases, and these proteases had similar expression kinetics as the C-X-C chemokines (Table 1) (36). Matrix metalloproteases also can enhance the potency of C-X-C ligands of CXCR2 through proteolytic processing as has been shown, for instance, for MMP9 and MMP13 acting on CXCL2 (37). The driving force for joint expression of CXCR2 ligands may be IL-1, a cytokine expressed in parallel with disease activity that is critical for serum-transferred arthritis and that among its many activities can stimulate C-X-C chemokine production (Table 1) (38;39). CXCR2 ligands may mediate the neutrophil recruitment that is observed when IL-1 is introduced into tissues such as joints (40). Supporting this, neutrophil infiltration into the hippocampus triggered by transgenic IL-1 expression has been shown to be CXCR2-dependent (41).
Table 1.
Selected genes from each of the four clusters of differentially expressed ankle genes are shown, divided by functional category. Genes that were also upregulated in SF are underlined. TTFC and p-values (P) are given for each gene. The p=0.05 significance level adjusted using the Bonferroni correction is p=1×10−4 (1E-04).
| Selected differentially expressed genes in ankle tissue during serum-transferred arthritis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CATEGORY | GENE | TTFC | P | CATEGORY | GENE | TTFC | P | CATEGORY | GENE | TTFC | P |
| Cluster 1 | Cytokine inhibitor | IL-1ra | 6.0 | 1E-06 | Cluster 4 | ||||||
| Cytokine receptor | IL-1R1 | 1.9 | 0.016 | ||||||||
| C-type lectin | DCIR | 2.0 | 6E-05 | IL-13Rα1 | 3.3 | 1E-04 | Antioxidant | Aldehyde dehydrogenase 2 | 1.9 | 0.005 | |
| Chemokine | CCL2 | 8.6 | 8E-06 | ST2 (IL-1 receptor-like 1) | 4.8 | 2E-07 | Glutathione peroxidase 3 | 2.7 | 0.001 | ||
| CCL6 | 1.8 | 0.002 | TNFR p75 | 1.8 | 0.021 | Glutathione S-transferase μ1 | 1.8 | 0.002 | |||
| CCL7 | 4.5 | 2E-06 | Cytoskeleton | Actinin a1 | 2.3 | 6E-04 | Paraoxonase 2 | 1.8 | 0.012 | ||
| CCL8 | 2.6 | 0.022 | MARCKS | 2.7 | 0.009 | Cell adhesion | Cadherin 13 | 3.1 | 9E-05 | ||
| CCL9 | 2.0 | 0.015 | Extracellular matrix | Aggrecan | 3.3 | 1E-05 | Integrin α6 | 2.0 | 0.028 | ||
| CXCL12 | 1.9 | 0.008 | Collagen Vα2 | 1.9 | 0.011 | Integrin β4 | 1.7 | 0.004 | |||
| Chemokine receptor | CCR2 | 2.1 | 0.008 | Lumican | 3.4 | 0.061 | Complement factor | Factor D | 2.8 | 2E-04 | |
| Cytokine receptor | IL-4Rα | 1.9 | 0.030 | Osteoblast-specific factor 2 | 3.9 | 0.003 | Complement inhibitor | C1 inhibitor | 1.8 | 0.015 | |
| Eicosanoids | PGE2 Synthase 3 | 1.7 | 0.026 | Tenascin C | 5.0 | 8E-07 | Clusterin | 2.5 | 0.004 | ||
| Signal transduction | SOCS3 | 3.4 | 0.006 | Thrombospondin 1 | 2.9 | 0.013 | Decay accelerating factor 2 | 2.4 | 0.039 | ||
| Versican | 2.5 | 0.005 | Eicosanoid synthesis | LTC4 synthase | 3.1 | 0.002 | |||||
| Cluster 2 | Growth factor | WISP-1 | 3.7 | 9E-10 | PGI2 synthase | 2.4 | 4E-04 | ||||
| Histamine | Histidine decarboxylase | 2.8 | 4E-04 | Extracellular matrix | Cartilage intermediate layer protein 2 |
3.4 | 4E-06 | ||||
| Acute phase reactant | SAA2 | 1.9 | 0.002 | L-arginine metabolism | Arginase 1 | 5.9 | 0.002 | ||||
| SAA3 | 49.8 | 2E-09 | Macrophage receptor | Macrophage C-type lectin | 9.9 | 6E-08 | Chondroadherin | 2.4 | 9E-05 | ||
| Angiogenesis | Angiopoietin-like 4 | 2.4 | 0.001 | MSR1 | 2.4 | 2E-05 | Fibromodulin | 2.4 | 0.002 | ||
| HIF1α | 2.8 | 6E-04 | Protease | Cathepsin S | 2.2 | 0.037 | Osteocalcin | 4.2 | 2E-06 | ||
| Cell adhesion | SELPLG | 1.7 | 0.005 | MMP3 | 14.6 | 9E-08 | Tenascin XB | 3.3 | 0.003 | ||
| VCAM-1 | 3.5 | 0.001 | MMP9 | 5.9 | 2E-04 | Growth factor | BMP4 | 1.7 | 3E-05 | ||
| VE-cadherin | 2.0 | 0.035 | MMP13 | 16.5 | 1E-09 | BMP6 | 1.7 | 4E-04 | |||
| Cell cycle | Cdc2a | 2.4 | 5E-05 | Protease inhibitor | SLPI | 13.4 | 2E-07 | Connective tissue growth factor |
2.2 | 0.013 | |
| Cdc3a | 1.7 | 2E-04 | TIMP1 | 5.0 | 4E-07 | ||||||
| Cdc20 | 1.8 | 0.001 | S100 proteins | S100A8 | 7.2 | 5E-08 | Growth factor receptor | Epidermal growth factor receptor |
1.7 | 0.008 | |
| Cyclin B1 | 1.9 | 0.060 | S100A9 | 8.3 | 8E-07 | ||||||
| Topoisomerase IIα | 3.2 | 8E-06 | Signal transduction | ROR2 | 1.8 | 3E-05 | Lipid metabolism | Adiponectin | 2.0 | 0.003 | |
| Chemokine | CXCL1 | 6.4 | 4E-04 | SOCS2 | 1.8 | 0.014 | Apolipoprotein D | 1.8 | 0.001 | ||
| CXCL2 | 4.3 | 4E-06 | Hormone-sensitive lipase | 1.9 | 0.001 | ||||||
| CXCL5 | 7.4 | 2E-05 | Cluster 3 | Lipoprotein lipase | 1.9 | 0.003 | |||||
| Chemokine receptor | CCR5 | 3.4 | 5E-04 | Monoglyceride lipase | 2.0 | 0.001 | |||||
| Complement | C1r | 1.7 | 0.017 | Cytokine inhibitor | IL-1R2 | 1.7 | 0.009 | PPARγ | 1.9 | 0.046 | |
| Complement receptor | C1qR1 | 2.0 | 6E-06 | Cytoskeleton | Synaptopodin | 2.0 | 0.007 | Protease | Cathepsin F | 2.9 | 0.002 |
| Cytokine | IL-1β | 3.4 | 6E-05 | Extracellular matrix | Collagen Vα1 | 1.7 | 0.005 | Signal transduction | ERK6 | 1.7 | 2E-05 |
| Osteopontin | 5.0 | 4E-05 | MHC class I | H2-D1 | 1.8 | 0.008 | R-ras | 1.8 | 0.012 | ||
| RANKL | 2.4 | 1E-04 | Growth factor regulation | IGFBP4 | 1.8 | 0.017 | RasD1 | 2.4 | 0.006 | ||
BM-cell transfer experiments revealed that CXCR2 needed to be displayed principally on hematopoietic cells. This receptor can mediate trafficking of multiple hematopoietic cell lineages involved in inflammatory arthritis, including neutrophils, monocytes/macrophages and mast cells (33;34;36). Mast cell CXCR2 is unlikely to account for the dependence on CXCR2 as this cell type is required only prior to the onset of measurable arthritis in this system (20). It is also unlikely that defective monocyte/macrophage recruitment explains CXCR2’s importance as only a limited number of SF macrophages expressed detectable CXCR2. Rather, our data support a role for CXCR2 in neutrophil recruitment to inflamed joints. This conclusion is consistent with findings in the adjuvant-induced arthritis model of reduced numbers of rolling, arrested and total neutrophils in synovial tissue after CXCR2 inhibition (42;43). These studies could not differentiate whether reduced neutrophil recruitment was due to a cell autonomous requirement for CXCR2 on neutrophils, themselves, or was an indirect effect of decreased disease severity from CXCR2 blockade. We demonstrated that CXCR2 does indeed have intrinsic activity on neutrophils as CXCR2-deficient neutrophils had a reduced capacity to migrate into inflamed joints when compared side-by-side with CXCR2-expressing neutrophils. A similar requirement for neutrophil CXCR2 has been demonstrated in experimental autoimmune encephalitis (44). Of note, the mild reduction in disease activity in mice lacking CXCR2 on non-hematopoietic cells may reflect a small secondary contribution of endothelial CXCR2, which can facilitate the transendothelial movement of neutrophils (35).
Although CXCR2 conferred an advantage in neutrophil migration to inflamed joints, large numbers of CXCR2-deficient neutrophils were still found in arthritic joints in the competition experiments (Fig. 5D). Their presence likely reflected the activity of other neutrophil chemoattractants including LTB4, which, along with its receptor BLT1, is required for serum-transferred arthritis and plays a sustained role after disease onset (21;22). BLT1-deficient neutrophils were abundant in the SF of BLT1-knockout mice that had received wild-type neutrophils to reconstitute arthritis susceptibility, similar to our observations on CXCR2-deficient neutrophils in the competition experiment (22). We suspect that CXCR2 and BLT1 can each mediate neutrophil recruitment in the absence of the other, but that neither mediator can fully compensate for the other. As neutrophils are the critical source of LTB4 in serum-transferred arthritis, CXCR2-mediated neutrophil recruitment may be amplified by LTB4 released from the newly arrived neutrophils (21). In accord with this scenario, neutrophil migration to joints in response to intra-articular injection of CXCL1 and CXCL5 was greatly reduced by co-administration of an LTB4 inhibitor (43). The complement receptor C5aR is also capable of mediating neutrophil chemotaxis and is required for serum-transferred arthritis (19). It may act in concert with CXCR2 and BLT1 to drive recruitment of neutrophils to inflamed joints. In human RA, C-X-C chemokines, LTB4, and C5a are all present in synovial fluid, although clinical trials of BLT1 and C5aR inhibitors have yielded disappointing results (45–47). We are unaware of any published clinical trials of a CXCR2 inhibitor in RA.
Perhaps our study’s most striking finding was the surprising dearth of chemokine receptors required for the effector phase of autoantibody-induced arthritis. Like both human RA and other animal models of arthritis, the K/BxN serum-transferred disease was accompanied by upregulation of signal chemokines, with the caveat that mRNA levels may not always correlate with protein levels. While the increased expression of ELR+ C-X-C chemokines in ankle tissue was reflected by reduced arthritis in the absence of their receptor, CXCR2, this induction-level to disease-severity correlation did not hold for the receptors of other upregulated C-C chemokines, nor for CCL2. It is possible that receptor and/or ligand redundancy permits other receptors to take the place of any single chemokine receptor targeted. Results on certain of the receptors tested, in particular CX3CR1, fell just below statistical significance, so we cannot exclude that they have a moderate contribution to disease that might be confirmed by further testing. Moreover, some chemokine receptors were not included in this study for technical reasons, most notably CXCR4 (the receptor for CXCL12), which could not be tested as deficiency in this molecule is lethal during embryogenesis.
However, these caveats do not explain why inhibition or deficiency of CCR1, CCR2, CCR7, CXCR3, CXCR5, CCL2 or CCL3 altered disease severity in other arthritis models but not serum-transferred arthritis (6;9;10;14;16;32). These disparate results cannot simply be attributed to inadequate genetic targeting as studies in other animal models used the same CCR7, CXCR5, and CCL3 knockout lines and the other lines used here have previously been clearly demonstrated to lack expression of the targeted gene (16;23;24;32;48;49). The discrepancy may reflect the contributions of these chemokines and chemokine receptors to adaptive immunity, consistent with evidence of altered T or B cell activity reported in many of these studies. In CIA, increased disease severity with CCR2 deficiency was associated with enhanced titers of anti-collagen antibodies, formation of rheumatoid factor (not detected in controls), and decreased activation-induced cell death (10). In a study demonstrating improvement in adjuvant-induced arthritis with anti-CXCR3 monoclonal antibody therapy, CXCR3−/− T cells had a reduced capacity to migrate to inflamed joints in comparison with wild-type T cells (14). CCR7−/− and CXCR5−/− mice with antigen-induced arthritis had reduced titers of antibodies to the inciting antigen, reduced T cell proliferation in response to antigen, and aberrant or absent lymphoid follicle formation within chronically inflamed synovial tissue (16). CCL3 represents a special case as it has been shown to have a role in anti-collagen-antibody-induced arthritis, which like serum-transferred arthritis is induced by passive antibody transfer (32). The primary difference between the two models besides their autoantibody specificity is the requirement for lipopolysaccharide in anti-collagen-antibody-induced arthritis. LPS induces CCL3 and promotes neutrophil accumulation in a CCL3-dependent manner (50). It may be that, in the absence of CCL3, LPS is unable to provide enough additional pro-inflammatory activity to induce disease upon transfer of anti-collagen antibodies, whereas antibodies alone are sufficient to induce disease in K/BxN serum-transferred arthritis.
This study demonstrates a necessary role for CXCR2 in sustaining autoantibody-induced arthritis, and identifies this receptor as a second critical element by which neutrophils can be recruited to inflamed joints, alongside BLT1. Our findings highlight CXCR2, and potentially also CXCR1 (a closely related human receptor without a known murine homolog that binds CXCL6 and CXCL8, a.k.a. IL-8), as an attractive therapeutic target for RA (2). Its importance in the arthritis effector phase, the stage when most patients present, enhances its appeal.
Acknowledgments
We thank Dr. Philip Murphy for the CX3CR1−/− mice, Dr. Martin Lipp for the CCR7−/− and CXCR5−/− mice, Dr. Sergio Lira for the CCR6−/− mice, Robert Saccone and Jennifer Johnson for Affymetrix Core services, and Vanessa Tran and Kimie Hattori for maintaining mouse colonies.
Supported by NIH grant R01AR055271 to CB and DM and by Joslin Diabetes Center’s National Institutes of Diabetes and Digestive and Kidney Diseases Diabetes and Endocrinology Research Center core laboratories. Dr. Jacobs’ work was supported by fellowships from the Howard Hughes Medical Institute, Arthritis Foundation Massachusetts Chapter, and Lupus Foundation of New England.
References
- 1.Mackay CR. Chemokines: immunology's high impact factors. Nat Immunol. 2001;2(2):95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 2.Koch AE. Chemokines and their receptors in rheumatoid arthritis: future targets? Arthritis Rheum. 2005;52(3):710–721. doi: 10.1002/art.20932. [DOI] [PubMed] [Google Scholar]
- 3.Haringman JJ, Ludikhuize J, Tak PP. Chemokines in joint disease: the key to inflammation? Ann Rheum Dis. 2004;63(10):1186–1194. doi: 10.1136/ard.2004.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vergunst CE, Gerlag DM, Lopatinskaya L, Klareskog L, Smith MD, van den BF, et al. Modulation of CCR2 in rheumatoid arthritis: a double-blind, randomized, placebo-controlled clinical trial. Arthritis Rheum. 2008;58(7):1931–1939. doi: 10.1002/art.23591. [DOI] [PubMed] [Google Scholar]
- 5.Haringman JJ, Gerlag DM, Smeets TJ, Baeten D, van den BF, Bresnihan B, et al. A randomized controlled trial with an anti-CCL2 (anti-monocyte chemotactic protein 1) monoclonal antibody in patients with rheumatoid arthritis. Arthritis Rheum. 2006;54(8):2387–2392. doi: 10.1002/art.21975. [DOI] [PubMed] [Google Scholar]
- 6.Ogata H, Takeya M, Yoshimura T, Takagi K, Takahashi K. The role of monocyte chemoattractant protein-1 (MCP-1) in the pathogenesis of collagen-induced arthritis in rats. J Pathol. 1997;182(1):106–114. doi: 10.1002/(SICI)1096-9896(199705)182:1<106::AID-PATH816>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Zheng B, Ozen Z, Zhang X, De Silva S, Marinova E, Guo L, et al. CXCL13 neutralization reduces the severity of collagen-induced arthritis. Arthritis Rheum. 2005;52(2):620–626. doi: 10.1002/art.20768. [DOI] [PubMed] [Google Scholar]
- 8.Nanki T, Shimaoka T, Hayashida K, Taniguchi K, Yonehara S, Miyasaka N. Pathogenic role of the CXCL16-CXCR6 pathway in rheumatoid arthritis. Arthritis Rheum. 2005;52(10):3004–3014. doi: 10.1002/art.21301. [DOI] [PubMed] [Google Scholar]
- 9.Amat M, Benjamim CF, Williams LM, Prats N, Terricabras E, Beleta J, et al. Pharmacological blockade of CCR1 ameliorates murine arthritis and alters cytokine networks in vivo. Br J Pharmacol. 2006;149(6):666–675. doi: 10.1038/sj.bjp.0706912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinones MP, Ahuja SK, Jimenez F, Schaefer J, Garavito E, Rao A, et al. Experimental arthritis in CC chemokine receptor 2-null mice closely mimics severe human rheumatoid arthritis. J Clin Invest. 2004;113(6):856–866. doi: 10.1172/JCI20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahrara S, Proudfoot AE, Park CC, Volin MV, Haines GK, Woods JM, et al. Inhibition of monocyte chemoattractant protein-1 ameliorates rat adjuvant-induced arthritis. J Immunol. 2008;180(5):3447–3456. doi: 10.4049/jimmunol.180.5.3447. [DOI] [PubMed] [Google Scholar]
- 12.Brodmerkel CM, Huber R, Covington M, Diamond S, Hall L, Collins R, et al. Discovery and pharmacological characterization of a novel rodent-active CCR2 antagonist, INCB3344. J Immunol. 2005;175(8):5370–5378. doi: 10.4049/jimmunol.175.8.5370. [DOI] [PubMed] [Google Scholar]
- 13.Barsante MM, Cunha TM, Allegretti M, Cattani F, Policani F, Bizzarri C, et al. Blockade of the chemokine receptor CXCR2 ameliorates adjuvant-induced arthritis in rats. Br J Pharmacol. 2008;153(5):992–1002. doi: 10.1038/sj.bjp.0707462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan K, Issekutz TB. Blockade of chemokine receptor CXCR3 inhibits T cell recruitment to inflamed joints and decreases the severity of adjuvant arthritis. J Immunol. 2007;179(12):8463–8469. doi: 10.4049/jimmunol.179.12.8463. [DOI] [PubMed] [Google Scholar]
- 15.Podolin PL, Bolognese BJ, Foley JJ, Schmidt DB, Buckley PT, Widdowson KL, et al. A potent and selective nonpeptide antagonist of CXCR2 inhibits acute and chronic models of arthritis in the rabbit. J Immunol. 2002;169(11):6435–6444. doi: 10.4049/jimmunol.169.11.6435. [DOI] [PubMed] [Google Scholar]
- 16.Wengner AM, Hopken UE, Petrow PK, Hartmann S, Schurigt U, Brauer R, et al. CXCR5- and CCR7-dependent lymphoid neogenesis in a murine model of chronic antigen-induced arthritis. Arthritis Rheum. 2007;56(10):3271–3283. doi: 10.1002/art.22939. [DOI] [PubMed] [Google Scholar]
- 17.Monach PA, Benoist C, Mathis D. The role of antibodies in mouse models of rheumatoid arthritis, and relevance to human disease. Adv Immunol. 2004;82:217–248. doi: 10.1016/S0065-2776(04)82005-4. [DOI] [PubMed] [Google Scholar]
- 18.Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87(5):811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 19.Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FMA, Boackle SA, et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 20.Nigrovic PA, Binstadt BA, Monach PA, Johnsen A, Gurish M, Iwakura Y, et al. Mast cells contribute to initiation of autoantibody-mediated arthritis via IL-1. Proc Natl Acad Sci U S A. 2007;104(7):2325–2330. doi: 10.1073/pnas.0610852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Lam BK, Kanaoka Y, Nigrovic PA, Audoly LP, Austen KF, et al. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J Exp Med. 2006;203(4):837–842. doi: 10.1084/jem.20052371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ND, Chou RC, Seung E, Tager AM, Luster AD. A unique requirement for the leukotriene B4 receptor BLT1 for neutrophil recruitment in inflammatory arthritis. J Exp Med. 2006;203(4):829–835. doi: 10.1084/jem.20052349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock WW, Lu B, Gao W, Csizmadia V, Faia K, King JA, et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med. 2000;192(10):1515–1520. doi: 10.1084/jem.192.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerard C, Frossard JL, Bhatia M, Saluja A, Gerard NP, Lu B, et al. Targeted disruption of the beta-chemokine receptor CCR1 protects against pancreatitis-associated lung injury. J Clin Invest. 1997;100(8):2022–2027. doi: 10.1172/JCI119734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humbles AA, Lu B, Friend DS, Okinaga S, Lora J, Al garawi A, et al. The murine CCR3 receptor regulates both the role of eosinophils and mast cells in allergen-induced airway inflammation and hyperresponsiveness. Proc Natl Acad Sci U S A. 2002;99(3):1479–1484. doi: 10.1073/pnas.261462598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chvatchko Y, Hoogewerf AJ, Meyer A, Alouani S, Juillard P, Buser R, et al. A key role for CC chemokine receptor 4 in lipopolysaccharide-induced endotoxic shock. J Exp Med. 2000;191(10):1755–1764. doi: 10.1084/jem.191.10.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook DN, Prosser DM, Forster R, Zhang J, Kuklin NA, Abbondanzo SJ, et al. CCR6 mediates dendritic cell localization, lymphocyte homeostasis, and immune responses in mucosal tissue. Immunity. 2000;12(5):495–503. doi: 10.1016/s1074-7613(00)80201-0. [DOI] [PubMed] [Google Scholar]
- 28.Wurbel MA, Malissen M, Guy-Grand D, Meffre E, Nussenzweig MC, Richelme M, et al. Mice lacking the CCR9 CC-chemokine receptor show a mild impairment of early T- and B-cell development and a reduction in T-cell receptor gammadelta(+) gut intraepithelial lymphocytes. Blood. 2001;98(9):2626–2632. doi: 10.1182/blood.v98.9.2626. [DOI] [PubMed] [Google Scholar]
- 29.Combadiere C, Potteaux S, Gao JL, Esposito B, Casanova S, Lee EJ, et al. Decreased atherosclerotic lesion formation in CX3CR1/apolipoprotein E double knockout mice. Circulation. 2003;107(7):1009–1016. doi: 10.1161/01.cir.0000057548.68243.42. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Pouw Kraan TC, van Gaalen FA, Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003;48(8):2132–2145. doi: 10.1002/art.11096. [DOI] [PubMed] [Google Scholar]
- 32.Chintalacharuvu SR, Wang JX, Giaconia JM, Venkataraman C. An essential role for CCL3 in the development of collagen antibody-induced arthritis. Immunol Lett. 2005;100(2):202–204. doi: 10.1016/j.imlet.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Boisvert WA, Rose DM, Johnson KA, Fuentes ME, Lira SA, Curtiss LK, et al. Up-regulated expression of the CXCR2 ligand KC/GRO-alpha in atherosclerotic lesions plays a central role in macrophage accumulation and lesion progression. Am J Pathol. 2006;168(4):1385–1395. doi: 10.2353/ajpath.2006.040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallgren J, Jones TG, Abonia JP, Xing W, Humbles A, Austen KF, et al. Pulmonary CXCR2 regulates VCAM-1 and antigen-induced recruitment of mast cell progenitors. Proc Natl Acad Sci U S A. 2007;104(51):20478–20483. doi: 10.1073/pnas.0709651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reutershan J, Morris MA, Burcin TL, Smith DF, Chang D, Saprito MS, et al. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J Clin Invest. 2006;116(3):695–702. doi: 10.1172/JCI27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weathington NM, van Houwelingen AH, Noerager BD, Jackson PL, Kraneveld AD, Galin FS, et al. A novel peptide CXCR ligand derived from extracellular matrix degradation during airway inflammation. Nat Med. 2006;12(3):317–323. doi: 10.1038/nm1361. [DOI] [PubMed] [Google Scholar]
- 37.Van LP, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol. 2007;82(6):1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 38.Ji H, Pettit A, Ohmura K, Ortiz-Lopez A, Duchatelle V, Degott C, et al. Critical roles for interleukin-1 and tumor necrosis factor-α in antibody-induced arthritis. J Exp Med. 2002;196:77–85. doi: 10.1084/jem.20020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed S, Pakozdi A, Koch AE. Regulation of interleukin-1beta-induced chemokine production and matrix metalloproteinase 2 activation by epigallocatechin-3-gallate in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2006;54(8):2393–2401. doi: 10.1002/art.22023. [DOI] [PubMed] [Google Scholar]
- 40.Pettipher ER, Higgs GA, Henderson B. Interleukin 1 induces leukocyte infiltration and cartilage proteoglycan degradation in the synovial joint. Proc Natl Acad Sci U S A. 1986;83(22):8749–8753. doi: 10.1073/pnas.83.22.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaftel SS, Carlson TJ, Olschowka JA, Kyrkanides S, Matousek SB, O'Banion MK. Chronic interleukin-1beta expression in mouse brain leads to leukocyte infiltration and neutrophil-independent blood brain barrier permeability without overt neurodegeneration. J Neurosci. 2007;27(35):9301–9309. doi: 10.1523/JNEUROSCI.1418-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coelho FM, Pinho V, Amaral FA, Sachs D, Costa VV, Rodrigues DH, et al. The chemokine receptors CXCR1/CXCR2 modulate antigen-induced arthritis by regulating adhesion of neutrophils to the synovial microvasculature. Arthritis Rheum. 2008;58(8):2329–2337. doi: 10.1002/art.23622. [DOI] [PubMed] [Google Scholar]
- 43.Grespan R, Fukada SY, Lemos HP, Vieira SM, Napimoga MH, Teixeira MM, et al. CXCR2-specific chemokines mediate leukotriene B4-dependent recruitment of neutrophils to inflamed joints in mice with antigen-induced arthritis. Arthritis Rheum. 2008;58(7):2030–2040. doi: 10.1002/art.23597. [DOI] [PubMed] [Google Scholar]
- 44.Carlson T, Kroenke M, Rao P, Lane TE, Segal B. The Th17-ELR+ CXC chemokine pathway is essential for the development of central nervous system autoimmune disease. J Exp Med. 2008;205(4):811–823. doi: 10.1084/jem.20072404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koch AE, Kunkel SL, Shah MR, Hosaka S, Halloran MM, Haines GK, et al. Growth-related gene product alpha. A chemotactic cytokine for neutrophils in rheumatoid arthritis. J Immunol. 1995;155(7):3660–3666. [PubMed] [Google Scholar]
- 46.Vergunst CE, Gerlag DM, Dinant H, Schulz L, Vinkenoog M, Smeets TJ, et al. Blocking the receptor for C5a in patients with rheumatoid arthritis does not reduce synovial inflammation. Rheumatology (Oxford) 2007;46(12):1773–1778. doi: 10.1093/rheumatology/kem222. [DOI] [PubMed] [Google Scholar]
- 47.Diaz-Gonzalez F, Alten RH, Bensen WG, Brown JP, Sibley JT, Dougados M, et al. Clinical trial of a leucotriene B4 receptor antagonist, BIIL 284, in patients with rheumatoid arthritis. Ann Rheum Dis. 2007;66(5):628–632. doi: 10.1136/ard.2006.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boring L, Gosling J, Chensue SW, Kunkel SL, Farese RV, Jr, Broxmeyer HE, et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100(10):2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu B, Rutledge BJ, Gu L, Fiorillo J, Lukacs NW, Kunkel SL, et al. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J Exp Med. 1998;187(4):601–608. doi: 10.1084/jem.187.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Standiford TJ, Kunkel SL, Lukacs NW, Greenberger MJ, Danforth JM, Kunkel RG, et al. Macrophage inflammatory protein-1 alpha mediates lung leukocyte recruitment, lung capillary leak, and early mortality in murine endotoxemia. J Immunol. 1995;155(3):1515–1524. [PubMed] [Google Scholar]