Abstract
There has long been interest in understanding the genetic basis of human adaptation. To what extent are phenotypic differences among human populations driven by natural selection? With the recent arrival of large genome-wide data sets on human variation, there is now unprecedented opportunity for progress on this type of question. Several lines of evidence argue for an important role of positive selection in shaping human variation and differences among populations. These include studies of comparative morphology and physiology, as well as population genetic studies of candidate loci and genome-wide data. However, the data also suggest that it is unusual for strong selection to drive new mutations rapidly to fixation in particular populations (the ‘hard sweep’ model). We argue, instead, for alternatives to the hard sweep model: in particular, polygenic adaptation could allow rapid adaptation while not producing classical signatures of selective sweeps. We close by discussing some of the likely opportunities for progress in the field.
Introduction
Within the past 100,000 years, anatomically modern humans have spread from sub-Saharan Africa to colonize most of the world’s land masses (see the other reviews in this special issue). Human populations live in an extraordinary variety of different habitats: hot and cold; wet and dry; in forests, grasslands, and tundra. Different human groups feed on a wide variety of food sources. For many populations, diets shifted further with the development of agriculture in the past 10,000 years.
To what extent have these, and other, factors led to genetic adaptation? If they have, can we identify the types of genes and phenotypes that have been most affected? With the recent availability of genome-wide single nucleotide polymorphism (SNP) data for many populations, and with the expectation that genome-wide sequence data will soon also be available for large numbers of individuals, there is now great interest in these questions. There has also been a great deal of recent work on methods for using genome-wide data to identify signals of selection. These methods generally make use of the idea that selective events distort patterns of neutral variation in predictable ways, depending on the model of selection: for example, reducing haplotype diversity, increasing the fraction of rare alleles and increasing the extent of allele frequency differences between populations. In this review, we provide a brief overview of some of the key findings thus far, and then focus on what we see as some of the major open questions. A number of other recent reviews discuss either the general principles for detecting selection or summarize the overall results in more detail than we attempt here [1–6].
Recent Human Adaptations
While human populations differ in various phenotypes, there is a considerable burden of proof to show that phenotypic differences have a genetic basis and are adaptive. However, we do now have reasonable evidence of differential adaptation of various traits. For example, it has long been known that mammals that live in cold climates tend to have larger, rounder bodies (‘Bergmann’s rule’) and shorter limbs (‘Allen’s rule’) than members of the same or closely related species in warm climates. These patterns — although noisy—do appear to also hold in humans, implying that population movements into colder climates were accompanied by adaptation to larger, stockier body shape, presumably to improve thermal efficiency [7]. At the other end of the spectrum is the striking ‘pygmy’ phenotype that has evolved convergently in rainforest populations in Africa, South-East Asia, and South America [8]. It has been suggested that the pygmy phenotype may be an adaptation to food limitations, high humidity or dense forest undergrowth [8].
Another impressive example of adaptation is provided by human populations living at high altitude, especially in the Himalayas and the Andes [9,10]. Compared to related lowland populations, these high-elevation populations show a suite of physiological adaptations to low oxygen [11]. These adaptations include markedly increased blood flow and oxygen delivery to the uterus during pregnancy, substantially reducing the risk of babies with low birthweight [12]. Current evidence suggests that these differences are not simply the result of recent acclimation, but are at least partly genetic, although the relevant loci are not known [9,10,12]. If this is the case, then the adaptation must have occurred rapidly, because these high altitude regions were settled within the last 10,000 years, and the adaptation occurred in spite of likely gene flow from lowland neighbors [10].
Skin pigmentation is perhaps the phenotype that varies most conspicuously among human populations. Dark pigmentation is strongly associated with tropical climates, and the spread of prehistoric humans into northern latitudes was accompanied by a shift to lighter skin color [13,14]. We now know of at least half a dozen different genes that affect skin, hair or eye pigmentation, and have strong genetic signals of selection based on low haplotype diversity or extreme frequency differences between populations [15–21]. There are surely additional selected loci yet to be found. In particular, the evolution of light skin color occurred largely in parallel in western Eurasia and east Asia, but we still know few of the relevant genes in east Asia [17,18,22]. Adaptation to lighter pigmentation may have been driven by a need to increase UV absorption for vitamin D synthesis at high latitudes or by sexual selection [14].
In addition to pigmentation, there are a handful of other genes for which there are both strong selection signals and compelling explanations for their adaptive significance. Several of these are involved in malaria resistance, including the Duffy antigen protein (DARC) [23] and Glucose-6-phosphate dehydrogenase (G6PD) [24], as well as rarer mutations in the α- and β-globin genes that can lead to sickle cell anemia or thalassemias. Another clear example of adaptation is provided by lactase, the enzyme that hydrolyzes lactose, the main sugar in milk. Lactase gene expression has evolved repeatedly to continue throughout life in dairy farming populations in Europe, east Africa, and the Middle East [25–27].
Genome-Wide Data
Moving beyond candidate loci, many researchers have made use of the new genome-wide SNP data to scan for signals of ongoing or recently completed selective sweeps. Some global trends have emerged that show clear evidence for abundant selection. In particular, various types of signals are consistently concentrated around genes, as opposed to in intergenic regions. These include signals of partial sweeps [20], reduced diversity at putatively neutral sites [28,29] and an excess of SNPs with extreme population differentiation (high FST) within genes (Figure 1A) [30]. Also consistent with the action of selection, there is reduced diversity in regions of low recombination, especially in gene-rich regions [28,29,31]. Taken together, these observations are most easily explained by either widespread positive selection, or possibly by background selection against mildly deleterious alleles [29,32]. The conclusion that adaptive selection may be widespread is further bolstered by similar results for other organisms, especially Drosophila [6], and also by recent estimates that 10–20% of amino acid replacements on the human lineage have been driven by positive selection [33].
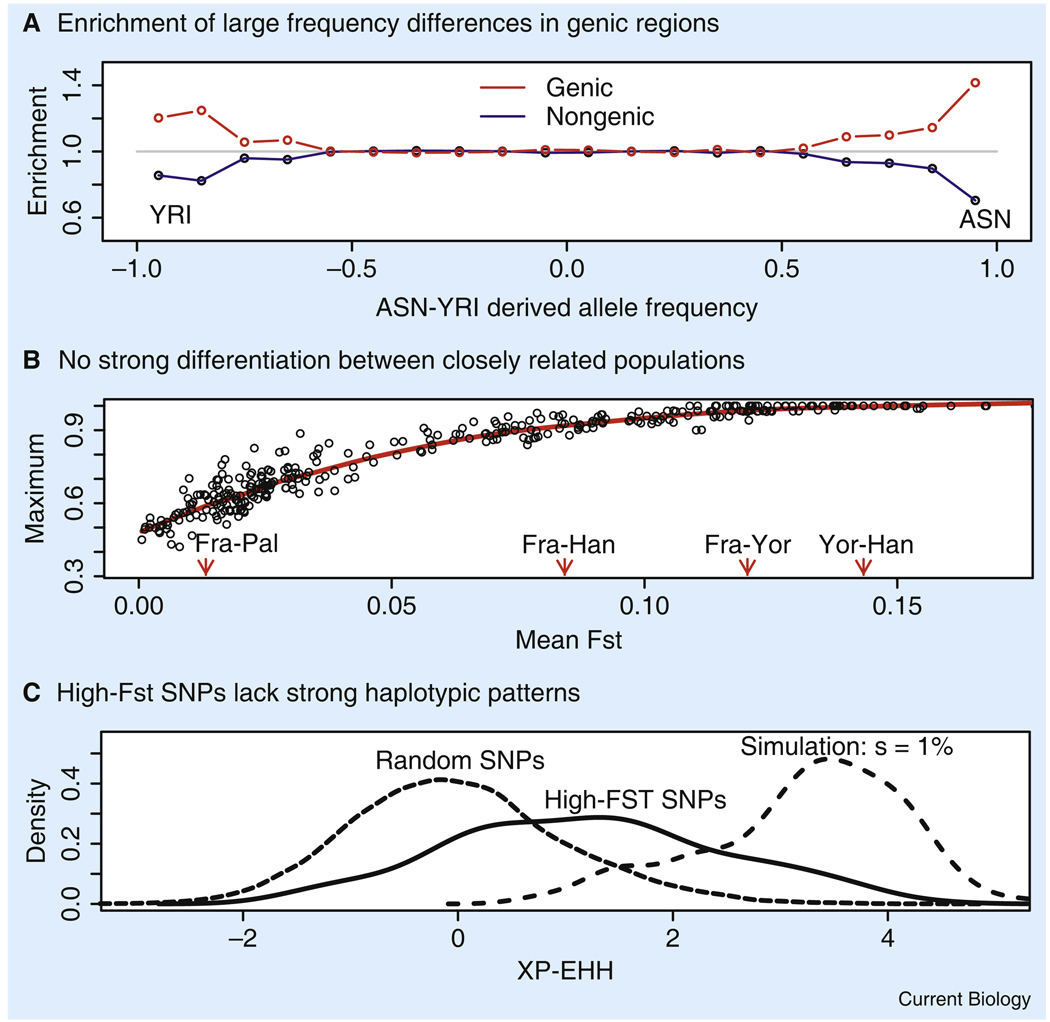
Figure 1. Conflicting evidence of population-specific selection.
(A) The x-axis shows the signed difference in derived allele frequency between HapMap Yoruba and east Asians. The y-axis shows the fractions of SNPs in each bin of frequency differences that are genic, and nongenic, respectively, divided by the total fraction of SNPs that are genic, and nongenic, respectively. (B) The x-axis shows the mean pairwise FST between all pairs of HGDP populations with sample sizes >15 individuals (the values of four arbitrary pairs comparing France, Palestine, Han and Yoruba are indicated to provide a sense of scale). The y-axis shows the value of the most extreme allele frequency difference for each population at any of the 640,000 genotyped SNPs. (C) The three curves show the distributions of XP-EHH, a measure of haplotype diversity [34] for (i) random SNPs in east Asians, (ii) SNPs with a frequency difference >90% between HapMap Yoruba and east Asians, and (iii) simulated SNPs with a selective advantage of 1% and a frequency difference >90%, assuming a uniform rate of input of favored mutations. In fact, the middle curve is most similar to data simulated under a neutral model, but conditioned on the frequency difference of >90% (not shown). All three plots are redrawn from [22].
There has also been a great deal of interest in identifying the particular loci that have been targets of positive selection. However, in some respects this has proven to be challenging. A recent review lists 21 genome-wide scans, using a variety of different methods [1]. Although each of these scans highlights potentially interesting signals, it is currently difficult to assess how much confidence should be placed in individual signals in the absence of further biological or functional information [34,35]. Indeed, there is poor agreement among the studies, even though many of them actually analyze the same data, frequently genome-wide SNP data from HapMap or Perlegen [36,37].
Perhaps consistent with some of the challenges of the genome-wide scans, recent work by Coop et al. [22] found that, as described below, some aspects of the human variation data do not show clear signals of widespread, strong selection (Figure 1). The authors studied three million SNPs genotyped in the Phase II HapMap samples, along with a data set of 640,000 SNPs genotyped in 927 individuals from the CEPH-Human Genome Diversity Panel [37,38]. They focused primarily on SNPs with high FST values, with the view that these should be particularly sensitive for detecting differential selection between populations.
Overall, however, the HapMap data show relatively few fixed or nearly fixed differences between populations from different continents, implying that new alleles have only rarely spread rapidly to fixation within populations, even though there has been sufficient time for strongly favored alleles (selection coefficient, s ≥ 0.5%) to spread from low to high frequency since these populations separated [22]. Nearly all of these rare fixation events have taken place outside Africa and, curiously, most are found in the east Asians, the group that has experienced the strongest genetic drift of the three HapMap groups [39]. For example, there are just 13 non-synonymous SNPs in Phase II HapMap with a frequency difference >90% between the Yoruba and east Asians. Of these, only one is due to a high frequency derived allele in the Yoruba. Additionally, few of the east Asian fixation events are associated with strong haplotype signals (Figure 1C), as measured by cross-population extended haplotype homozygosity (XP-EHH) [34]. This indicates that few of these alleles were fixed very rapidly. Instead, the XP-EHH data are more consistent with a steady, slow increase in frequency during the time since the out-of-Africa migration roughly 60,000 years ago. Finally, these putatively selected alleles can be grouped in a small number of geographic patterns that reflect neutral population structure; these geographical patterns have been described as non- African, West Eurasian and East Asian sweep patterns (Figure 3) [22]. The observation that sweep patterns mimic neutral population structure is not what might have been expected if the frequencies of individual alleles were strongly determined by environmental factors, such as climate or diet, that likely vary over different geographic scales. Additionally, looking across all populations, and all SNPs, there is not a single example of a SNP with very extreme allele frequency differences between closely related populations (Figure 1B). At the level of individual SNPs, there is thus no clear evidence for extreme differential adaptation between closely related populations.
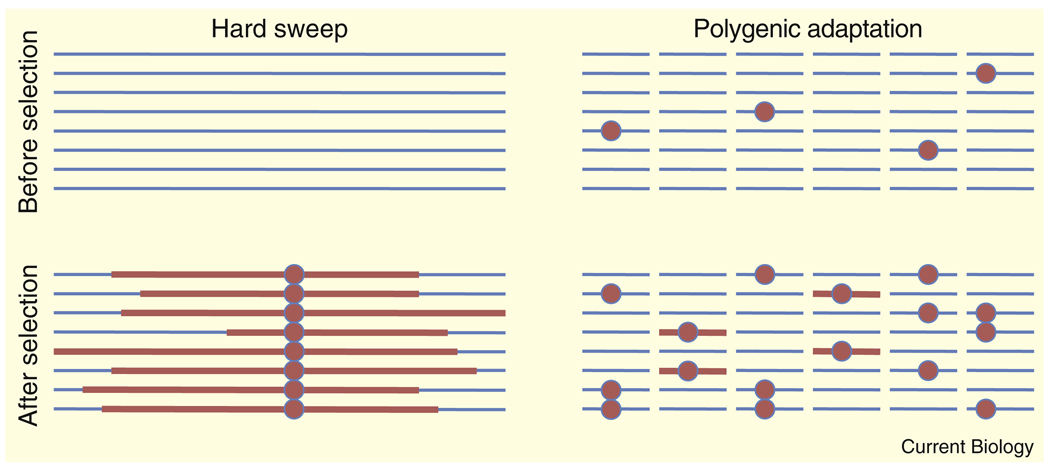
Figure 3. A cartoon illustration of the hard sweep and polygenic adaptation models.
The horizontal blue lines represent haplotypes, and the red lines indicate regions that are identical by descent (IBD). The red circles indicate alleles that are favored following an environmental change. In the monogenic (hard selection) model, selection drives a new mutation to fixation, creating a large region of IBD. In the polygenic model, prior to selection red alleles exist at modest frequencies at various loci across the genome. (The red alleles can be thought of as being alleles that all shift a particular phenotype in the same direction, e.g., alleles that increase height.) After selection, the genome-wide abundance of favored alleles has increased, but in this cartoon they have not fixed at any locus. In this example, at some loci selection has acted on new variants, creating signals of partial sweeps at those loci (the x-axis scale is not necessarily the same in the left- and right-hand plots).
The question, then, is how to make sense of these apparent discrepancies. On one hand there are examples of apparent physiological and morphological adaptations in modern human populations, strong signals of selection at candidate loci and genome-wide patterns showing clear differences between genic and non-genic regions that are difficult to explain by neutral processes. On the other hand, genome-wide data suggest that there are relatively few fixed (or nearly fixed) differences between HapMap populations, that those that do exist have generally become fixed relatively slowly and that the geographic distributions of putatively selected alleles are strongly influenced by the historical relationships among populations.
Based in part on the scarcity of high-FST SNPs with strong haplotype signals, Coop et al. argued that few hard sweeps (i.e., sweeps of new mutations) with selection coefficients of more than 1% have swept to fixation in the time since the out-of-Africa migration [22]. A number of possible explanations were proposed for the overall patterns in the data, including that most selection on individual alleles may be relatively weak so that these alleles have not had time to sweep to fixation within continental populations; that the strength of selection may vary temporally, and it may be rare for selection to be consistently strong for the 10,000 years or more required to drive an allele to near fixation; and that much of human adaptation may proceed by either polygenic adaptation or soft sweeps that can be difficult to detect using standard methods.
Hard Sweeps, Soft Sweeps and Mutational Target Size
The standard approaches to detecting selection in population genetic data have been strongly shaped by the classical hitch-hiking model explored in detail by Maynard Smith and Haigh [40]. In this model, new advantageous mutations spread rapidly to fixation, purging variation at linked sites as they spread. This type of process has been referred to as a ‘hard sweep’ (Box 1) [41]. Most of the genome-wide scans in humans have aimed to detect signals of this type of selective sweep. However, there is good reason to believe that other modes of adaptation are important. For example, recent empirical [23,27,42–44] and theoretical [41,45–48] work has highlighted the potential importance of ‘soft sweeps’, i.e., sweeps from standing variation, or sweeps in which multiple mutations start to sweep simultaneously at a single locus (if the favored mutations are roughly equivalent, then no single allele sweeps rapidly to fixation). Simulations have shown that soft sweeps typically have weaker effects on linked sites and, therefore, may be more difficult to detect than hard sweeps [35,48,49].
Box 1.
Glossary
Background selection: refers to a process in which weakly deleterious mutations drift up to low frequencies and are then purged from the population. This causes a reduction in diversity, especially around conserved regions. In some respects, the signals of background selection can mimic patterns produced by positive selection [29,32].
FST: a classical measure of the amount of allele frequency differentiation between two or more populations. FST can take values between 0 and 1, with 0 corresponding to identical allele frequencies in both (all) populations, and 1 corresponding to a fixed difference: i.e., that the allele is absent in one population, and fixed in the other. High FST values for particular SNPs may sometimes provide evidence that those SNPs are under selection.
Hard sweep: the classical selective sweep model in which a new advantageous mutation arises, and spreads quickly to fixation due to natural selection [40]. Under this model, neutral variation near to the favored site “hitch-hikes” along with the favored allele. This impacts patterns of variation around the selected site in ways that can be detected using a variety of tests of selection [5].
Mutational target size: refers to the number of sites at a locus that, if appropriately mutated, could generate a particular favored phenotype. For example, it appears that several mutations in an upstream enhancer of lactase cause lifelong expression of the lactase gene [27,76]. The size of the mutational target affects the probability that standing variation will be available to allow rapid evolution following an environmental change.
Polygenic adaptation: here, we use this term to describe a process in which adaptation occurs by simultaneous selection on variants at many loci (perhaps tens or hundreds or more). We envisage that a common scenario of polygenic adaptation would be that there is a shift in the optimal phenotype for a quantitative trait that is affected by hundreds of alleles of small effect. In this case, we can anticipate a response to selection that is due to small frequency shifts of many alleles. Polygenic adaptation might also occur from new mutations at many loci, following a shift in the optimal phenotype. This latter scenario would be most likely if the newly favored phenotype had previously been strongly disfavored.
Partial sweep: an event in which a favored allele increases rapidly from low frequency, but has not yet reached fixation (perhaps because the sweep is still in progress, or because the selective advantage of the favored allele has weakened).
Selective sweep: an event in which the frequency of a favored allele increases rapidly due to selection. This term is often understood to refer to complete hard sweeps, but may also refer to partial sweeps or soft sweeps (see below), depending on the context.
Soft sweep: this term was introduced to describe two slightly different scenarios that both contrast with the standard hard sweep model [41]. In one scenario, due to a change in selection, an allele that is already segregating in the population (i.e., standing variation) becomes selectively favored, and sweeps up in frequency. It is usually assumed that the allele is neutral or mildly deleterious prior to the change in selection. In the second scenario, multiple independent mutations at a single locus are all favored and all increase in frequency simultaneously until the sum of the frequencies is 1. If the favored alleles are all similarly advantageous, then typically none of the favored mutations would fix during the selective event. Both scenarios tend to be more difficult than hard sweeps to detect using standard tests of selection.
Standing variation: variants that are polymorphic in a population. The term is used here in the context of a selective force that is turned on so that variants that had been drifting (nearly) neutrally suddenly become favored.
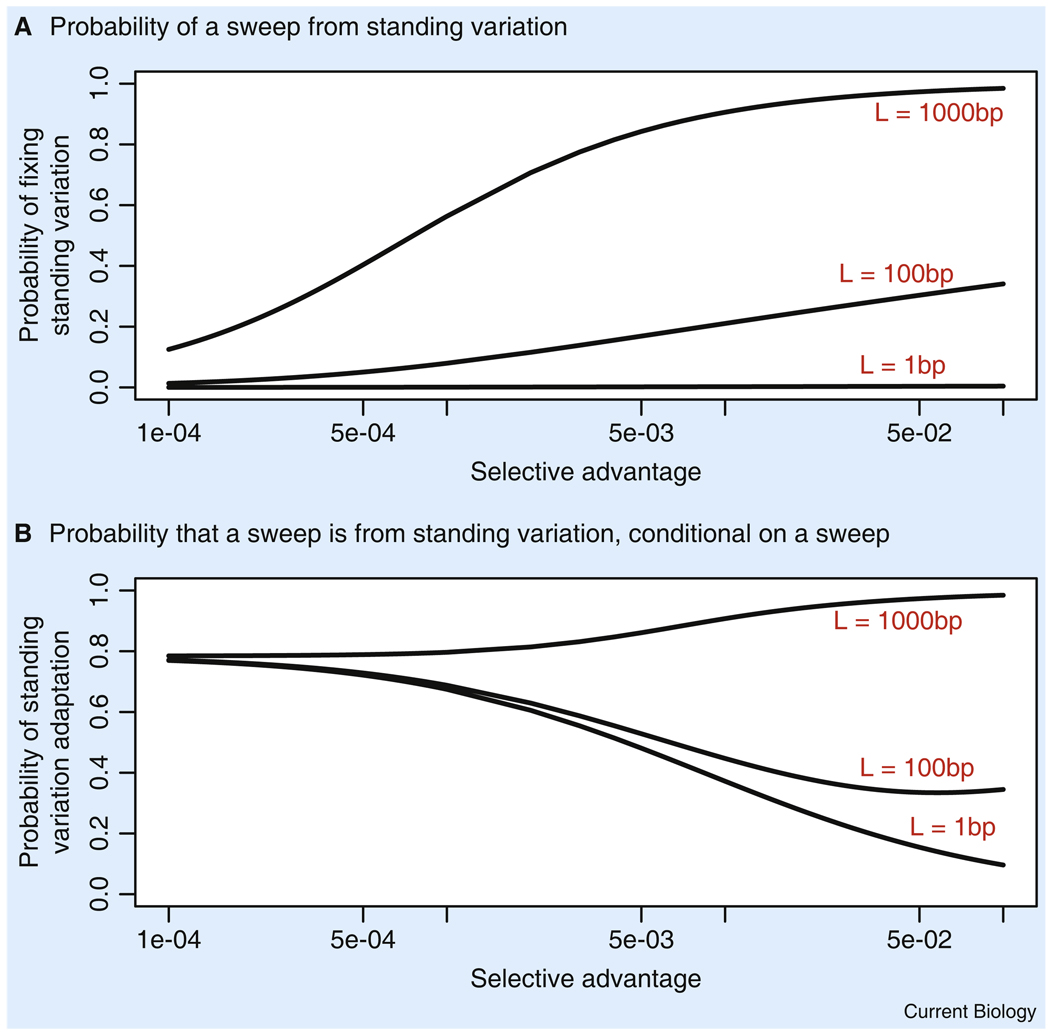
Although their prevalence is ultimately an empirical question, simple models suggest that, for plausible parameter values, soft sweeps are likely to be widespread [41,49]. Consider the process of adaptation to a sudden environmental change — for example, think of the onset of dairy farming, or a sudden spread of Plasmodium vivax. Suppose that mutant alleles at any of L base pairs in the genome would change a particular aspect of the organism’s phenotype to yield higher fitness in the new environment. This mutational target size of L base pairs might represent the size of a regulatory enhancer or a set of amino acids that could change a protein’s function. Prior to the environmental change, we might consider that SNPs in the mutational target are either neutral or mildly deleterious, allowing standing variation to be present at low to intermediate frequencies within the population. How likely is it that adaptation occurs from standing variation? Under this type of model, Hermisson and Pennings [41] showed that if the size of the mutational target is on the order of 100 base pairs, then there is a substantial probability that a sweep occurs from standing variation present in the population at the time of the environmental change (Figure 2) [41] — these calculations assume an effective population size of 10,000 individuals. If L is as large as 1000 base pairs, then it is extremely likely that substantial variation exists within the target region prior to the environmental change, and hence most adaptation will occur from standing variation.
Figure 2. The probability of sweeps from standing variation following an environmental change.
(A) Probability that a full sweep occurs from standing variation, as a function of the mutational target size and the strength of selection after the environmental switch. (B) Probability that a sweep occurs from standing variation, conditional on a sweep occurring either from standing variation or from new variation that arises within the first 1000 generations after the environmental change. The model is as described in the text and in [41]. Parameters: Prior to the environmental switch, variation is deleterious with 2Nes = −10. We assume Ne = 10,000 and µ = 2 × 10−8. All selection is assumed additive. Modified with permission from [41].
Conversely, if the mutational target is very small, or if prior to the environmental change the now-beneficial alleles were strongly deleterious, then sweeps from standing variation are unlikely. If sweeps occur under these circumstances, then they are somewhat more likely to be hard sweeps than soft sweeps. However, the flip side of this is that when the mutational target is small, the waiting time for new mutations can be extremely long. The expected waiting time to the first new mutation that goes to fixation is (4NeµLs)−1, where Ne is the effective population size, µ is the mutation rate per site, and s is the selective advantage of each copy of the favored allele, assuming an additive model (e.g., from [50], assuming 2NµL favored mutations per generation). Consider the worst case, where only one possible mutation at a single site will work, so that L = 1/3. Then, if s = 1% and 4Neµ = 10−3, the expected waiting time is extremely long, namely 300,000 generations. In this extreme case, adaptation to the new environment is essentially ineffective. It has been proposed that recent population growth of humans could have supplied a greater input of new mutations [51,52]; additionally, even modest population growth can greatly increase fixation probabilities of favored alleles [53]. The latter effect could substantially reduce the waiting time for new favored mutations to start spreading at loci with very small mutational targets, although it should be noted that it is still unclear when growth in census population size began to increase the effective population size [54].
These models assume that environmental change, broadly defined, is a primary driver of adaptation. If this is the case, then the results would argue that soft sweeps are likely to be common, and perhaps the main mode of adaptation. The exact balance between hard and soft sweeps would depend on the distribution of mutational target sizes, which we do not yet know. For traits where the mutational target size is hundreds of base pairs or more, we can expect that adaptation from standing variation is likely to be the rule and, additionally, that very often multiple favored mutations may sweep up simultaneously, with none reaching fixation during the selective sweep [47].
Polygenic Adaptation from Standing Variation
Most of the recent literature in human population genetics focuses on models of selection at one, or a small number of loci, as in the previous section. This is in contrast to classical models of natural and artificial selection in quantitative genetics, where it is assumed that most traits of interest are highly polygenic, and are influenced to a small degree by standing variation at many loci [55]. The quantitative genetics view is supported both by classical breeding and selection experiments and occasionally field observations [56–58], as well as by recent genome-wide association studies showing that many traits are highly polygenic.
We would argue that for many traits, the quantitative perspective may be closer to reality: that is, that short-term adaptation takes place by selection on standing variation at many loci simultaneously (e.g., [22,59–61]). Consider a trait that is affected by a large (finite) number of loci. If the environment shifts so that there is a new phenotypic optimum, then the population will adapt by allele frequency shifts at many loci (Figure 3). Once the typical phenotype in the population matches the new optimum, selection will weaken. This means that it may be very common for selection to push alleles upwards in frequency, but generally not to fixation [22,62,63]. In principle, this type of process could allow very rapid adaptation, yet be difficult to detect using most current population genetic methods.
The example of human height illustrates these issues. Height has long been a textbook example of a polygenic trait [64]; recently, three genome-wide association studies identified a total of around 50 loci that contribute to adult height in Europeans [65–68]. Each associated allele affects total height by about 3–6 mm, and together these loci explain about 5% of the population variation in height, after controlling for sex. Since height is extremely heritable [69], many more loci remain to be found. If there were a sudden onset of strong selection for increased height, we could expect a rapid upward shift in average height [55]. However, the response to selection would be generated by modest allele frequency shifts at many loci that are already polymorphic. Even with very strong selection, and a strong phenotypic response, standard methods for detecting selective sweeps would have little power. In the final section of this review, we will discuss possible approaches to studying polygenic adaptation.
The idea that polygenic adaptation from standing variation is important could help to explain key aspects of the data. This would allow rapid phenotypic adaptation (for example, to high altitude, as described above) without necessarily generating any large differences in allele frequencies between populations. This could also help to explain the apparent scarcity of rapid, hard sweeps. Qualitatively, increased drift of neutral alleles within genes due to effects of polygenic selection on nearby sites [70] should create the types of differences that are observed between genic and non-genic regions. However, it is not yet clear whether the magnitude of this effect could explain the observed patterns (background selection may also contribute [29]).
Of course, we do not mean to imply that all adaptation occurs in this way. Indeed, it is notable that some of the most impressive selection signals involve loci that act in a monogenic fashion. For example, the mutations that modify expression of lactase and the African Duffy/DARC mutation are fundamentally single-gene traits. A number of other loci, such as the pigmentation loci SLC24A5 and KITLG [15,16], which are not clearly monogenic, do show striking sweep signals that match the qualitative predictions of the hard-sweep model. It may be that pigmentation has a genetic architecture that makes hard sweeps more likely: perhaps because this trait is less polygenic than many other traits, or because the relevant loci tend to have less functional standing variation prior to selection (as would occur if functional variants tend to be highly deleterious in the “wrong” environment).
The polygenic model also suggests an interesting decoupling of short-term adaptation from the fixation of alleles. A shift in the optimal phenotype would cause allele frequency changes at many loci. Once the population reaches the new optimum, all the relevant alleles would be subject to a sort of weak frequency-dependent selection, where downward drift of some allele frequencies would have to be balanced by upward drift of other alleles elsewhere in the genome. If some alleles with negative pleiotropic effects had initially increased in frequency due to the selection, then these could now be eliminated, to the benefit of other alleles. At larger time scales, species differences are generally due to fixed differences; this might occur by the somewhat random fixation of an appropriate constellation of alleles to maintain the preferred phenotype [71].
Finding the Real Selection Signals
Ultimately, a comprehensive model of the nature of selection would tell us how much adaptation occurs by any of a variety of different models and mechanisms. For example, what are the relative contributions of hard sweeps, soft sweeps and polygenic adaptation; or of coding and non-coding changes? How important are pleiotropic or epistatic effects on selected variants? Additionally, we would want to know the typical geographic range of selection events, the actual target loci and the relevant phenotypes. These questions all focus on positive selection, but a truly comprehensive model would also tell us how much phenotypic change is instead due to drift of neutral alleles, and the relative importance of background selection.
To make real progress on these problems will require much greater integration of selection studies with biological information. Except for the strongest, clearest sweep signals, it is difficult to confidently distinguish true signals from false positives using population genetic data alone [35]. However, as we learn more about gene functions, we will surely find that there are some — perhaps many — more great biological candidate loci lurking near the top of the selection scan lists. Fortunately, advances in genomics should allow real progress on these problems in the coming years. We are now seeing dramatic improvements in functional annotation of the genome. This now includes information on thousands of variants that impact risk for diseases, affect quantitative traits or alter gene expression levels. Through a variety of experimental and computational methods, we can expect that the annotation of regulatory elements will be greatly improved. Our knowledge of gene functions and gene pathways continues to improve steadily. Finally, there will soon be genome sequences for large numbers of individuals, and these will offer numerous advantages over the current SNP data.
In the future, therefore, we expect to see more success at identifying phenotypes or pathways that are enriched for selection signals [4,18,34,72,73], which will also bolster the support for the action of selection in general. At present, these approaches are somewhat underpowered since we generally know only a few of the relevant genes, and it is difficult to predict functional variants within each locus. One would want to place more weight on sweep signals that include variation at likely functional sites. Improved external information will likely help greatly in the coming years.
It is of particular interest to think about how to make progress in detecting polygenic adaptation, where traditional population genetic methods are likely to fail. One way forward would rely on combining information over large numbers of variants that have been mapped for a trait of interest. One could then test whether, looking across many loci, there is a significant tendency for alleles that increase the phenotype value to increase (or decrease) in frequency together more than might be expected under a model of drift; note that such tests must account for whether the trait was ascertained to differ between the populations in question [74,75]. For example, one might predict that alleles that increase body mass are, collectively, at higher frequencies in arctic populations than in related warmer weather populations, consistent with this phenotypic shift being selectively driven. Similarly, it may be possible to search the genome for alleles that respond consistently to particular environmental pressures [4]. In summary, we argue that broadening the search for positive selection in the human genome to a wider range of selective models will be a fruitful avenue for progress on these problems in the coming years.
Acknowledgements
We thank Anna Di Rienzo, Colleen Julian, Chuck Langley, George Perry, Molly Przeworski, Michael Turelli, and an anonymous reviewer for helpful discussions or comments. This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health RO1 MH084703-01 (J.K. Pritchard), a National Institutes of Health training grant to the University of Chicago (J.K. Pickrell), and the Sloan Foundation (G.C.).
References
- 1.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 6.Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruff C. Morphological adaptation to climate in modern and fossil hominids. Am. J. Phys. Anthropol. 1994;37:65–107. [Google Scholar]
- 8.Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol. Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Beall CM. Tibetan and Andean contrasts in adaptation to high-altitude hypoxia. Adv. Exp. Med. Biol. 2000;475:63–74. doi: 10.1007/0-306-46825-5_7. [DOI] [PubMed] [Google Scholar]
- 10.Moore LG. Human genetic adaptation to high altitude. High Altitude Med. Biol. 2001;2:257–279. doi: 10.1089/152702901750265341. [DOI] [PubMed] [Google Scholar]
- 11.Brutsaert TD. Population genetic aspects and phenotypic plasticity of ventilatory responses in high altitude natives. Resp. Physiol. Neurobiol. 2007;158:151–160. doi: 10.1016/j.resp.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Julian CG, Wilson MJ, Moore LG. Evolutionary adaptation to high altitude: a view from in utero. Am. J. Hum. Biol. 2009;21:614–622. doi: 10.1002/ajhb.20900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jablonski NG, Chaplin G. The evolution of human skin coloration. J. Hum. Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 14.Parra EJ. Human pigmentation variation: evolution, genetic basis, and implications for public health. Am. J. Phys. Anthropol. 2007;45 Suppl.:85–105. doi: 10.1002/ajpa.20727. [DOI] [PubMed] [Google Scholar]
- 15.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 16.Miller C, Beleza S, Pollen A, Schluter D, Kittles R, Shriver MD, Kingsley DM. Cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norton H, Kittles R, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 18.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 20.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamblin M, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 25.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF, et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. Convergent adaptation of human lactase persistence in Africa and Eur. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai JJ, Macpherson JM, Sella G, Petrov DA. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 2009;5:e1000336. doi: 10.1371/journal.pgen.1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 31.Hellmann I, Mang Y, Gu Z, Li P, De La Vega FM, Clark AG, Nielsen R. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 2008;18:1020–1029. doi: 10.1101/gr.074187.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teshima K, Coop G, Przeworski MF. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 37.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 39.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 41.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 43.Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological diverence between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Scheinfeldt LB, Biswas S, Madeoy J, Connelly CF, Schadt EE, Akey JM. Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Mol. Biol. Evol. 2009;26:1357–1367. doi: 10.1093/molbev/msp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Nat. Acad. Sci. USA. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orr HA, Betancourt AJ. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pennings P, Hermisson J. Soft sweeps II-molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 2006;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- 48.Przeworski M, Coop G, Wall J. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- 49.Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2006;2:e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 51.Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc. Natl. Acad. Sci. USA. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed FA, Aquadro CF. Mutation, selection and the future of human evolution. Trends Genet. 2006;22:479–484. doi: 10.1016/j.tig.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Otto SP, Whitlock MC. The probability of fixation in populations of changing size. Genetics. 1997;146:723–733. doi: 10.1093/genetics/146.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. New York: Longman; 1996. [Google Scholar]
- 56.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat. Rev. Genet. 2002;3:11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 57.Grant PR, Grant BR. Predicting microevolutionary responses to directional selection on heritable variation. Evolution. 1995;49:241–251. doi: 10.1111/j.1558-5646.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- 58.Hill WG, Caballero A. Artificial selection experiments. Annu. Rev. Ecol. Sys. 1992;23:287–310. [Google Scholar]
- 59.Bulmer M. The effect of selection on natural variability. Am. Nat. 1971;105:314–331. [Google Scholar]
- 60.Turelli M, Barton NH. Dynamics of polygenic characters under selection. Theor. Popul. Biol. 1990;38:1–57. [Google Scholar]
- 61.Turelli M, Barton N. Genetic and statistical analyses of strong selection on polygenic traits: what, me normal? Genetics. 1994;138:913–941. doi: 10.1093/genetics/138.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chevin LM, Hospital F. Selective sweep at a quantitative trait locus in the presence of background genetic variation. Genetics. 2008;180:1645–1660. doi: 10.1534/genetics.108.093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lande R. The response to selection on major and minor mutations affecting a metrical trait. Heredity. 1983;50:47–65. [Google Scholar]
- 64.Fisher RA. On the correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinburgh. 1918;52:399–433. [Google Scholar]
- 65.Aulchenko YS, Struchalin MV, Belonogova NM, Axenovich TI, Weedon MN, Hofman A, Uitterlinden AG, Kayser M, Oostra BA, van Duijn CM, et al. Predicting human height by Victorian and genomic methods. Eur. J. Hum. Genet. 2009;17:1070–1075. doi: 10.1038/ejhg.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 67.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visscher PM, Medland SE, Ferreira MA, Morley KI, Zhu G, Cornes BK, Montgomery GW, Martin NG. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barton NH. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B. 2000;355:1553–1562. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barton NH. The divergence of a polygenic system subject to stabilizing selection, mutation and drift. Genet. Res. 1989;54:59–77. doi: 10.1017/s0016672300028378. [DOI] [PubMed] [Google Scholar]
- 72.Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Myles S, Somel M, Tang K, Kelso J, Stoneking M. Identifying genes underlying skin pigmentation differences among human populations. Hum. Genet. 2007;120:613–621. doi: 10.1007/s00439-006-0256-4. [DOI] [PubMed] [Google Scholar]
- 74.Anderson EC, Slatkin M. Orr’s quantitative trait loci sign test under conditions of trait ascertainment. Genetics. 2003;165:445–446. doi: 10.1093/genetics/165.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]