Abstract
We report a case of hemolytic anemia that was subsequently identified to be a case of α-thalassaemia harboring the common rightward 3.7 kb deletion/HbH. The diagnosis was based on sequential analyses using BioRad D10 HPLC, Alkaline gel electrophoresis, GPO α THAL-IC strips and the identification of the specific genetic lesion using an α Globin reverse dot blot hybridization assay. Supravital stain of RBCs helped in identifying classical HbH inclusions. In a background of a variable clinical presentation, lack of definitive hematological markers, and general under-diagnosis of α-thalassaemias we have used this case to highlight the features and sequence of techniques involved in identifying and characterizing an α-globin chain mutation, starting from a diffuse clinical history and presentation up to the identification of a specific genetic lesion involved.
Keywords: HbH, α THAL-IC strip, Reverse dot blot hybridization, HPLC, Alkaline hemoglobin electrophoresis
Introduction
Thalassaemia disorders in humans caused by the deficiency of the alpha chain of hemoglobin have been well documented and validated throughout the world, by various stepwise diagnostic strategies and techniques [1–3]. The condition is caused by the disruption of one or more of the four resident homologous hemoglobin alpha genes, α1α2//α1α2 clustered in chromosome 16p. One gene (α+/silent carriers) or two gene (α0/thal trait) disruptions out of the four may lead to mild anemia and microcytosis and do not present a problem to the carriers, only predisposing them to a mild α-thalassaemia trait. This genetic condition remains an important segment of α-thalassaemias largely for the potential risk the carriers have in bearing offspring with more severe forms of the disease.
The two remaining categories of alpha thalassaemia disease are Hemoglobin H (HbH) disease and α-thalassaemia major. α-Thalassaemia major is a severe genetic condition where all four of the α alleles are defunct and the crisis is overtly manifested as Hb Bart’s hydropsis fetalis syndrome. In this case, with the absence of alpha chains, fetal γ-globin tetramers (Hb Bart’s) partition oxygen far too effectively to be compatible with life and in the absence of transfusion or transplant intervention carriers of α-thal major die perinatally.
However, triple gene disruptions leads to the condition known as HbH disease, where the single functional gene permits carriers to survive but a large reduction of α-globin chain production consequently disturbs the associative stoichiometry of the α2β2 chains in hemoglobin. In this cellular environment of reduced α-chain availability, the excess β-globin chains often form aggregating unstable HbH (β4-tetramers) in addition to some Hb Bart’s (γ-globin tetramers) and traces of the embryonic Hb Portland (ζ2γ2). These can be microscopically visualized as numerous dot size inclusions (Heinz bodies) in the erythrocyte cytoplasm using supravital brilliant cresyl blue (BCB) stains. Anywhere from 30–100% of the RBCs may contain these typical inclusions, but brisk reticulocytosis can make identification and enumeration difficult. Individuals with hemoglobin H disease experience events of hemolytic anemia that range from mild to moderate severity and, along with a range of MCH, MCHC and MCV indices, there is very little that indicates the underlying genetic condition. As a result of this while HbH remains the predominant clinical cases presented for α-thalassaemias they have been an under-diagnosed entity in India.
The present study describes a case of a 24 year old Bengali Muslim patient suffering from hemolytic anemia referred to Manipal Hospital, Bangalore. The communication presents the sequence of tests which have been conducted to diagnose the occurrence of HbH and identification of an associated genetic lesion in the patient suffering from α-thalassaemia.
Materials and Methods
Blood sample from the patient was collected in an EDTA tube and was analyzed on a Sysmex XT 1800i Hematology analyzer. A serum sample collected in a BD vacutainer gel tube was used for biochemical investigations on a COBAS 6000 modular system. Investigations conducted included Lactate Dehydrogenase, Iron, Ferritin, Liver function tests, Bilirubin, Liver enzymes, Total Protein and Albumin. Serum Haptoglobin was assayed on the Siemens BN-Prospec Nephelometer.
Based on these findings from the above analyses the hemoglobin variant analysis was initially conducted on a BioRad D10 HPLC instrument as per standard procedures using the β-Thal short (BTS) program.
Hemolysates for electrophoresis was prepared as per Helena SAS-Mx Alkaline Hb-10 protocol, and electrophoresis carried out on HELENA SAS-II analyzer at an alkaline pH.
Whole blood was used for screening of α-thalassaemia carrier status by using the GPO α-Thal sandwich-type immunochromatographic (IC) strip. The strip contains: (1) monoclonal anti Hb Bart’s antibody in the test line, (2) Anti mouse immunoglobulin antibody in the control line, (3) Monoclonal anti Hb Bart’s colloidal gold conjugate in the absorbent area. The α-thal specific protein used is a γ4 protein. A GPO α THAL-IC strip was dipped in 200 μl of EDTA blood with 200 μl of lysis buffer which was added and vortexed. After 5 min, the strip was washed with the supplied buffer.
To identify presence of HbH inclusions 1% brilliant cresyl blue (BCB) was used by observing the in vitro treated cells for 4–24 h.
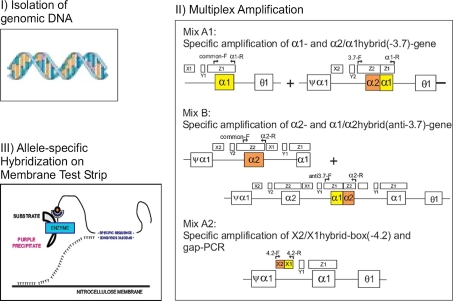
For the Reverse dot blot hybridization assay genomic DNA was extracted from the blood sample using the DNEasy kit from Qiagen. In order to identify the α-thalassaemia mutation an α-Globin strip Assay kit (cat 4-160/4-161) from Vienna lab (Labor diagnostika GmbH) was used. The method is based on multiplex DNA amplification (including gap-PCR) and involves three steps: (1) DNA isolation, (2) PCR amplification using biotinylated primers, and (3) hybridization of amplification products to test strips containing allele specific oligonucleotide probes immobilized as an array of parallel lines (Fig. 1).
Fig. 1.
Schematic representation of the Test Principle using the Vienna Lab Reverse Dot Blot Hybridization technique (Courtesy: Vienna Lab, Labordiagnostika, GmbH)
Results and Discussion
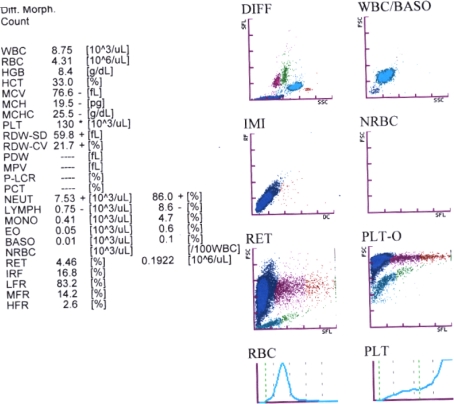
The 24 year old patient was admitted to hospital suffering with fever for 10 days and had a history of intermittent jaundice for the past 4–5 years. His physical examination showed mild hemolytic facies and a palpable spleen. On admission his hemoglobin was 8.4 gm/dl, MCV 76.6 fl, MCH 19.5 pg, MCHC 25.5 gm/dl, with an abnormal platelet distribution. Retic preparations showed 11% reticulocytes (Fig. 2).
Fig. 2.
Printed output for the analysis of EDTA-Blood sample from patient on a Sysmex XT 1800i Hematology analyzer
The peripheral smear showed presence of microcytes, macrocytes, elliptocytes and tear drop cells.
Biochemical investigations showed Total Bilirubin to be 5.0 mgs/dl and Direct Bilirubin 0.3 mgs/dl, serum LDH was 305 U/l, Iron was 150 μg/dl, Ferritin 377 ng/ml, SGOT 35 U/L, SGPT 45 U/L, Alkaline phosphatase 82 U/L and Haptoglobin which was tested on the Siemens BN-Prospec Nephelometer was <0.25 g/l.
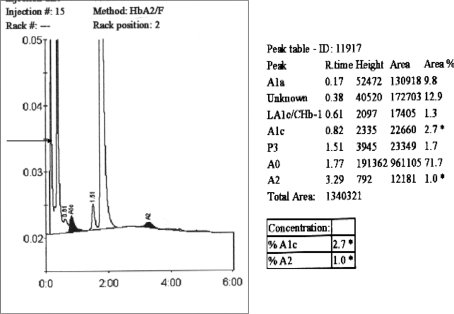
BioRad D10 HPLC profile (reverse phase HPLC) (Fig. 3) showed 2 peaks with Retention time of 0.17 and 0.38 min with area percentage of 9.8 and 12.9% respectively. HbA2 with RT at 3.29 min was reduced at only 1.0%.
Fig. 3.
Print out-put from the BioRad D10 HPLC machine showing (arrow) unidentified proteins eluting at retention times of 0.17 and 0.38 min. HbA2 levels are also reduced (at 1%)
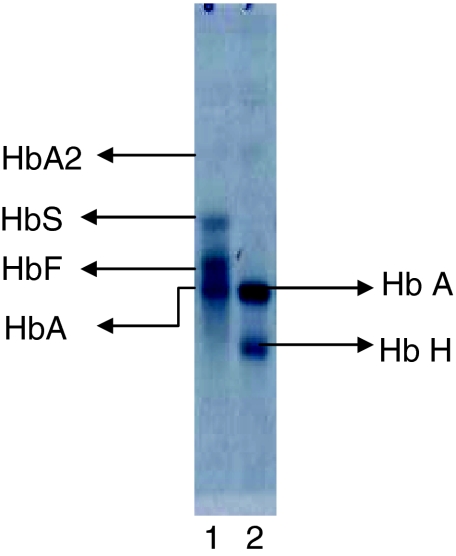
α-Thalassaemia is suggested when eluted proteins are detected at a retention time between 0 and 1 min. A retention peak less than 1 min on a BTS program of the BioRad HPLC system is suggestive of Hb Bart’s and HbH [4]. In addition, a reduced level of HbA2 < 1.5% is also indicative of α-thalassaemia. In order to pursue this further, an alkaline hemoglobin electrophoresis at pH 8.6 was conducted using the HELENA SAS II system. A band which moved ahead of the adult HbA band was seen highly suggestive of HbH (Fig. 4).
Fig. 4.
Electropherogram of the alkaline hemoglobin electrophoresis (pH 8.6) conducted on the HELENA SASII system for the patient sample. Lane 1: AFSA2 pack Control, Identities of the migrating bands are indicated on the figure. Lane 2: Patient sample shows the presence of a putative HbH band migrating ahead of HbA
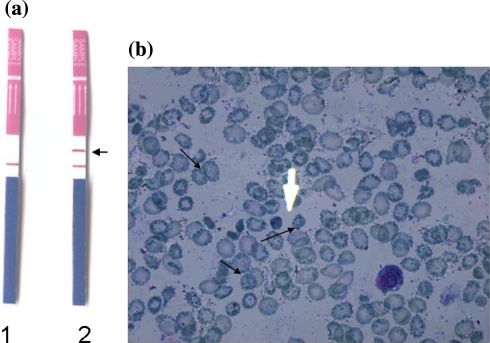
A GPO α THAL-IC strip was used for the qualitative confirmation of α-thalassaemia as described in materials and methods. This method remains as a convenient confirmatory tool for screening of α-thalassaemia [5]. The presence of two colour lines, one in the control region and other in that region confirmed it to be case of α-thalassaemia (Fig. 5a).
Fig. 5.
a Results for the GPO α THAL-IC strip test conducted on control and patient samples. Strip 1: Normal sample, Strip 2: Patient sample showing positive for an extra band (arrow) indicating the presence of α-thalassaemia disorder. b 1% brilliant cresyl blue (BCB) precipitated HbH inclusions within typical ‘golf ball’ cells (arrows) confirming the presence of HbH
The qualitative confirmation of α-thalassaemia was verified by microscopically examining slides for HbH inclusions. In this case smears prepared from the patient sample were stained with 1% brilliant cresyl blue (BCB) and kept for observation between 4 and 24 h. Presence of “golf ball” cells with classical HbH inclusions confirmed the findings (Fig. 5b). HbH tetramers, when oxidized in vitro, precipitate and hence can be visualized microscopically and therefore staining unfixed cells with an oxidative dye such as BCB generate HbH inclusions. It is important to consider that batch to batch variability in dye occurs, so the use of positive and negative control slides for this visualization is critical.
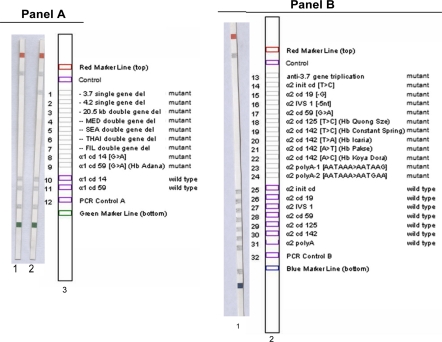
The above series of tests have shown unambiguously that the patient carried an α-thalassaemia disorder. In order to identify the associated mutation(s) of α thalassaemia we made use of the Alpha Globin strip Assay from Vienna Lab. The test is a mutiplex PCR reverse dot blot hybridization based procedure developed for screening and detection of 21 known alpha-globin mutations (Fig. 6). The developed strips revealed the presence of a 3.7 kb single gene deletion for the patient, confirming the association of a genetic lesion with the α-thalassaemia band detected on the GPO α THAL-IC strip.
Fig. 6.
Vienna Lab α-thal mutation screening strips for the deletion (A) and non-deletion (B) mutations reported for α-thalassaemia. A Strip 1: normal control sample, Strip 2: patient sample showing the −3.7 kb gene deletion positive band, Strip 3: Mutation key strip. B Strip 1: No mutations detected in the non-deletional set of mutations for the patient sample, Strip 2: Mutation key strip
It is predicted that in addition to the 3.7 kb-single gene deletion identified by the strip, there are more α-gene disabling mutations lurking within the α-globin gene cluster of this patient on the short arm of chromosome 16. In all probability the patient is homozygous for the −3.7 deletion with an additional inactivating mutation (+α3.7/−α3.7). The deletional mutations are the most prevalent α-thal mutation in India [6] and −3.7 homozygosity has been described as among the more prevalent genetic conditions in a population survey in Kolkata and the Middle east [7, 8] where 80% of inhabitants are of subcontinental and Arab origin. Efforts are underway to sequence the relevant α-globin regions of this patient to identify the associated mutations.
Conclusion
Though the aspect of gene disruptions in HbH α-thalassaemia is well described, the clinical picture presents a somewhat messier reality with considerable variability in its presentation. The non-equivalence of the alpha globin alleles and their mutations is gradually being established and the clinical severity in HbH disease is now understood to be subject to the nature of the mutations that come together in any patient [9–11]. A genotype–phenotype correlation has been reported with patients harboring non deletional mutations presenting symptoms at an earlier age, with more severe hemolytic anemia and required more transfusions than patients with deletional HbH disease. There may be a day when sufficient correlations will be established to predict, with some accuracy, the nature of the underlying mutations in α-thal patients based on phenotype alone, however for the moment the vigilance of the clinicians and laboratories testing for the disorder is called for.
The standard practice of excluding iron deficiency and beta thalassemia and using cell counter indices as a marker for alpha thalassemia remains the first step to ascertaining alpha globin deficiencies followed by support of HPLC and electrophoresis procedures [12].We have relied on accessory clues for the condition using the BioRad D10-BTS program to qualitatively indicate common α-thalassaemia diseases including HbH and Bart’s. We have followed this up by a series of investigations to test this suspicion with electrophoresis, qualitative α-thal strips and microscopy. Though the routine diagnosis of HbH disease may not need the specific mutation(s) to be identified as finally conducted for this study, the complex genetics of α-thalassaemia means that population based diagnostic studies would need to screen rapidly and accurately for common α-thal mutations [2, 5, 13]. Testing formats such as reverse dot hybridization which empower laboratories to identify specific mutations using multiplex PCR techniques in conjunction with standard hybridization protocols may be the way ahead [8, 14].
References
- 1.Weatherall DJ, Clegg JB. Inherited hemoglobin disorders: an increasing global health Problem. Bull World Health Organ. 2001;79(8):704–712. [PMC free article] [PubMed] [Google Scholar]
- 2.Baysal E, Huisman THJ. Detection of common deletional α thalassaemia-2 determinants by PCR. Am J Hematol. 1994;46:208–213. doi: 10.1002/ajh.2830460309. [DOI] [PubMed] [Google Scholar]
- 3.Clarke GM, Higgins TN. Laboratory investigation of hemoglobinopathies and thalassaemias: review and update. Clin Chem. 2000;46(8):1284–1290. [PubMed] [Google Scholar]
- 4.Fucharoen S, Winichagoon P, Wisedpanichkij R, Sae-Ngow B, Sriphanich R, Oncoung W, et al. Prenatal & postnatal diagnoses of thalassaemias and hemoglobinopathies by HPLC. Clin Chem. 1998;44(4):740–748. [PubMed] [Google Scholar]
- 5.Tayapiwatana C, Kuntaruk S, Tatu T, Chiampanichayakul S, Munikongdee T, Winichagoon P, et al. Simple method for screening of α-thalassaemia-1 carriers. Int J Hematol. 2009;89:559–567. doi: 10.1007/s12185-009-0331-4. [DOI] [PubMed] [Google Scholar]
- 6.Nadkarni A, Phanasgaonkar S, Colah R, Mohanty D, Ghosh K. Prevalence and molecular characterization of α-thalassaemia syndromes among Indians. Genet Test. 2008;12(2):177–180. doi: 10.1089/gte.2007.0080. [DOI] [PubMed] [Google Scholar]
- 7.Baysal E. Hemoglobinopathies in the United Arab Emirates. Hemoglobin. 2001;25(2):247–253. doi: 10.1081/HEM-100104033. [DOI] [PubMed] [Google Scholar]
- 8.Hellani A, Fadel E, El-Sadadi S, El-Sweilam H, El-Dawood A, Abu-Amero KK. Molecular spectrum of α-thalassaemia mutations in microcytic hypochromic anemia patients from Saudi Arabia. Genet Test Mol Biomark. 2009;13(2):219–221. doi: 10.1089/gtmb.2008.0123. [DOI] [PubMed] [Google Scholar]
- 9.Li DZ, Liao C, Li J, Xie XM, Huang YN, Wu QC. Hemoglobin H hydrops fetalis syndrome resulting from the association of the – –SEA deletion and the αQuong Szeα mutation in a Chinese woman. Eur J Haematol. 2005;75(3):259–261. doi: 10.1111/j.1600-0609.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 10.Chui DH, Fucharoen S, Chan V. Hemoglobin H disease: not necessarily a benign disorder. Blood. 2003;101:791–800. doi: 10.1182/blood-2002-07-1975. [DOI] [PubMed] [Google Scholar]
- 11.Fodde R, Losekoot M, Brock MH, Oldenburg M, Rashida N, Schreuder A, et al. Prevalence and molecular heterogeneity of α+ thalassaemia in two tribalpopulations from Andhra Pradesh, India. Hum Genet. 1988;80:157–160. doi: 10.1007/BF00702860. [DOI] [PubMed] [Google Scholar]
- 12.Hegde UM, White JM, Hart GH, Marsh GW. Diagnosis of alpha-thalassemia trait from Coulter counter ‘S’ indices. Clin Chem Lab Med. 2007;45(5):605–610. doi: 10.1515/CCLM.2007.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaji RV, Eunice SE, Baidya S, Srivastava A, Chandy M. Determination of the breakpoint and molecular diagnosis of a common α-thalassaemia-1 deletion in the Indian population. Br J Haematol. 2003;123:942–947. doi: 10.1046/j.1365-141.2003.04704.x. [DOI] [PubMed] [Google Scholar]
- 14.Puehringer H, Najmabadi H, Law HY, Krugluger W, Viprakasit V, Pissard S, Baysal E, et al. Validation of a reverse-hybridization strip Assay for the simultaneous analysis of common alpha-thalassemia point mutations and deletions. Clin Chem Lab Med. 2007;45(5):605–610. doi: 10.1515/CCLM.2007.125. [DOI] [PubMed] [Google Scholar]