Summary
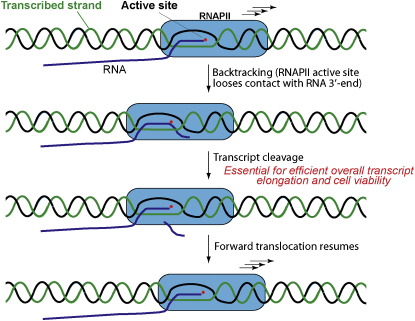
During transcript elongation in vitro, backtracking of RNA polymerase II (RNAPII) is a frequent occurrence that can lead to transcriptional arrest. The polymerase active site can cleave the transcript during such backtracking, allowing transcription to resume. Transcript cleavage is either stimulated by elongation factor TFIIS or occurs much more slowly in its absence. However, whether backtracking actually occurs in vivo, and whether transcript cleavage is important to escape it, has been unclear. Using a yeast TFIIS mutant that lacks transcript cleavage stimulatory activity and simultaneously inhibits unstimulated cleavage, we now provide evidence that escape from backtracking via transcript cleavage is essential for cell viability and efficient transcript elongation. Our results suggest that transcription problems leading to backtracking are frequent in vivo and that reactivation of backtracked RNAPII is crucial for transcription.
Keywords: DNA, RNA
Graphical Abstract
Highlights
► A TFIIS mutant that inhibits intrinsic RNAPII transcript cleavage has been isolated ► Transcript cleavage by RNAPII is essential for transcription and viability ► RNAPII backtracking must be a frequent event in vivo ► Irreversibly backtracked RNAPII is a target for ubiquitylation/degradation
Introduction
RNA polymerase II (RNAPII) is responsible for transcription of all protein-encoding genes and a large number of stable and unstable noncoding RNAs in eukaryotes. Interestingly, during transcript elongation RNAPII moves by Brownian motion rather than by the ATP-driven powerstrokes typical of DNA helicases (Bar-Nahum et al., 2005; Nudler, 2009). This means that although it moves rapidly forward on average, the polymerase can also perform retrograde motion (backtracking). Indeed, transcript elongation by RNAPII in vitro is known to be a highly discontinuous process, involving frequent pausing, backtracking, and transcriptional arrest, and a plethora of elongation factors are required to stimulate its progression (Reines et al., 1999; Arndt and Kane, 2003; Galburt et al., 2007; Svejstrup, 2007).
The accessory factor TFIIS (also called SII) reactivates backtracked elongation complexes by directing transcript cleavage—by RNAPII—which serves to realign the polymerase active site with the new RNA transcript end so that transcription can continue (Izban and Luse, 1992; Reines, 1992). As a result, TFIIS action can result in dramatic enhancement of overall transcription in reconstituted transcription reactions (Sekimizu et al., 1976; Reinberg and Roeder, 1987; Bengal et al., 1991; Jeon et al., 1994). TFIIS can be detected on the coding region of active genes in vivo (Pokholok et al., 2002; Ghavi-Helm et al., 2008), but often only when transcription is compromised, for example by nucleotide depletion (Pokholok et al., 2002). The apparently discontinuous nature of transcript elongation in vitro makes it surprising that the single gene encoding TFIIS (called DST1, or PPR2) is not essential in yeast (Hubert et al., 1983) and that the consequences of its absence for the efficiency of transcript elongation even through a long gene appear to be relatively minor (Mason and Struhl, 2005). This could imply that transcription is a much more smooth, continuous process in vivo than it is in vitro (i.e., that backtracking rarely occurs inside cells under normal conditions), so that transcript cleavage and escape from arrest are not important. Alternatively, the ability to escape backtracking by using the slower, intrinsic (uncatalyzed) transcript cleavage activity of RNAPII might be important and sufficient for transcription and cell viability in the absence of TFIIS. Here, we report on a study aimed at assessing the importance of transcript cleavage for transcript elongation, in vitro and in vivo. Our results indicate that transcript cleavage is essential for transcription and cell viability.
Results
A TFIIS Mutant that Inhibits Intrinsic Transcript Cleavage In Vitro
The highly conserved acidic loop in TFIIS interacts with the active site of RNAPII to direct transcript cleavage, probably by positioning a metal ion and water to promote hydrolytic cleavage (Jeon et al., 1994; Kettenberger et al., 2003; Wang et al., 2009). TFIIS-stimulated transcript cleavage depends on an Asp290-Glu291 dipeptide in this loop (Jeon et al., 1994), so in order to study the role of transcript cleavage in elongation, we changed both these residues to alanine (generating TFIISMut). In another mutant (TFIISAcDel), the same residues were deleted, leading to shortening of the loop and its predicted partial withdrawal from the RNAPII active site.
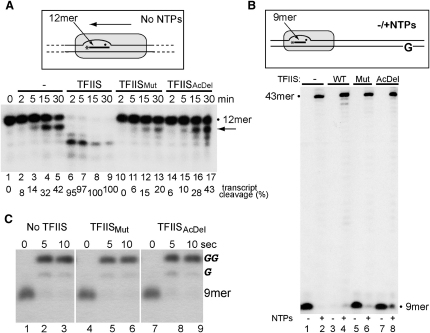
To study the effect of the mutations on TFIIS-stimulated transcript cleavage, reconstituted RNAPII elongation complexes containing a radioactively labeled 12-mer RNA were incubated for different times before analysis of the RNA by denaturing PAGE (Figure 1A). On this substrate (in the absence of NTPs), RNAPII spontaneously backtracks, allowing transcript cleavage to be detected. Even in the absence of TFIIS, slow but easily detectable cleavage activity was observed in this assay (Figure 1A, lanes 2–5). This cleavage was not due to contamination of the highly purified RNAPII preparation with TFIIS, as polymerase purified from dst1Δ cells had similar activity (see Figure S1A available online). Transcript cleavage was, however, strongly stimulated by wild-type TFIIS, with full-length RNA in the elongation complex disappearing quickly (Figure 1A, lanes 6–9). As expected, TFIISMut failed to stimulate transcript cleavage but, surprisingly, instead actually inhibited the intrinsic RNAPII transcript cleavage reaction (Figure 1A, lanes 10–13). TFIISAcDel also failed to stimulate transcript cleavage but in contrast to TFIISMut did not significantly affect intrinsic RNAPII transcript cleavage (Figure 1A, compare lanes 14–17 with 2–5). We speculate that TFIISAcDel has lost the ability to direct the correct placement of a metal ion and water to promote hydrolytic cleavage, while TFIISMut has not only lost this ability but at the same time inhibits the intrinsic cleavage reaction (which is thought to occur via a different mechanism [Wang et al., 2009]).
Figure 1.
TFIISMut Inhibits Intrinsic Transcript Cleavage but Does Not Affect Nucleotide Addition
(A) (Box) Schematic of experiment. Radioactive label is denoted by asterisk and the RNAPII active site as a filled sphere. (Lower) Characterization by denaturing PAGE of transcript cleavage in RNAPII elongation complexes assembled with a labeled 12-mer RNA oligonucleotide and then incubated with different TFIIS forms for the times indicated. Migration of full-length (12-mer) RNA and the major intrinsic cleavage product (arrow) is indicated. Quantification of transcript cleavage is shown below the autoradiograph.
(B) (Box) Schematic. Nucleotides, except CTP (NTPs), allow transcription of a reconstituted RNAPII elongation complex to the first guanine (G) in the template strand. (Lower) Characterization by denaturing PAGE of transcription for 5 min from the end-labeled 9-mer RNA oligonucleotide. Wild-type TFIIS induces transcript cleavage and thereby degradation of the RNA primer in the absence of nucleotides (lane 3).
(C) Nucleotide addition by RNAPII elongation complexes preincubated on ice without TFIIS, or in the presence of TFIISMut or TFIISAcDel, as indicated. GTP was added to allow nucleotide addition for 5 and 10 s. Migration of bands representing starting material (9-mer) and the addition of one or two G's are indicated to the right. See also Figure S1.
Because the insertion of TFIIS's hairpin loop into the active site of RNAPII results in the rearrangements of protein-RNA interactions (Wang et al., 2009), TFIISMut might also conceivably affect intrinsic transcript cleavage indirectly, by binding and fixating RNAPII in a configuration which is not conducive to transcript cleavage. However, in agreement with previous reports showing that binding to RNAPII is independent of TFIIS's C-terminal acidic loop (part of TFIIS domain III) but requires domain II (Awrey et al., 1998; Kettenberger et al., 2003), we found no evidence for a difference in the overall binding affinity of TFIISMut for RNAPII elongation complexes (Figure S1B). The TFIIS mutants also failed to affect transcription to the end of a short, easily transcribed DNA substrate on which no significant transcription pause/arrest sites are detected under normal conditions (precluding transcriptional arrest and the need for transcript cleavage) (Figure 1B), indicating that neither mutant significantly affects the process of nucleotide addition either.
This conclusion was further supported by an experiment in which it was additionally investigated whether TFIISMut might be pushing RNAPII back or fixating it in the backtracked state. In this experiment (Figure 1C), RNAPII elongation complexes were preincubated on ice for 10–15 min in transcription buffer (TB) without TFIIS, or in the presence of TFIISMut or TFIISAcDel. Previous experiments had shown that substantial transcript cleavage takes place already within 5 min of incubation with wild-type TFIIS under these conditions (data not shown), showing that RNAPII moves backward and forward on this substrate and can be “caught” by TFIIS in the backtracked state. After preincubation, GTP was added (time = 0), allowing chain elongation at low temperature. All starting material was rapidly converted to extended product, with no significant difference observed in the presence of TFIISMut (Figure 1C, compare lanes 2–3 with 5 and 6, and 8 and 9), indicating that this TFIIS mutant does not drive backtracking, immobilize RNAPII in a backtracked position, or inhibit nucleotide addition.
TFIISMut Inhibits Transcription through Pause Sites
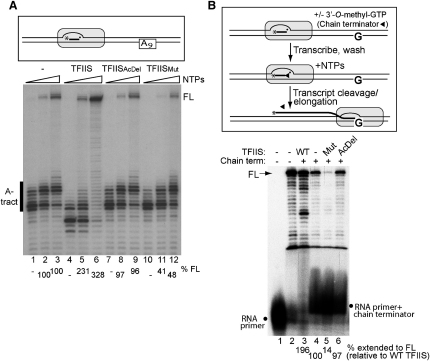
Hoping that TFIISMut would be useful for studying what happens when both types of transcript cleavage activity are inhibited, we proceeded to further characterize its effect on a transcription template containing an adenine tract, which acts as a potent pause site (Saeki and Svejstrup, 2009). Transcription of this substrate results in substantial polymerase stalling and backtracking and therefore a need for transcript cleavage for efficient readthrough. Only a relatively small fraction of RNAPII was capable of transcribing through the tract (Figure 2A, lanes 1–3), but the addition of wild-type TFIIS resulted in an increase in full-length product, with the majority of RNAPII reaching the end of the template at high nucleotide concentration (lanes 5 and 6). The addition of TFIISAcDel had no such effect (lanes 7–9), suggesting that stimulated transcript cleavage was responsible for the increase in A/T tract readthrough observed with wild-type TFIIS. In contrast, addition of TFIISMut had an inhibitory effect: compared to RNAPII alone (or in the presence of TFIISAcDel), fewer RNAPII molecules now reached the end of the template (Figure 2A, compare lanes 10–12 with 1–3 and 7–9). Since TFIISMut inhibits intrinsic transcript cleavage, this result is consistent with the idea that intrinsic transcript cleavage has a positive effect on transcription through the A/T tract by allowing reactivation of backtracked elongation complexes.
Figure 2.
TFIISMut Inhibits Transcript Elongation through Pause Sites
(A) (Box) Schematic of experiment. A9 designates 9-nucleotide A-tract (pause site). (Lower) Characterization, by denaturing PAGE, of transcription for 5 min as in Figure 1B, but with DNA substrate containing an A tract. FL, full-length product. Quantification of FL transcripts, relative to RNAPII alone (set to 100%), are shown below the autoradiograph where meaningful.
(B) (Box) Schematic of experiment. After extension of the RNA primer with chain terminator nucleotide, transcription for 10 min with normal nucleotides is allowed to occur. Such transcription absolutely requires transcript cleavage, removing the chain terminating nucleotide (shown as filled triangle). (Lower) Characterization by denaturing PAGE of transcription reconstituted as in Figure 1B, but adding 3′-O-methyl-GTP (and washing it away), prior to transcription with the normal NTPs (-CTP). Quantification of the full-length transcripts, relative to RNAPII alone (lane 4, set to 100%) is shown below the autoradiograph where meaningful. Note that TFIIS actually stimulates transcription more than 2-fold (196%), but this reaction runs out of nucleotides because of repeated cycles of backtracking/transcript cleavage/forward motion at FL, resulting in some polymerases ending up backtracked from FL. See also Figure S2.
This conclusion was further supported by the use of the nucleotide analog 3′-O-methyl-GTP, which acts as a transcript chain terminator. Elongation complexes having incorporated this nucleotide in the transcript cannot continue transcription unless the analog is removed by transcript cleavage (Figure 2B, schematic). On the DNA template whose transcription was otherwise unaffected by the different forms of TFIIS (see Figure 1B), wild-type TFIIS increased transcription from chain-terminated transcripts, but even without addition of TFIIS considerable transcript cleavage-dependent transcription occurred (Figure 2B, compare lanes 3 and 4). Significantly, while the addition of TFIISAcDel had little or no effect on such transcription, TFIISMut significantly inhibited it (compare lane 5 with lanes 4 and 6).
Together, these results further support the idea that TFIISMut reduces transcription by inhibiting transcript cleavage, rather than by inhibiting nucleotide addition, pushing RNAPII back, or fixating it in the backtracked state.
TFIISMut Inhibits Transcript Elongation through a Long Gene In Vitro
The data presented above indicate that TFIISMut inhibits the reactivation of RNAPII elongation complexes undergoing transcriptional arrest at individual artificial pause or blockage sites. Obviously, during transcript elongation through a normal, much longer gene, even a modest inhibitory effect on individual cleavage events (such as that observed in Figure 1A, lanes 10–13) could have a great effect overall. If inhibition of the individual cleavage event were 2-fold, for example, the fold overall inhibition of transcription would thus be 2N (where N is the number of cleavage events). We therefore now tested the effect of TFIISMut on promoter-specific transcription in crude cell-free yeast extracts (Schultz et al., 1991; Kong and Svejstrup, 2002). The DNA template in these experiments contained an RNAPII promoter driving transcription through two G-less cassettes separated by ∼1500 nucleotides (Figure 3, upper panel), and has previously been used to investigate the effect of different factors on transcript elongation (Lee and Greenleaf, 1997; Rondon et al., 2003). Treatment of the resulting transcripts with RNase T1 (which cleaves at G residues) results in the elimination of RNA surrounding these cassettes, so that the relative efficiency of transcription close to the promoter (proximal cassette) and far from it (distal cassette) can be evaluated. Even in the absence of TFIIS (extract from dst1Δ cells was used), transcription through both cassettes was observed (Figure 3, lanes 1, 4, and 7), but addition of wild-type TFIIS enhanced transcript elongation, so that approximately 40% of the polymerases that transcribed the proximal cassette also passed through the distal cassette (versus ∼22% in its absence) (lanes 2 and 3). In contrast, TFIISMut severely inhibited elongation, with much fewer polymerases (∼1%) also making it through to the distal cassette (lanes 5 and 6). TFIISAcDel had little effect (lanes 8 and 9). Note that TFIIS also plays positive roles in the transcription cycle prior to transcript elongation, some of which are independent of its transcript cleavage activity (Wery et al., 2004; Adelman et al., 2005; Guglielmi et al., 2007; Kim et al., 2007). This might explain the stimulation of transcription observed through the short proximal cassette observed with the different versions of TFIIS, including TFIISMut.
Figure 3.
TFIISMut Inhibits Promoter-Driven Transcript Elongation
(Box) Schematic of DNA substrate for promoter-driven transcription. Angled arrow indicates transcription start site; boxes indicate G-less cassettes of different sizes (84 and 376 nucleotides, respectively [Lee and Greenleaf, 1997; Rondon et al., 2003]). (Lower) Characterization by denaturing PAGE of promoter-specific transcription reconstituted with crude WCEs from a dst1Δ strain in the absence (lanes 1, 4, and 7) and presence of the TFIIS forms indicated. Migration of the RNA bands representing transcription of the proximal (prox) and distal (dis) cassettes is indicated on the right. Quantification of transcription through the distal cassette (in percent of that observed through the proximal cassette) is shown below the autoradiograph.
Expression of TFIISMut Is Lethal in Yeast
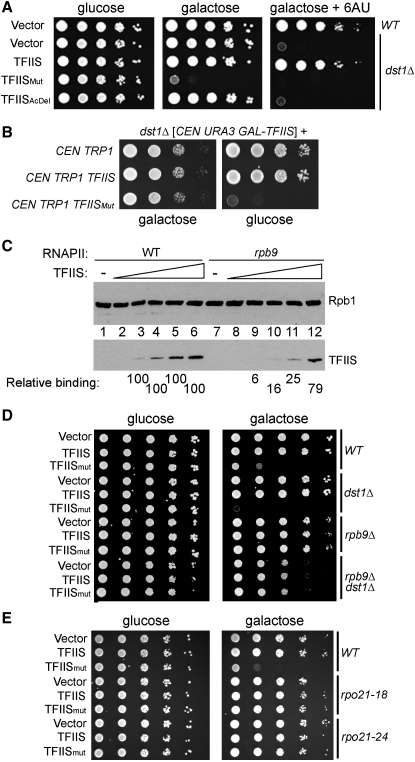
The data presented above suggest that TFIISMut significantly affects the ability of RNAPII to escape transcriptional arrest via inhibition of transcript cleavage. We therefore reasoned that TFIISMut would provide a unique tool for investigating the relevance of these processes for transcription in vivo, which has not previously been feasible. The DST1 gene is nonessential (Hubert et al., 1983), and though dst1Δ cells are 6-azauracil (6AU) sensitive (6AU depletes nucleotide pools and inhibits elongation [Exinger and Lacroute, 1992]), they have no significant growth defects under normal conditions (see below). Nevertheless, when a CEN (single-copy) plasmid encoding TFIISMut was transformed into a dst1Δ strain, we repeatedly failed to obtain a significant number of viable colonies, while numerous colonies were obtained with wild-type control plasmid and vector alone (Figure S3A; data not shown).
Expression of wild-type TFIIS, TFIISMut, and TFIISAcDel was therefore now put under control of the inducible GAL promoter. As expected, no effect on cell growth was observed with any of these constructs under repressing conditions (Figure 4A, “glucose”). Moreover, while expression of wild-type TFIIS rescued the 6AU sensitivity of dst1Δ cells, expression of TFIISAcDel did not, and its expression had no effect on growth (Figure 4A; “galactose + 6AU,” and “galactose,” respectively). In marked contrast, cells expressing TFIISMut failed to grow on galactose (Figure 4A), and TFIISMut even functioned as a dominant-negative, strongly inhibiting growth when expressed in DST1 (WT) cells (Figure 4D). The fact that TFIISAcDel (which abolishes TFIIS-mediated transcript cleavage stimulation but does not inhibit intrinsic RNAPII transcript cleavage) is not lethal to cells is important, as it correlates the inhibition of transcript cleavage with inhibition of cell growth.
Figure 4.
Inhibition of RNAPII Transcript Cleavage Is Lethal in Yeast
(A) Dilution series of yeast cells (of the genotype indicated on the right, and carrying the GAL-driven plasmid indicated on the left) plated on yeast minimal plates containing glucose, galactose, or galactose + 6-azauracil (6AU), as indicated.
(B) Colonies resulting from dst1Δ cells carrying the plasmids indicated on the left (vector control, and TFIIS versions expressed from the native DST1 promoter) as well as a plasmid expressing wild-type TFIIS under the control of the GAL promoter.
(C) Binding of TFIIS (0, 80, 160, 240, 320, or 480 ng) to immobilized RNAPII elongation complexes, reconstituted using either wild-type (WT) RNAPII, or RNAPII lacking the Rpb9 subunit (rpb9). Quantification of binding relative to wild-type RNAPII (set to 100% in every case, see lanes 3–6) is shown below where meaningful.
(D and E) Legend as for (A). See also Figure S3.
We observed that viable cells could be obtained if they simultaneously expressed TFIISMut (from the DST1 promoter) and overexpressed wild-type TFIIS (from the GAL promoter) (Figure 4B, “galactose”). However, these cells ceased growing when replated on glucose (on which overexpression of wild-type TFIIS ceased, so that only physiological levels of TFIISMut were maintained) (Figure 4B, “glucose”), further showing that TFIISMut causes growth cessation.
In the absence of the nonessential Rpb9 subunit, TFIIS-directed transcript cleavage and transcription through pause sites are significantly affected (Awrey et al., 1997; see also Figure S3B), and rpb9 cells have phenotypes consistent with a role for the subunit in transcript elongation (Hemming et al., 2000). It has been reported that TFIIS binds to RNAPII in an Rpb9-independent manner (Awrey et al., 1997). However, previous studies used “free” RNAPII, not RNAPII elongation complexes, for such binding experiments. Using reconstituted elongation complexes, we found that normal interaction of RNAPII with TFIIS requires the Rpb9 subunit (Figure 4C, compare lanes 3–6 with 9–12). Besides helping to explain the well-known effect of rpb9 mutation on transcript elongation and TFIIS activity, these data also allowed us to determine whether the detrimental effect of TFIISMut in vivo requires normal interaction with RNAPII elongation complexes. Strikingly, in rpb9Δ cells TFIISMut expression had virtually no deleterious effect (Figure 4D, “galactose,” lower half; see also Figure S3A [lower]). Moreover, point mutations in the largest RNAPII subunit, RPB1 (RPO21), have been isolated that greatly affect the ability of the polymerase to bind TFIIS (Wu et al., 1996). These mutants were also unaffected by TFIISMut expression (Figure 4E).
Expression of TFIISMut Inhibits Transcript Elongation In Vivo
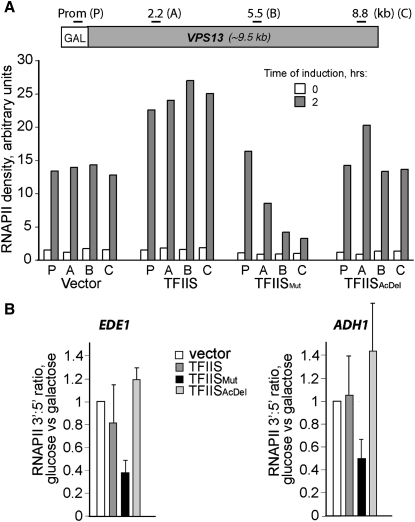
Together, the experiments described so far indicate that the activity inhibited by TFIISMut is intrinsic to the RNAPII elongation complex, and support the idea that RNAPII transcript cleavage and efficient escape from backtracking is required for cell growth. To more directly investigate the effect of inhibiting transcript cleavage on RNAPII transcription in vivo, we now used chromatin immunoprecipitation in dst1Δ cells to measure polymerase density across a long GAL-driven gene (Kristjuhan and Svejstrup, 2004) (Figure 5A). In cells lacking TFIIS (vector), the density of RNAPII on this model gene was similar at the promoter and across the coding region. Expression of wild-type TFIIS modestly increased RNAPII density in all regions tested, while TFIISAcDel had little or no effect. By contrast, in cells expressing TFIISMut, promoter recruitment was similar to that of control cells, but the density of polymerases became progressively lower through the coding region, reaching a level close to that of the nonactivated control at the end of the gene (Figure 5A).
Figure 5.
Inhibition of RNAPII Transcript Cleavage Has Severe Consequences for Transcript Elongation
(A) (Upper) Schematic of GAL-VPS13 fusion gene and location of primers used for ChIP. (Lower) RNAPII density at the promoter and through the coding region of GAL-VPS13 was measured by ChIP, before (white bars) and upon (gray bars) gene activation and expression of the indicated TFIIS variants. The result is representative of two biological replicate experiments.
(B) RNAPII density at the promoter (5′) and at the end of the coding region (3′) of the indicated genes was measured by ChIP, before and after expression of the indicated TFIIS variants. The 3′:5′ ratios of RNAPII density were calculated in the two conditions. For simplicity, the results are presented by setting the ratio in the vector control to one and then expressing the corresponding ratios in the others relative to that. Error bars represent standard deviation.
Although the effects were less dramatic, TFIISMut (but not TFIISAcDel) expression also negatively affected transcript elongation at other genes tested, as indicated by the lower RNAPII density at the 3′ end relative to the 5′ end in these genes (Figure 5B). Together, these data support the idea that TFIISMut selectively inhibits transcript elongation in vivo.
Expression of TFIISMut Elicits RNAPII Polyubiquitylation
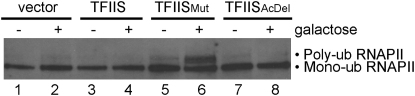
We previously reported that ubiquitylation/degradation of RNAPII functions as a last resort when arrested RNAPII complexes cannot otherwise be reactivated (Somesh et al., 2005). Using a matrix which specifically binds ubiquitylated proteins and western blotting with RNAPII-specific antibodies to measure the level of RNAPII ubiquitylation (Anindya et al., 2007), we found that upon expression of TFIISMut, but none of the other TFIIS forms, a dramatic increase in cellular RNAPII polyubiquitylation was observed (Figure 6, compare lane 6 with lanes 2, 4, and 8). This result may explain why RNAPII does not accumulate to a higher level in the beginning of the coding region in TFIISMut-expressing cells (see Figure 5): arrested polymerases are removed via ubiquitylation and proteasome-mediated degradation in these cells.
Figure 6.
Inhibition of Transcript Cleavage Induces RNAPII Polyubiquitylation
Western blot showing mono- and polyubiquitylation of RNAPII (Rpb1) before (−) and 1 hr after induction (+) of TFIIS variant expression. Ubiquitin protease was used to show that RNAPII isolated this way is ubiquitylated, altering its mobility (Anindya et al., 2007; and data not shown).
Discussion
Somewhat surprisingly, the gene encoding transcript cleavage stimulatory factor TFIIS is not essential for transcription or viability, suggesting that transcript cleavage is not important for transcription. However, RNAPII also has an intrinsic transcript cleavage activity, whose importance for transcription, if any, has been unclear. Here we provide evidence to support the idea that transcription obstacles resulting in RNAPII backtracking are frequent in vivo, and that the ability of RNAPII to cleave its transcript in response to such backtracking is essential for transcription and cell viability.
Using a variety of in vitro assays, we found that a TFIIS double point mutant (TFIIS Asp290Ala, Glu291Ala [TFIISMut]) not only fails to support TFIIS-stimulated transcript cleavage but also inhibits the slow intrinsic cleavage activity of RNAPII itself. This protein inhibits transcription through artificial pause sites and dramatically decreases the efficiency of transcript elongation through a long test gene in vitro. In contrast, a control protein (TFIISAcDel, in which Asp290 and Glu291 are deleted), which fails to support TFIIS-stimulated transcript cleavage, but does not inhibit the intrinsic cleavage activity of RNAPII, has virtually no effect in these transcription assays. Strikingly, when constructs expressing these proteins were studied in vivo in yeast, we found that TFIISMut, but not TFIISAcDel, is lethal, and that TFIISMut inhibits transcript elongation and elicits dramatic RNAPII polyubiquitylation in cells.
Mechanism of TFIISMut Action
The movement of RNA polymerases is not generated via the ATP-dependent “powerstrokes” typical of DNA helicases. Rather, it is governed by a Brownian ratchet mechanism, with one of the forward translocated states being stabilized by binding of the correct incoming nucleotide (Bar-Nahum et al., 2005; Herbert et al., 2008; Nudler, 2009). One consequence of this type of mechanism is that although the polymerase moves rapidly forward on average, it also frequently moves backward. So, although the formation of a new phosphodiester bond is likely to be immediately followed by forward translocation, the enzyme can also instead backtrack for one or several nucleotides so that the 3′ RNA terminus comes out of alignment with the active site. Brownian motion can bring the end of the RNA back in register with the active site again, but the polymerase is also capable of endonucleolytic cleavage of the transcript, a process which is highly stimulated by TFIIS (Izban and Luse, 1992; Reines, 1992).
We found that TFIISMut, but not the closely related TFIISAcDel mutant, inhibits the intrinsic ability of RNAPII to cleave its nascent transcript, and that this correlates with lethality in yeast. We also tested the possibility that TFIISMut might be capable of “driving” RNAPII backtracking and/or that it might be very slow to dissociate from the elongation complex, perhaps precluding RNAPII from returning (forward translocating) to the catalytically competent configuration. In this scenario, inhibition of intrinsic transcript cleavage might be an indirect effect. However, we were unable to detect any change in the RNAPII-binding characteristics of TFIISMut compared to wild-type, and the mutant also did not significantly affect the rate of nucleotide addition in experiments (Figure 1C and Figure S1B). Most importantly, transcription of a DNA template without prominent pause sites was largely unaffected by the presence of TFIISMut (Figure 1B), but in the presence of a chain terminator where transcription of the same template could only be restored by transcript cleavage, TFIISMut significantly inhibited transcription (Figure 2B), strongly arguing that its effect on transcript elongation is due to inhibition of transcript cleavage. Thus, although we cannot completely rule out the possibility that TFIISMut might under certain circumstances affect pause escape or forward translocation as well, our data are best consistent with the idea that it affects transcription and cell viability through its inability to support/allow transcript cleavage.
The precise mechanism of inhibition of intrinsic transcript cleavage by TFIISMut remains to be resolved, but results from recent structural studies support the idea that intrinsic RNAPII transcript cleavage and TFIIS-stimulated cleavage occur via different mechanisms (Wang et al., 2009). In “intrinsic cleavage mode,” the base of the backtracked nucleotide in the “P site” coordinates magnesium to facilitate transcript cleavage. By contrast, in “TFIIS-dependent cleavage mode” structural changes in the RNAPII active site upon insertion of TFIIS's acidic loop mean that the two acidic residues in the TFIIS loop tip provide the coordination of magnesium instead of the backtracked nucleotide, supporting a much more efficient way of cleavage (Wang et al., 2009). It thus seems reasonable to suggest that TFIISMut results in structural changes to backtracked RNAPII that inhibit intrinsic transcript cleavage, but without supporting TFIIS-stimulated transcript cleavage.
An Essential Role for Transcript Cleavage in Transcription and Viability
The gene encoding TFIIS in yeast is dispensable for viability (Hubert et al., 1983). Interestingly, bacteria such as Escherichia coli encode two distinct transcript cleavage stimulatory factors, GreA and GreB. However, even the greA− greB− double mutant is viable (Orlova et al., 1995). This, as in eukaryotes, may suggest either that transcript cleavage is not important, or that the intrinsic transcript cleavage activity of RNAP suffices for bacterial transcription and survival. Our data in the eukaryotic system support the second possibility and certainly provide a likely explanation for why DST1 is not an essential gene in yeast: the intrinsic transcript cleavage activity of RNAPII may suffice for transcription and survival in the absence of stimulated transcript cleavage.
Obstacles for Transcription Are Frequent In Vivo
The evidence described here also supports the idea that transcription problems leading to transcriptional backtracking are frequent in vivo. This is most clearly indicated by the lack of polymerases making it to the end of GAL-VPS13 in the presence of TFIISMut (Figure 5A). This effect is particularly striking given that transcript cleavage inhibition by TFIISMut is not dramatic in vitro (Figure 1) and given that it requires direct association with elongating RNAPII (Figure 4), meaning that, in all likelihood, transcript cleavage in only a fraction of polymerases that backtrack and require transcript cleavage can be inhibited by TFIISMut expression.
The precise nature of the obstacles resulting in RNA backtracking is unknown, but as backtracking is an intrinsic consequence of RNAPII moving via Brownian motion rather than ATP-driven powerstrokes, it seems reasonable to expect that all kinds of obstacles, including natural pause sites, secondary DNA structures, nucleosomes, and other DNA-binding proteins, are likely to cause transcription problems and backtracking.
Expression of TFIISMut in cells results in a dramatic increase in RNAPII polyubiquitylation. Ubiquitylation/degradation of RNAPII occurs in response to transcription obstacles, such as UV-induced DNA damage, but is also triggered by 6AU (Somesh et al., 2005) and the RNAPII-specific inhibitor α-amanitin (Nguyen et al., 1996; Anindya et al., 2007). It thus seems highly likely that the signal for ubiquitylation/degradation is the backtracked polymerase, and that expression of TFIISMut gives rise to a dramatic increase in the number of polymerases caught in this state.
In summary, our data suggest that RNAPII transcript elongation in vivo is surprisingly problematic and involves frequent retrograde motion, with a constant need to rescue backtracked elongation complexes by transcript cleavage. Without efficient reactivation of such complexes, overall transcript elongation is severely compromised, with polymerases ending up “dead in their tracks” and being removed in a process at least partly dependent on RNAPII ubiquitylation/degradation.
Experimental Procedures
Yeast Strains, Plasmids, and Proteins
All Saccharomyces cerevisiae strains used in this study were grown and manipulated using standard techniques (Sherman, 1991) and were congenic with W303-1A (Thomas and Rothstein, 1989). Please see the Supplemental Experimental Procedures for details about strains, plasmids, and proteins used.
Formation and Purification of Transcriptionally Active Ternary Complexes
The formation of ternary complexes has been described previously (Somesh et al., 2005), but in order to enrich for transcriptionally competent complexes, further purification steps were introduced in most experiments. A P32-5′-end-labeled RNA oligonucleotide (9-mer, or 12-mer, in 1:1 molar ratio) was annealed to a DNA template strand (which was biotinylated at the 3′ end). Typically, 1.5 μg RNAPII was incubated with 2.5 pmol of the DNA/RNA hybrid, followed by the addition of 5 pmol of the nontemplate DNA strand. Stable and transcriptionally active RNAPII elongation complexes were enriched by incubating in wash buffer (50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 2 mM MgCl2) at 30°C for 90 min, followed by binding to M2 (anti-Flag) agarose (Sigma) to isolate Flag-tagged RNAPII and remove free DNA and RNA. M2-associated RNAPII was released by once repeated incubation for 5 min at 4°C with 0.2 mg/ml “3xFlag” peptide (N-MDYKDHDGDYKDHDIDYKDDDDK-C). RNAPII incorporated into elongation complexes was then separated from free RNAPII by binding elongation complexes to streptavidin-paramagnetic beads (Promega) via the biotin tag on the template DNA strand. This was followed by three 2 min wash steps at room temperature (RT) in TB (20 mM Tris-HCl [pH 7.9], 40 mM KCl, 5 mM MgCl2). Oligonucleotide sequences are shown in the Supplemental Experimental Procedures, and a schematic of reconstituted elongation complexes is shown in Figure S2.
Transcription of Purified Ternary Complexes
Transcription using purified ternary complexes was typically performed in TB for 5 min at RT in the presence of RNase inhibitor (RNasin, Promega) and 500 μM each of ATP, GTP, and UTP. Reactions were stopped by the addition of stop buffer (0.5% SDS, 20 mM EDTA, and 0.8 mg/ml Proteinase K) followed by incubation at 37°C for 30 min. After phenol/chloroform extraction and ethanol precipitation, samples were resuspended in 95% formamide buffer containing 5 mM EDTA and 0.1% SDS and were resolved by 12% denaturing PAGE. Radioactive bands were visualized by autoradiography. Where relevant, results were also quantitated by PhosphorImager.
For transcription on A/T tract oligonucleotides, ternary complexes were assembled as described above. After the formation of ternary complexes and binding to streptavidin-paramagnetic beads (nontagged RNAPII was used, so the Flag-binding step was omitted), the ternary complexes were washed in TB buffer, followed by wash with TB buffer containing 1 M KCl for 20 min at 37°C. After washing again in TB buffer to reduce the salt concentration, the ternary complexes were transcribed in TB buffer, using 500 μM each of ATP, GTP, and CTP and various concentrations of UTP (5, 50, and 500 μM). After 5 min incubation, the reactions were stopped and processed as described above.
For chain termination experiments, elongation complexes were bound to streptavidin beads via biotin on the transcribed strand followed by washing in TB. The immobilized ternary complexes were then incubated with 1 mM chain terminator (3′-O-methylguanosine 5′-triphosphate, Amersham Biosciences) in TB for 5 min at RT. Following extensive washing to remove the terminator, the ternary complexes were incubated with 500 mM ATP, GTP, and UTP for 10 min and treated as described above.
Intrinsic Transcript Cleavage Experiment
Ternary complexes using a 12-mer RNA oligonucleotide were assembled and purified in the absence of MgCl2 (precluding transcript cleavage). The complexes were then incubated for 5 min with the relevant version of TFIIS (120 ng). Reactions were started by adding MgCl2 to 10 mM final concentration, terminated at the indicated time points by the addition of stop buffer, and processed as described above.
Promoter-Specific Transcription
Promoter-specific transcription was carried out as described using a dst1Δ yeast whole-cell extract (WCE) and the double G-less cassette template plasmid pGCYC1-402 (Kong and Svejstrup, 2002; Rondon et al., 2003). Briefly, reactions were performed in a final volume of 25 μl (40 mM HEPES [pH 7.5]; 15 mM MgCl2; 8 mM DTT and 100 mM KOAc; 0.8 mM CTP, ATP, GTP; 12 μM UTP; 10 μCi 32P-UTP [3000 Ci/mMol; Perkin Elmer]; 30 mM phosphocreatine; 60 μg creatine kinase; and 40 units RNasin). In each reaction, ∼50 μg WCE, 100 ng Gal4-VP16 (Kim et al., 1994), and 400 ng pGCYC1-402 DNA were incubated at RT for 1 hr in the presence of different concentrations of TFIIS (80 or 160 ng). Reactions were stopped by addition of 200 μl stop buffer (10 mM Tris [pH 7.5], 0.3 M NaCl, and 5 mM EDTA) and 100 units of RNase T1 (Roche) for 15 min at RT. Samples were then treated with Proteinase K, phenol/chloroform extracted, and samples run on a sequencing gel as described (Sayre et al., 1992). Results were visualized by autoradiography. Quantification of transcription through the proximal and distal cassettes by Phosphorimager took into account the different number of potential radioactive labels in the respective cassettes.
Recruitment of TFIIS to Ternary Complexes
Purified elongation complexes (WT RNAPII or RNAPII Δrpb9) were incubated with 500 mM of GTP, ATP, and UTP in TB to transcribe to the first guanine in the template. After washing away free nucleotides (leading to substantial RNAPII backtracking [data not shown]), ternary complexes were incubated with increasing concentration of TFIIS or TFIISMut (40, 100, or 200 ng) for 30 min at 30°C in TB buffer (containing 1 mg/ml BSA but lacking MgCl2) (Figure S1B). For comparing binding of TFIIS to WT and rpb9Δ RNAPII (Figure 4C), 0, 80, 160, 240, 320, or 480 ng of WT TFIIS was incubated for 30 min at RT with elongation complexes formed with WT RNAPII or RNAPII Δrpb9, respectively. In both types of experiments, samples were subsequently washed extensively in wash buffer (50 mM Tris-HCl [pH 7.5], 200 mM KOAc, 0.5% Triton X-100, 10% glycerol, 1 mM β-mercaptoethanol), resuspended in SDS-PAGE loading buffer, and fractionated on a 12% SDS-PAGE gel. TFIIS was visualized by western blotting using anti-his antibodies (Novagen).
For RNAPII-TFIIS binding-competition assays (Figure S1B), TFIISMut (350 ml of 2 mg/ml) was biotinylated using 20-fold molar excess of the biotin reagent as recommended by the manufacturers (EZ-Link NHS-Biotin, Thermo Scientific) for 2 hr on ice, followed by dialysis. Based on SDS-PAGE mobility shift analysis, all treated TFIISMut protein was modified. Biotinylated TFIISMut (315 ng) was bound to streptavidin beads for 10 min at RT followed by washing in TB. The RNAPII elongation complexes were then added and incubated with the immobilized TFIISMut for 10 min on ice and then washed in TB. Increasing concentration of nonmodified TFIIS or TFIISMut proteins (20–400 ng) were then incubated for 10 min on ice to compete the ternary complex off the biotinylated TFIISMut and into the supernatant. SDS-PAGE followed by western blotting, probed with the Rpb1 antibody 4H8, was used to assess the amount of ternary complexes released into the supernatant.
Chromatin Immunoprecipitation
To study the occupancy of RNAPII on GAL-VPS13, formaldehyde-crosslinked chromatin was immunoprecipitated with 4H8 antibody from dst1Δ cells expressing different versions of TFIIS, as described (Kristjuhan and Svejstrup, 2004).
Immunoprecipitation of Ubiquitylated RNAPII
Yeast cells (W303) expressing TFIIS, TFIISMut, or TFIISAcDel from the galactose-inducible vector pYC2/CT were grown in minimal media containing raffinose. Samples were taken before and 1 hr after galactose induction. Samples were resuspended in lysis buffer (150 mM Tris-acetate [pH 7.8], 50 mM KOAc, 20% glycerol, 3 mM EDTA, 5 mM DTT) containing 2 mM N-ethylmaleimide (Sigma), 20 mM lactocystine (Boston Biochemicals), 50 U benzonase (Novagen), 0.1% Triton X-100, and protease inhibitors [Otero et al., 1999]). Cells were disrupted using glass beads in a FastPrep FP120 Ribolyzer (Bio 101, Thermo). After clearing the extract by centrifugation, the supernatant was mixed with glutathione Sepharose 4B beads containing GST-Dsk2 (Anindya et al., 2007). Beads were washed in TENT buffer (50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 150 mM NaCl, 0.5% Triton X-100) and bound proteins eluted using 10 mM reduced glutathione (in 50 mM Tris-HCl [pH 8.0]). Ubiquitylated proteins were subjected to denaturing SDS-PAGE, and western blotting, probing with 4H8 (anti-Rpb1) antibody.
Acknowledgments
This work was supported by an EMBO long-term postdoctoral fellowship to S.S., and by grants from Cancer Research UK, Association of International Cancer Research, and the European Community (Integrated Project DNA repair, LSHG-CT-2005-512113) to J.Q.S. Caroline Kane is thanked for the His-TFIIS expression plasmid; David Jansma and Jim Friesen are thanked for yeast strains; and Dong Wang, Dave Bushnell, Craig Kaplan, and Roger Kornberg are thanked for helpful discussions and advice on RNAPII structure, as well as for comments on the manuscript. Peter Verrijzer and members of the Svejstrup laboratory are thanked for discussions and helpful comments on the manuscript.
Published: April 22, 2010
Footnotes
Supplemental Information includes three figures, Supplemental Experimental Procedures, and Supplemental References and can be found at doi:10.1016/j.molcel.2010.02.026.
Supplemental Information
References
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Anindya R., Aygun O., Svejstrup J.Q. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Arndt K.M., Kane C.M. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Awrey D.E., Weilbaecher R.G., Hemming S.A., Orlicky S.M., Kane C.M., Edwards A.M. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- Awrey D.E., Shimasaki N., Koth C., Weilbaecher R., Olmsted V., Kazanis S., Shan X., Arellano J., Arrowsmith C.H., Kane C.M. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J. Biol. Chem. 1998;273:22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Bengal E., Flores O., Krauskopf A., Reinberg D., Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol. Cell. Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F., Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Galburt E.A., Grill S.W., Wiedmann A., Lubkowska L., Choy J., Nogales E., Kashlev M., Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Michaut M., Acker J., Aude J.C., Thuriaux P., Werner M., Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B., Soutourina J., Esnault C., Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming S.A., Jansma D.B., Macgregor P.F., Goryachev A., Friesen J.D., Edwards A.M. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 2000;275:35506–35511. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- Herbert K.M., Greenleaf W.J., Block S.M. Single-molecule studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert J.C., Guyonvarch A., Kammerer B., Exinger F., Liljelund P., Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO J. 1983;2:2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M.G., Luse D.S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′—5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Jeon C., Yoon H., Agarwal K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl. Acad. Sci. USA. 1994;91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H., Armache K.J., Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kim B., Nesvizhskii A.I., Rani P.G., Hahn S., Aebersold R., Ranish J.A. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl. Acad. Sci. USA. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S.E., Svejstrup J.Q. Incision of a 1,3-intrastrand d(GpTpG)-cisplatin adduct by nucleotide excision repair proteins from yeast. DNA Repair (Amst.) 2002;1:731–741. doi: 10.1016/s1568-7864(02)00080-0. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A., Svejstrup J.Q. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Greenleaf A.L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Mason P.B., Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Nguyen V.T., Giannoni F., Dubois M.F., Seo S.J., Vigneron M., Kedinger C., Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M., Newlands J., Das A., Goldfarb A., Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M.G., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Hannett N.M., Young R.A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Roeder R.G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J. Biol. Chem. 1987;262:3331–3337. [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Reines D., Conaway R.C., Conaway J.W. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon A.G., Garcia-Rubio M., Gonzalez-Barrera S., Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H., Svejstrup J.Q. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre M.H., Tschochner H., Kornberg R.D. Purification and properties of Saccharomyces cerevisiae RNA polymerase II general initiation factor a. J. Biol. Chem. 1992;267:23383–23387. [PubMed] [Google Scholar]
- Schultz M.C., Choe S.Y., Reeder R.H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc. Natl. Acad. Sci. USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K., Kobayashi N., Mizuno D., Natori S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976;15:5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C., Fink G.R., editors. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc.; San Diego: 1991. pp. 3–20. [Google Scholar]
- Somesh B.P., Reid J., Liu W.F., Sogaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M., Shematorova E., Van Driessche B., Vandenhaute J., Thuriaux P., Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Awrey D.E., Edwards A.M., Archambault J., Friesen J.D. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc. Natl. Acad. Sci. USA. 1996;93:11552–11557. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.