Abstract
Cystic fibrosis is a common autosomal recessive disorder usually found in population of white Caucasian descent. Now it is well documented the presence of CF disease in India with the advancement of laboratory testing. As once it was thought non existence of this disease in our population. Most of the phenotype of CF disease was in accordance of western population. Genetic analysis of CFTR gene in Indian CF patients revealed that most common mutation was delta F508 mutation. However, it was less than Caucasian population. CFTR mutations are also a causative factor in the pathogenesis of male infertility due to obstructive azoospermia. There are two most common mutation viz. IVS8-T5 and delta F508 which are responsible for congenital absence of vas deferens in male infertility patients. Elevated levels of sweat chloride at two occasions along with the presence of two mutations in CFTR gene was gold standard method for diagnosis of CF disease. It is noteworthy here that due to magnitude of Indian population, the total CF disease load would be more than many European countries. Clinical data demonstrate the prevalence of both classical and genetic form of CF in India.
Keywords: Cystic fibrosis, CFTR, Delta F508, Congenital absence of vas deferens (CAVD)
Introduction
In the last two decades, biochemical genetic disorders of metabolism are of great importance due to improved molecular genetic analysis and biochemical testing. Other reason for emphasis on genetic disorders is due to more admission of children at Pediatric Ward. Presently, children deaths due to infectious diseases are reduced to great extent at Indian scenario. We established the relative frequencies of various genetic disorders at PGIMER, Chandigarh. Cystic fibrosis was found with 7.56 frequency [1]. Notwithstanding, CF is a common autosomal recessive disorder usually found in population of White Caucasian descent, such as those of Europe, North America and Australia. CF like other genetic disorders has geographical variation in the epidemiology, clinical presentation and course. Before seventies, CF was considered as non existent in India. First case of CF disease was reported in 1968 from PGIMER, Chandigarh [2]. Later, suggested incidence of cystic fibrosis (CF) in Asian migrants (Indians and Pakistanis) in the United Kingdom was reported about 1 in 10,800 to 12,000 [3, 4]. Nevertheless, there have been many publications describing diverse and unique manifestations of the disease in Indian population. The present review is intended to provide a point of reference at national organization and to help them in planning, diagnostic and genetic analysis for affected patients and their families. Since a better awareness of CF and the increasing availability diagnostic tests—the sweat chloride and/or DNA tests frequently leads to the identification of a higher number of affected individuals.
History
Cystic fibrosis was first recognized by Dorothy Andersen in 1938 [5] as cystic fibrosis of the pancreas and its relation to celiac disease. Farber in 1945 [6] recognized the implication of sticky mucus in manifestation of symptoms, and termed as mucoviscidosis. Acute salt loss caused by excessive sweating in babies with CF during heat waves was demonstrated by Di Sant’Agnese in 1953 [7]. Subsequently, Pilocarpine ionotophoresis method was developed for sweat collection and Chloride measurement. Altered electrical properties with abnormalities of both sodium and chloride transport was described by Knowles et al. [8]. CF gene was mapped to a small segment of long arm of chromosome 7q using restriction fragment length polymorphism. Linkage disequilibrium was observed between the markers XV2C/KM19 [9] and cystic fibrosis [10]. Later CF gene was cloned and C-DNA sequence was documented [11].
Prevalence and Clinical Manifestations of CF Disease
The estimated gene frequency of cystic fibrosis in the Caucasians is approximately 1 in 2,500. It also affects other ethnic groups such as black population with an incidence of 1 in 17,000 and the native American population with an approximate incidence of 1 in 80,000. The estimated incidence of CF in Asians (Indians and Pakistanis) migrated to UK is about 1:10,000 to 1:12,000. However a Similar data for Indian population is not available. In an extensive literature search, we could find a few case reports from India (Table 1). Cystic fibrosis was thought to be extremely rare in India. However, Published reports indicate that CF is probably far more common in people of Indian origin than previously thought but is under diagnosed or missed in majority of cases.
Table 1.
Cystic fibrosis in India
| Year | Author | Place | Number of patients |
|---|---|---|---|
| 1968 | Bhakoo | Chandigarh | 1 |
| 1969 | Mehta | Chandigarh | 13 |
| 1970 | Reddy | Andra | 2 |
| 1990 | Prasad | New Delhi | 2 |
| 1990 | Devanayagam | Madras | 68 |
| 1992 | Sarkar | Calcutta | 1 |
| 1996 | Kabra | New Delhi | 15 |
| 1998 | Singh | Chandigarh | 12 |
| 1999 | Kabra | New Delhi | 62 |
| 2000 | Kabra | New Delhi | 24 |
| 2009 | Prasad | Chandigarh | 98 |
At PGIMER Chandigarh 100 cases of classical CF were diagnosed by us according to clinical criteria sweat testing and molecular diagnosis over the last 7 years out of which 50 were adult CF infertile males [12]. The incidence in migrant Indian population in USA has been estimated to be 1 in 40,000 [4] and in the UK between 1 in 10,000 to 12,000 [13]. The median age of diagnosis among Indian American is 12 months compared with 6 months among Caucasians American children. Kabra et al. suggested that genetic and clinical profile on Indian children with CF may be different [14]. Kapoor et al. reported a carrier rate as 0.4% on 955 cord samples and estimated its incidence to be 1 in 40,000 [15].
Classical CF characterized by progressive lung disease, pancreatic dysfunction, elevated sweat chloride electrolyte, meconiumileus and male infertility [16]. However a wide variability in clinical expression is found among patients. Upto 20% of affected infant are born intestinal obstruction. Other patients are diagnosed with various mode of presentation from birth to adulthood and with considerable variability in the severity and rate of disease progression. Notwithstanding progressive lung disease is the most common cause mortality in CF. There is a great variability in the age of onset and severity of lung disease in different age groups.
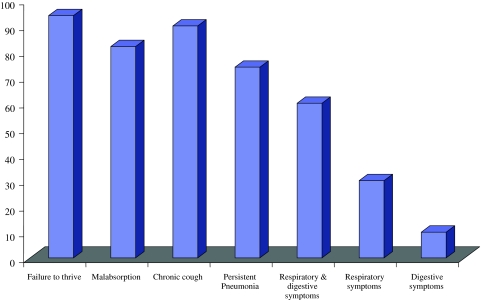
Our data on Clinical profile in Indian patients of classical CF shows that 94% had history of failure to thrive, 82% malabsorption, 90% chronic cough and 74% had recurrent or persistent pneumonia at the time of presentation, both respiratory and digestive symptoms were 60% more common as compared to either respiratory 30% or digestive symptoms 10% (Fig. 1). Congenital bilateral absence of vas deferens (CBAVD) occurs in 1–2% of infertile but otherwise healthy men [17]. CBAVD is present in more than 95% of cystic fibrosis (CF) males. Different studies have shown a high frequency of cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in CBAVD patients [18–21]. In intron 8 of the CFTR gene, the T5 allele, in contrast with the other two alleles, T7 and T9, leads to a high proportion of mRNA transcripts lacking exon 9. Consequently, the T5 variant produces abnormally low levels of CFTR protein. The T5 variant is the most frequent mutation associated with the CBAVD phenotype [22]. Low frequencies of CFTR mutations have been detected in patients with unilateral absence of vas deferens (CUAVD) [23]. Between 11 and 26% of patients with congenital absence of vas deferens (CAVD) have renal agenesis in association with it and initial negative results in the analysis of CFTR mutations in these patients suggested that urogenital anomalies have a different etiology to isolated CAVD [24]. Little is known about the spectrum and frequency of CFTR gene mutations in India. Our recent study as well as few other investigations have provided evidence for an extensive allelic heterogeneity in Indian patients with classic CF [25–27]. Congenital absence of vas deferens, as a distinct clinical entity with regard to CFTR mutations, has never been investigated in our population. We recently investigated the family history of infertility among CF subjects and characterized mutations in infertile members [12].
Fig. 1.
Clinical phenotype of CF patients [25]
Molecular Genetics of CFTR Gene
CF represents the first genetics disorder elucidated strictly by the process of positional cloning [11] succeeded in covering the CF region on long arm of chromosome 7 (7q31.2) by the chromosome walking and jumping. Screening of c DNA library constructed from cultured sweat gland cells proved that the CF gene was about 250 kb long [28]. Isolated overlapping c DNA clones from epithelial cell libraries with a genomic DNA segment containing apportion of putative CF gene. Transcript approximately 6,500 nucleotide in size were detectable. Riordan et al. [28] identified 24 exons in CF gene. The predicted protein has 1,480 amino acid with molecular mass of 1,68,138 Da. To avoid confusion with previously named CF antigen [28] referred to the protein as cystic fibrosis transmembrane conductance regulator gene (CFTR).
Structure of CFTR
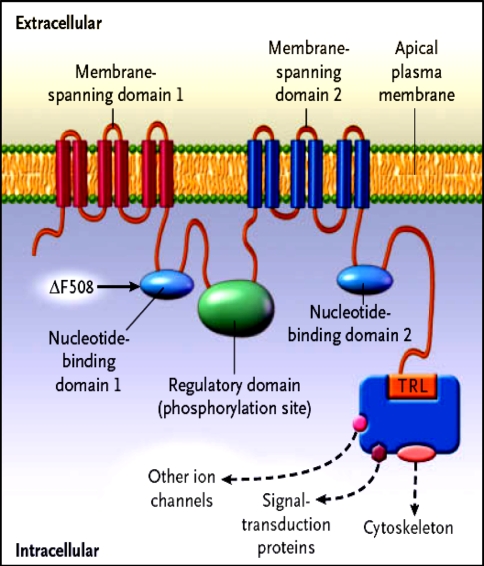
The CFTR is a unique member of the ABC transporter family that forms a novel Chloride channel. After identification of the primary amino acid sequence of CFTR, Riordan et al. proposed a structure (Fig. 1) composed of two motifs, each containing a membrane spanning domain (MSD) that is composed of six transmembrane segments and a nucleotide binding domain (NBD) that contains sequence predicted to interact with ATP [28, 29]. In CFTR, the two MSD–NBD motifs are linked by a unique domain called the R (regulatory domain) that contains multiple consequence phosphorylation sites and many charged amino acids. The carboxyl terminal (consisting of threonine, arginine, and leucine (TRL) of CFTR is anchored through a PDZ-type–binding interaction with the cytoskeleton and is kept in close approximation (Fig. 2) to a number of important proteins [30].
Fig. 2.
Hypothesized structure of CFTR showing proposed domain structure of CFTR protein. MSD membrane spanning domain, NBD nucleotide binding domain, R regulatory domain, PKA cAMP dependent protein kinase
Spectrum of CFTR Mutation
CF causing mutations have existed for more than 50,000 years [31]. To date more than 1,300 different alleles have been reported as proven or putative disease causing mutations to the Cystic Fibrosis Genetic Analysis Consortium (CFGAC). http://www.genet.sickkids.on.ca/cftr/.Worldwide there are 24 relatively common mutations identified in more than 50 chromosomes. The most common mutant allele is the delta F508 mutation which is a three-nucleotide deletion of a phenylalanine residue [32]. It is responsible for approximately two thirds (66%) of all CF chromosomes; however there is a great mutational heterogeneity in the remaining one-third of all alleles depending on populations and geographical locations [33].
Frequency of Delta F508 Mutation in Indian CF Population
The most common mutation in CF is the deletion of a phenylalanine residue at position 508 (delF508) in NBD1, accounting for about 66% of all CF alleles worldwide (CFGAC, http://www.genet.sickkids.on.ca/cftr/). However, it is not the predominant mutation in Asian CF patients [34]. While the delta F508 mutation is very common in number of different populations, its relative frequency varies for example between the north to south of Europe (delta F508 accounts for 90% of CF chromosomes in Denmark, and 30% in Turkey (Cystic Fibrosis Genetic Analysis Consortium, 1994). Schwarz et al. reported six affected Pakistani children, of whom three were homozygous for the delta F508 mutation [35].
By Kabra et al. [14, 36, 37], CF patients in India have been little studied. In the present investigation, we document that delta F508 mutation represents only 24% of the analyzed CF alleles. Comparison of delta F508 allele frequency with that reported from the Western countries shows that the Indian group has low percentage of the delta F508 frequency. A review of all genotyped South Asian patients, shows that delta F508 was identified in 19–44% of CF alleles [4, 14, 35, 36, 38–41], considerably lower than the reported frequency of 66% in the worldwide CF population. The frequency of delta F508 mutation in Indian children is reported to be between 19 and 44% [14, 36, 37]. In a report of 120 Indian children with CF [36], the delta F508 mutation was identified in 45 chromosomes (19%) out of the 240 tested, 19 patients (16%) were homozygous and 7 (6%) were heterozygous for the mutation. The frequency of delta F508 mutation was higher in patients whose parental origins were from Pakistan. Out of 23 patients whose origin was traced to Pakistan, 13 (56%) were positive for the delta F508 mutation. The figure for patients originating from other parts of India was 13 (13%) out of 97% patients. The spectrum of mutations other than delta F508 in Indian patients is highly variable and some rare and new mutations have been observed [25, 36].
In our study it is documented delta F508 as the most common CFTR gene mutation in Indian population [12, 25]. This observation supports the hypothesis that immigration patterns between US, Europe and the Indian subcontinent are contributing to the evolution of CF in the Indian subcontinent.
Spectrum of Non-Delta F508 CFTR Mutations in Indian CF Population
The striking characteristic of CF alleles in Indian population was the lack of common CF mutations in comparison with other Mediterranean and European population [31]. Apart from delta F508 mutation, twenty one other mutations were identified of which 13 were already known and 8 new mutations were characterized (Table 2). 25 mutation panel were detected in our population at a combined frequency of (10%). The other seven known but rare mutations (1161delC, Y517C, V520F, S549N, Y1381H, L218X and 1525-1G-A) were identified at a combined frequency of (17%), and eight new mutations (3986delC, 1792InsA, L69H, S158N, Q493L, I530L, E1329Q and 876-8del4) identified in our CF population represented (15%) of the total CF alleles analyzed. Substitution mutation, S549N and splice site mutation, 1525-1G-A were the two most common mutations identified with a frequency of (4%) followed by another splice site mutation 3849+10kbC-T (3%). However, we failed to identify CFTR gene mutations in 34% of the CF alleles in our study.
Table 2.
CFTR mutation identified in Indian population with classical CF [25]
| Genotype | No. of subjects |
|---|---|
| Delta F508/Delta F508 | 5 |
| Delta F508/3849+10kb C-T | 1 |
| Delta F508/S549 | 2 |
| Delta F508/Y138H | 1 |
| Delta F508/15251G-A | 1 |
| V520F/R117H | 2 |
| 1530L/1530L | 1 |
| 876-6del4/876-6del4 | 1 |
| 1792insA/1792insA | 1 |
| 3986-3987delC/3986-3987delC | 1 |
| Delta F508/U | 10 |
| 1161delC/U | 2 |
| L69H/U | 1 |
| R117H/U | 1 |
| Q493L/U | 1 |
| Y517C/U | 1 |
| S549N/U | 3 |
| G551D/U | 1 |
| E1329Q/U | 1 |
| N1303K/U | 1 |
| Y1381H/U | 1 |
| L218X/U | 1 |
| R553X/U | 1 |
| 1525-1G-A/U | 3 |
| 3120+1G-A/U | 2 |
| 3849+10kbC-T/U | 2 |
| U/U | 1 |
U unidentified
In contrast, Kabra et al. characterized ten mutations on 46% of the chromosomes by screening 16 of 27 CFTR exons. Less number exons screened is probably the reason for non-conformity [36]. Shastri and Kabra reported 1161delC, 3849+10kbC-T and S549 N as other most common mutations in Indian population [26]. It is in accordance with our study. G542X, second most common mutation (5%) in Hispanic Caucasians [42], was not found in our population. W1282X and 621+1G-T, that occur with a frequency greater than 1% in Non-Hispanic Caucasians [42] were also not detected.
It reinforces our hypothesis that spectrum of mutations in India is different. Thus, it is emphasized that mutations specific for Indian population should be included in the panel for the genetic diagnosis of CF.
CFTR Dysfunction and Male Infertility Due to Obstructive Azoospermia
CF and obstructive azoospermia causing male infertility due to bilateral/unilateral absence of vas deferens/absence of seminal vesicles are such different phenotypes that they appear to be distinct clinical entities. CF is a life threatening disorder that is usually diagnosed by pediatricians during the first few years of life because of gastrointestinal and pulmonary manifestations and failure to thrive. Obstructive azoospermia is diagnosed by urologists in adult males presenting with infertility; some of these patients have additional renal malformations, which is not a symptom of CF. However, despite of these differences, there exists a genetic commonality for both diseases in the majority of patients. Infertility in males, which is generally accepted as an almost inevitable component of cystic fibrosis (CF), results from congenital absence of vas deferens in about 95% of male patients [43, 44]. However, CBAVD may also occur in the absence of cystic fibrosis [24]. The question of whether obstructive azoospermia and cystic fibrosis are the “extreme” of one disease because such patients can be offered opportunity to have children. From this point of view, we searched for mutations in CFTR gene of obstructive azoospermia cases and their female partners.
In total, among 50 cases of genital CF (obstructive azoospermia) were studied by Prasad and coworkers, number of CBAVD and CUAD were 40 and 10 subjects respectively [25]. CFTR gene mutations were identified in 48 subjects. The mechanism by which the vas deferens is affected in cystic fibrosis is that the embryonic vas deferens needs an environment with electrolytes and fluid balance for potency. This balance is usually regulated by the CFTR present on vassal epithelial cells. However, in cystic fibrosis the function of this protein is defective and this balance is not attained, resulting in obliteration of vas deferens [45]. The seminal vesicles, body of epididymis are also affected but the testicular efferent ducts tend to be spared and some may be dilated [46].
A total of 12 mutations were identified on 48 of 100 alleles, as listed in Table 3. Two mutations were found to be very common: first, the IVS8-T5 allele was observed on 25 chromosomes; thus confirming the association of this splice site variant with CAVD in Indian patients, second the F508del mutation was detected in 11 chromosomes. In the CAVD patients with normal renal development, the initial screening identified one extra, R117H mutation in three chromosomes. SSCP analysis performed in patients with only one or no mutation revealed nine further mutations on one allele each including seven new sequence alterations: L69H, F87I, G126S, F157C, E543A, Y852F and D1270E (Table 3). Other identified mutations R117H, 3120+1>G-A and P1021S have been described previously in studies of patients with CAVD. None of the other screened mutations were identified in our population.
Table 3.
CFTR mutations identified and characterized in the Indian patients with CAVD [12]
| Mutation | Nucleotide change | No. of alleles |
|---|---|---|
| T5 | Reduction of oligo T tract to 5T at 1342-6 | 25 |
| F508del | Deletion of 3 bp(CTT or TTT) between 1652 and 1655 | 11 |
| L69H | T to A at 338 | 1 |
| F87I | T to A at 391 | 1 |
| R117H | G to A at 482 | 3 |
| G126S | G to A at 508 | 1 |
| F157C | T to G at 602 | 1 |
| E543A | A to C at 1760 | 1 |
| Y852F | A toT at 2687 | 1 |
| 3120+1G-A | G to A 3120+1 | 1 |
| P1021S | CtoT at 3193 | 1 |
| D1270E | T to A at 3942 | 1 |
We documented NBD1 and NBD2 as the hotspot identified in the CFTR protein in Indian CF population, whereas the regions known to alter chloride permeability (transmembrane regions) and delta F508 mutation in NBD1 are the hot spot for mutation identification in our genital form of CF cases (obstructive azoospermia).
Conclusion
CF was thought to be extremely rare in India. However a review of all the reports from Indian subcontinent suggests that CF is probably far more common in people of India/Indian origin than previously thought but is under diagnosed or missed in the majority of cases. Our demographic and mutation data together indicate that 6% of patients were born to consanguineous couples. Therefore, carrier frequency is expected to be high among Indian. The carrier frequency of delta f508 in the Indian population was calculated to be 1 in 238 [14].
High genetic heterogeneity in our population accompanies a much higher carrier frequency than expected. The precise incidence of CF among Indians is unknown. As a result of the widespread belief that CF does not occur in Indians, the disease is rarely suspected and even if it is suspected the diagnosis is not confirmed due to the poor availability of facilities for diagnosis. The proper availability of mutation analysis, searching for mutations appropriate for the population or whole gene sequencing may help in better characterization of CFTR gene mutation in Indian population.
References
- 1.Prasad R, Marwaha RK, Kaur G, Walia BNS. Spectrum of biochemical genetic diseases in North India. Med Sci Res. 1998;26:455–6.
- 2.Bhakoo ON, Kumar R, Walia BNS. Mucoviscidosis of the lung. Indian Pediatr. 1968;35:183–185. doi: 10.1007/BF02808629. [DOI] [PubMed] [Google Scholar]
- 3.Goodchild MC, Hansley J, Rushton DI, Gaze H. Cystic fibrosis in three Pakistani children. Arch Dis Child. 1974;49:739–741. doi: 10.1136/adc.49.9.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers CA, Potter EM, Wessel HU, Lloyd-Still JD. Cystic fibrosis in Asian Indians. Arch Pediatr Adolesc Med. 1996;150:554–555. doi: 10.1001/archpedi.1996.02170300108024. [DOI] [PubMed] [Google Scholar]
- 5.Andersen DH. Cystic fibrosis of the pancreas and its relation to celiac disease: a clinical and pathological study. Am J Dis Child. 1938;56:344–399. [Google Scholar]
- 6.Farber S. Some organic digestive disturbances in early life. J Mich Med Soc. 1945;44:587–594. [Google Scholar]
- 7.Di Sant’Agnese PA, Darling RC, Perera GA, Shea E. Abnormal electrolytic composition of sweat in cystic fibrosis of the pancreas. Clinical significance and relationship of the disease. Pediatrics. 1953;12:549–563. [PubMed] [Google Scholar]
- 8.Knowles M, Gatzy J, Boucher R. Increased bioelectric potential difference across respiratory epithelia in cystic fibrosis. N Engl J Med. 1981;305:1489–1495. doi: 10.1056/NEJM198112173052502. [DOI] [PubMed] [Google Scholar]
- 9.Tsui LC, Buchwald M, Barker D, Braman JC, Knowlton R, Schumm JW, et al. Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science. 1985;230:1054–1057. doi: 10.1126/science.2997931. [DOI] [PubMed] [Google Scholar]
- 10.Estivill X, Scambler PJ, Wainwright BJ, Hawley K, Frederick P, Schwartz M, et al. Patterns of polymorphism and linkage disequilibrium for cystic fibrosis. Genomics. 1987;1:257–263. doi: 10.1016/0888-7543(87)90052-8. [DOI] [PubMed] [Google Scholar]
- 11.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 12.Sharma N, Acharya N, Singh SK, Singh M, Sharma U, Prasad R. Hetrogenous spectrum of CFTR gene mutation in Indian patients with congenital absence of vas deferens. Hum Reprod. 2009;1:1–8. doi: 10.1093/humrep/den500. [DOI] [PubMed] [Google Scholar]
- 13.Spencer DA, Venkataraman M, Higgins S, Stevenson K, Weller PH. Cystic fibrosis in children from ethnic minorities in the West Midlands. Respir Med. 1994;88:671–675. doi: 10.1016/S0954-6111(05)80065-4. [DOI] [PubMed] [Google Scholar]
- 14.Kabra M, Ghosh M, Kabra SK, Khanna A, Verma IC. Delta F508 molecular mutation in Indian children with cystic fibrosis. Indian J Med Res. 1996;104:355–358. [PubMed] [Google Scholar]
- 15.Kapoor V, Shastri SS, Kabra M, Kabra SS, Ramachandran V, Arora S, et al. Carrier frequency of F508del mutation of cystic fibrosis in Indian population. J Cyst Fibros. 2005;23:1631–1652. doi: 10.1016/j.jcf.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Dequeker E, Cuppens H, Dodge J, Estivill X, Goossens M, Pignatti PR, et al. Recommendations for quality improvement in genetic testing for cystic fibrosis. European concerted action on cystic fibrosis. Eur J Hum Genet. 2000;8:S2–S24. doi: 10.1038/sj.ejhg.5200487. [DOI] [PubMed] [Google Scholar]
- 17.Holsclaw DS, Perlmutter AD, Jockin H, Shwachman H. Genital abnormalities in male patients with cystic fibrosis. J Urol. 1971;106:568–574. doi: 10.1016/s0022-5347(17)61343-0. [DOI] [PubMed] [Google Scholar]
- 18.Costes B, Girodon E, Ghanem N, Flori E, Jardin A, Soufir JC, et al. Frequent occurrence of the CFTR intron 8 (TG)n5T allele in men with congenital bilateral absence of vas deferens. Eur J Hum Genet. 1995;3:285–293. doi: 10.1159/000472312. [DOI] [PubMed] [Google Scholar]
- 19.Dork T, Dworniczak B, Aulehla-Scholz C, Wieczorek D, Bohm I, Mayerova A, et al. Distinct spectrum of CFTR mutations in congenital absence of vas deferens. Hum Genet. 1997;100:365–377. doi: 10.1007/s004390050518. [DOI] [PubMed] [Google Scholar]
- 20.Casals T, Nunes V, Palacios A, Gimenez J, Gaona A, Ibanez N, et al. Cystic fibrosis in Spain: high frequency of mutation G542X in the Mediterranean coastal area. Hum Genet. 1993;91:66–70. doi: 10.1007/BF00230225. [DOI] [PubMed] [Google Scholar]
- 21.Grangeia A, Sá R, Carvalho F, Martin J, Girodon E, Silva J, et al. Molecular characterization of the cystic fibrosis transmembrane conductance regulator gene in congenital absence of the vas deferens. Genet Med. 2007;9:163–172. doi: 10.1097/GIM.0b013e3180318aaf. [DOI] [PubMed] [Google Scholar]
- 22.Chillon M, Palacio A, Nunes V, Casals T, Gimenez J, Estivill X. Identification of a frameshift mutation (1609delCA) in exon 10 of the CFTR gene in seven Spanish cystic fibrosis patients. Hum Mutat. 1992;1:75–76. doi: 10.1002/humu.1380010113. [DOI] [PubMed] [Google Scholar]
- 23.Casals T, Bassas L, Ruiz-Romero J, Chillón M, Giménez J, Ramos MD, et al. Extensive analysis of 40 infertile patients with congenital absence of the vas deferens: in 50% of cases only one CFTR allele could be detected. Hum Genet. 1995;95:205–211. doi: 10.1007/BF00209403. [DOI] [PubMed] [Google Scholar]
- 24.Augarten A, Yahav Y, Keremn BS, Halle D, Laufer J, Szeinberg A, et al. Congenital bilateral absence of vas deferens in the absence of cystic fibrosis. Lancet. 1994;344:1473–1474. doi: 10.1016/S0140-6736(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 25.Sharma N, Singh M, Kaur G, Thapa BR, Prasad R. Identification and characterization of CFTR gene mutation in Indian CF patients. Ann Hum Genet. 2009;76:26–33. doi: 10.1111/j.1469-1809.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- 26.Shastri SS, Kabra SS. Mutation spectrum in India. In: Respicon 2006. Pre conference workshop on cysic fibrosis and fourth CF conference of India. Organised by Cystic Fibrosis Working Group, Respiratory Chapter, Indian Academy of Pediatrics, Nov 23, 2006.
- 27.Sharma N, Singh M, Acharya N, Singh SK, Thapa BR, Kaur G, Prasad R. Implication of cystic fibrosis transmembrane conductance regulator gene in infertile family members of Indian CF patients. Biochem Genet. 2008;46:847–56. [DOI] [PubMed]
- 28.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 29.Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, et al. Structural model of ATP-binding protein associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 30.Short DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, et al. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 31.Morral N, Bertanpetit J, Estivill X, Nunes V, Casals T, Gimenez J, et al. The origin of the major cystic fibrosis mutation (delta F508) in European populations. Nat Genet. 1994;7:169–175. doi: 10.1038/ng0694-169. [DOI] [PubMed] [Google Scholar]
- 32.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 33.Estivill X, Bancells C, Ramos C, Biomed CF. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. Hum Mutat. 1997;10:135–154. doi: 10.1002/(SICI)1098-1004(1997)10:2<135::AID-HUMU6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Yamashiro Y, Shimizu T, Oguchi S, Shioya T, Nagata S, Ohtsura Y, et al. The estimated incidence of cystic fibrosis in Japan. J Pediatr Gastroenterol Nutr. 1997;24:544–554. doi: 10.1097/00005176-199705000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz MJ, Super M, Wallis C, Beighton P, Newton C, Heptinstall LE, et al. Delta 508 testing of the DNA bank of the royal Manchester Children’s Hospital. Hum Genet. 1990;85:428–430. doi: 10.1007/BF02428298. [DOI] [PubMed] [Google Scholar]
- 36.Kabra M, Kabra SK, Ghosh M, Khanna A, Arora S, Menon PS, et al. Is the spectrum of mutations in Indian patients with cystic fibrosis different? Am J Med Genet. 2000;93:161–163. doi: 10.1002/1096-8628(20000717)93:2<161::AID-AJMG15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 37.Kabra SK, Kabra M, Lodha R, Shastri S, Ghosh M, Pandey RM, et al. Clinical profile and frequency of delta F508 mutation in Indian children with cystic fibrosis. Indian Pediatr. 2003;40:612–619. [PubMed] [Google Scholar]
- 38.Bowlers IM, Estelin EJ, Littlewood JM. Cystic fibrosis in Asians. Arch Dis Child. 1993;68:120–122. doi: 10.1136/adc.68.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtis A, Richardson RJ, Boohene J, Jackson A, Nelson R, Bhattacharya SS. Absence of cystic fibrosis mutations in a large Asian population sample and occurrence of a homozygous S549N mutation in an inbred Pakistani family. J Med Genet. 1993;30:164–166. doi: 10.1136/jmg.30.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhutta ZA, Moattar T, Shah U. Genetic analysis of cystic fibrosis in Pakistan: a preliminary report. J Pakistan Med Assoc. 2000;50:217–219. [PubMed] [Google Scholar]
- 41.Mei-Zahav M, Durie P, Zielenski J, Solomon M, Tullis E, Tsui LC, et al. The prevalence and clinical characteristics of cystic fibrosis in South Asian Canadian immigrants. Arch Dis Child. 2005;90:675–679. doi: 10.1136/adc.2003.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan E, Swachman H, Permutter AD, Rule A, Khaw KT, Holsclaw DS. Reproductive failure in males with cystic fibrosis. N Engl J Med. 1968;279:65–69. doi: 10.1056/NEJM196807112790203. [DOI] [PubMed] [Google Scholar]
- 44.Jequier AM, Ansell ID, Bullimore NJ. Congenital absence of the vas deferens presenting with infertility. J Androl. 1985;6:15–19. [PubMed] [Google Scholar]
- 45.Oates RD, Amos JA. Congenital bilateral absence of vas deferens and cystic fibrosis: a genetic commonality. World J Urol. 1993;11:82–88. doi: 10.1007/BF00182034. [DOI] [PubMed] [Google Scholar]
- 46.Landing BH, Wells TR, Wang CI. Abnormality of the epididymis and vas deferens in cystic fibrosis. Arch Pathol. 1969;88:569–580. [PubMed] [Google Scholar]