Abstract
Expression and activity of activation-induced cytidine deaminase (AID) encoded by the aicda gene are essential for immunoglobulin (Ig) gene somatic hypermutation (SHM) and class switch DNA recombination (CSR). SHM and CSR unfold in general in germinal centers and are central to the maturation of effective antibody responses. AID expression is induced by activated B cell CD40 signaling, which is critical for the germinal center reaction, and is further enhanced by other stimuli, including interleukin-4 (IL-4) secreted from CD4+ T cells or Toll-like receptor (TLR)-activating bacterial and/or viral molecules. Integration of different intracellular signal transduction pathways, as activated by these stimuli, leads to a dynamic aicda-regulating program, which involves both positively acting trans-factors, such as Pax5, HoxC4, E47 and Irf8, and negative modulators, such as Blimp1 and Id2, to restrict aicda expression primarily to germinal center B cells. The phosphatidylinositol 3-kinase (PI 3-K), which functions downstream of activated B cell receptor (BCR) signaling, likely plays an important role in triggering the downregulation of aicda expression in post-germinal center B cells and throughout plasmacytoid differentiation. In B cells undergoing SHM and CSR, AID activity and, possibly, AID targeting to the Ig locus are regulated at a post-translational level, including AID dimerization/oligomerization, nuclear/cytoplasmic AID translocation and phosphorylation of the AID Ser38 residue by protein kinase A (PKA). Here, we will discuss the role of B cell activation signals, transcription regulation programs and post-translational modifications in controlling aicda expression and AID activity, thereby delineating an integrated model of modulation of SHM and CSR in the germinal center reaction.
Keywords: activation-induced cytidine deaminase (AID), B cell, class switch DNA recombination (CSR), enhanceosome, germinal center reaction, immunoglobulin (Ig), plasmacytoid differentiation, post-translational modifications, signal transduction pathways, somatic hypermutation (SHM), transcription factors
2. Introduction
The ability of vertebrates to mount specific and effective responses to a broad range of pathogens and develop long-term memory is underpinned by the broad diversity and exquisite specificity of adaptive immunity. During B cell development in the bone marrow of mammals, rearrangement of germline V (variable), D (diversity) and J (joining) gene segments in the immunoglobulin heavy (H) chain locus and V and J gene segments in the Igλ or Igκ light (L) chain locus yields a highly diverse BCR repertoire, which is available for antigen recognition in the primary antibody response. Such V(D)J recombination events are dependent on the expression of the recombination-activating genes-1 (RAG-1) and RAG-2. The impact of antigen on the primary BCR repertoire leads to the antibody affinity maturation, i.e., emergence of high-affinity IgG, IgA and/or IgE antibodies through SHM and CSR, two unique genetic processes fostered by the specialized microenvironment of the germinal center1. During the germinal center reaction, B cells are activated to undergo high-rate proliferation, SHM and CSR, positive selection of B cells expressing high-affinity BCR and, subsequently, differentiation to plasma cells and memory B cells. SHM inserts mainly point-mutations in Ig V(D)J DNA at a rate of approximate 10−3 change per base per cell division, which is about a million-fold higher than the spontaneous somatic mutation rate of the genome at large. CSR replaces the expressed IgH constant (CH) exon cluster, as juxtaposed to the VHDJH region, from the initial Cμ to a downstream Cγ, Cα or Cε. The intervening DNA between the switch μ (Sμ) region, lying at upstream of Cμ, and a downstream S region, lying at upstream of the Cγ, Cα or Cε exon cluster, is “looped-out” as a switch (Sx-Sμ) circle. While SHM depends on Ig V(D)J DNA transcription, CSR is dependent on the germline IH-CH transcription initiated by the IH promoter that lies upstream of each S region2–6.
Both SHM and CSR depend on expression of AID7,8. AID shares significant sequence similarities with the RNA cytidine deaminase apolipoprotein-B editing complex catalytic subunit 1 (Apobec1)9, the founding member of the recently annotated Apobec/AID family of enzymes10. Many Apobec/AID family proteins, including AID, are involved in innate antiviral response in the human and the mouse11,12 (Table I). In spite of the possibility that AID may deaminate cytidine in pre-RNA13, it is now generally accepted that AID acts as a DNA deaminase, deaminating dC residues to dU in single-stranded DNA, as existing transiently in the non-template DNA strand during V(D)J transcription or as the R-loop structure within transcribed S region repeats14–18. Repair of dU by Ung-mediated base excision repair (BER) and Msh2-mediated mismatch repair (MMR) leads to SHM and CSR14–18, as shown by studies in ung−/−msh2−/− double knockout mice19. Such DNA repair pathways, which are otherwise high-fidelity and critical for maintenance of genome integrity, are diverted by the SHM and/or CSR machinery to become mutagenic/recombinogenic through a subversion of the “polymerase switch”, by which the high-fidelity DNA polymerase (pol) δ and pol ε are replaced by error-prone translesion DNA synthesis (TLS) pol θ, pol η, pol ζ and Rev1 to effect faulty DNA synthesis20–30.
Table 1.
Apobec/AID cytidine deaminase family of proteins in the human and the mouse
| Group | Chromosomal location | Enzymatic activities | Putative function |
|---|---|---|---|
| Apobec1 | hu Chr. 12 mo Chr. 6 |
C6666→U in ApoB mRNA192; dC→dU in single-stranded DNA (weak)12 | Determines tissue-specific expression of the truncated ApoB protein192 |
| Apobec2 | hu Chr. 6 mo Chr. 17 |
Unknown; No deamination activity | Unknown |
| Apobec3a | |||
| Apobec3B | hu Chr. 22 | Unknown | Inhibits HIV infection moderately |
| Apobec3C | hu Chr. 22 | dC→dU in DNA (weak)193 | Inhibits HIV infection193 |
| Apobec3F | hu Chr. 22 | dC→dU in nascent HIV minus-strand DNA; deamination-independent12,194 | Inhibits HIV infection; Inhibits HBV pregenomic RNA reverse transcription12,194 |
| Apobec3G | hu Chr. 22 | dC→dU in nascent HIV minus-strand DNA; deamination-independent12,194 | Inhibits HIV infection; Inhibits HBV pregenomic RNA reverse transcription12,194 |
| Apobec3 | mo Chr. 15 | deamination-independent195 | Inhibits MMTV infection195 |
| AID | hu Chr. 12 mo Chr. 6 |
dC→dU in single-stranded DNA178,179; dC→dU in transcribed double-stranded DNA174; C→U in putative pre-RNA13; deamination-independent196 | Mediates SHM and CSR7,8; Limits infection by transforming retroviruses196 |
In the human, there are at least eight Apobec3 group proteins, from Apobec3A to Apobec3H, and their genes are clustered; only the four best studied Apobec3 proteins are reported here. In the mouse, there is only one Apobec3 protein.
As suggested by the induction of the aicda gene9 and TLS polymerase genes20,21,28 in B cells upon engagement of CD40 by its ligand, CD154, and BCR crosslinking, two crucial signals in germinal center B cell differentiation31,32, multiple signal transduction pathways are activated and integrated to drive AID expression and many of the events that lead to SHM and CSR. However, the Ig locus- and B cell-specificity of SHM and CSR can be altered if appropriate and specific pathways are diverted or bypassed, as implied by the hypermutation in non-Ig transgenes and/or DNA recombination in plasmid substrates induced by enforced AID expression in fibroblasts33,34. Here we will discuss recent advances in the understanding of specific regulation of aicda gene expression and AID activity in the context of their relevance to SHM and CSR. We will use the terms “AID expression” and “aicda expression” to discuss findings stemming from AID protein analyses and from RT-PCR- or microarray-based aicda transcript analyses, respectively.
3.1. AID INDUCTION BY B CELL SIGNAL TRANSDUCTION PATHWAYS
The aicda gene was identified in the murine B lymphoma cell line CH12F3-2 as specifically upregulated upon stimulation by CD154, IL-4 and tumor growth factor-β (TGF-β), three signals that induce CSR to IgA at a high rate in CH12F3-2 cells9. CD154:CD40 engagement and IL-4 synergize to induce aicda expression in both human and mouse B cells9,35–38. Such aicda induction may be further enhanced by BCR crosslinking, as effected by physically-linked antigenic determinants. AID expression, B cell proliferation and CSR are also induced or enhanced by microbial lipopolysaccharides (LPS) and/or DNA containing multiple unmethylated CpG motifs (CpG DNA), which activate TLR4 and TLR9 signal transduction pathways, respectively. It is conceivable that, in vivo, these pathogen-associated molecules activate intracellular signal transduction pathways that cross-talk with pathways activated by host immune system-generated B cell stimuli, such as CD154 and IL-4, to induce optimal AID expression, eventually leading to robust anti-bacterial/anti-viral immune response. Here, we will focus on the role of CD40, IL-4 receptor (IL-4R), BCR and TLRs in activating aicda expression in germinal center B cells in vivo or B cells stimulated to undergo germinal center-like differentiation in vitro (Figure 1). TGF-β induces aicda expression in CH12F3-2 B cells and is essential in mediating CSR to IgA31,39,40, while inhibiting B cell proliferation and CSR to other Ig isotypes at a high concentration. TGF-β, in general, downregulates the activity of Stat6 and NF-κB, two positively acting trans-factors for aicda expression (see below), and upregulates the activity of Id3, a negative aicda modulators41. However, in the mucosal environment, where B cells undergo CSR to IgA at a high frequency, TGF-β may trigger significantly different signaling events that lead to AID induction. Finally, expression of aicda is also induced by mucosal stromal cell- or dentritic cell (DC)-secreted B cell-activating factor of the tumour-necrosis factor (TNF) family (BAFF, BLyS) and a proliferation-inducing ligand (APRIL)42–45, two B cell survival and activation factors that induce CSR to IgA in vitro and in vivo. Due to the space limitation, the induction of aicda and/or CSR by BAFF and APRIL through B cell maturation antigen (BCMA) and transmembrane activator and CAML interactor (TACI), two receptors common for BAFF and APRIL, and BAFF receptor (BAFF-R) (Figure 1) is not discussed here; readers are referred to these excellent reviews46–51 and original papers cited therein.
FIGURE 1.
Induction of aicda expression by activated B cell surface molecules. From left to right: (i) the TLR4-MD2-CD14 complex is the receptor for LPS or other bacterial glycoproteins; TLR9 is located in endosomes and recognizes CpG DNA from microbial pathogens. TLR4 and TLR9 associate with adaptor protein MyD88 or Trif through the Tir domain, depicted in yellow; (ii) the BCR complex containing BCR, Igα and Igβ is crosslinked by physically-linked antigens. The CD19/CD21/CD81 co-receptor is activated by the C3d(g) complement cleavage product and modulates the BCR signaling threshold. CD81 is a co-receptor for HCV E2 envelope protein; (iii) cytokine receptors IL-4R and TGF-βR mediate aicda induction and CSR. IL-4R-triggered signaling can be inhibited by activated CD45; and (iv) among the TNF receptor family of B cell surface molecules, CD40 is engaged by CD154 expressed on CD4+ T cell surface and exists as a trimer at B cell surface. CD30, when engaged by CD153 expressed CD40+ TH cell surface, negatively regulates CD40 signaling. TACI, BAFF-R and BCMA are receptors for BAFF and/or APRIL secreted by dentritic cells and can mediate CSR in B cells of mucosal areas. Activated intracellular signal transduction pathways likely converge to assemble an enhanceosome, which would include NF-κB, Stat6, Pax5, E47, the Irf8/SpiB complex and the HoxC4/Oct1/2/OcaB complex, at the aicda locus to induce efficient aicda expression. The arrows depict the transcription initiation of aicda or other germinal center B cell-specific genes.
3.1.1. CD40
CD40 is a 48 kDa transmembrane glycoprotein constitutively expressed on the surface of B cells, DCs, macrophages, epithelial cells and activated T cells52. Like other members of the TNF family of receptors and ligands, CD40 trimerizes upon engagement by the trimeric CD154, a type II integral membrane TNF family protein transiently expressed on activated CD4+ T cell surface. In addition to the mouse CH12F3-2 B cell line, primary murine B cells9,35,38, primary human B cells36,37 as well as human IgM+IgD+ B cell lines36 all significantly upregulate aicda expression upon CD154:CD40 engagement in vitro. In vivo, CD154:CD40 engagement is essential to trigger the T-dependent germinal center reaction, as shown by impaired germinal center development in mice deficient in CD40 or CD154 or in patients with X-linked hyper IgM syndrome (X-HIGM), which is caused by a defective CD154 molecule (Table 2)53–57. In cd154−/− or cd40−/− mice, serum levels of IgG1, IgG2a, IgG2b and IgE are severely decreased, similar to what occurs in X-HIGM patients, suggesting that the CD40 signaling is essential for CSR, which is, at least partially, due to the role of CD40 signaling in the AID induction. The normal or moderately decreased IgG3 level in cd154−/− or cd40−/− mice possibly reflects a T-independent B cell response triggered by chronic exposure to bacterial LPS, which induces both AID expression (see below) and CSR to IgG3.
Table 2.
Molecular basis of HIGM syndrome
| Molecular deficiency | aicda expression | AID activity | Phenotypes |
|---|---|---|---|
| CD154 (CD40L) | Impaired | Not determined | Defective CSR; defective germinal center formation and SHM |
| CD40 | Impaired | Not determined | Defective CSR; defective germinal center formation and SHM |
| NEMOa | Not affected | Not determined | Defective CSR; normal SHM; ectodermal dysplasia |
| AIDb | Not affected | No AID protein expression; loss of deaminase activity; loss of binding to CSR cofactorsc | Defective CSR; defective SHMc; lymphoid hyperplasis; defective plasmacytoid differentiation |
| UNG | Not affected | Not affected | Defective CSR; altered SHM spectrum |
| ATM | Not affected | Not affected | Defective CSR; normal SHM; accelerated lymphomagenesis |
| MRE11-NBS1 | Not determined | Not determined | Defective CSR; likely defective SHM; accelerated lymphomagenesis |
NEMO deficiency leads to a defective NF-κB activation;
More than 20 AID mutations have been identified. They can be segregated into three groups: autosomal recessive type (AR-AID), AID with carboxyl-terminal mutations and AID with carboxyl-terminal deletions;
AID with carboxyl-terminal deletions cannot bind to putative CSR co-factors, but displays normal dC deamination activity and mediates SHM.
In germinal centers, B cells proliferate vigorously, thus termed centroblasts, in a microanatomical region referred to as the “dark zone”, due to its high density of nuclei. Centroblasts transition from the dark zone into “light zone” centrocytes, which proliferate58,59, but at a significantly lower rate. In the light zone, centrocytes undergo positive selection, as effected by the antigen driving the ongoing response, and plasmacytoid differentiation. As part of the germinal center reaction, SHM primarily occurs in centroblasts in the dark zone. The onset of SHM is earlier than that of CSR60,61. In the human, CSR likely initiates in centroblasts and unfolds to completion while centroblasts transition to centrocytes, as indicated by the ready detection of germline IH-CH transcripts in centroblasts, appearance of S circle DNA, a marker of newly completed S-S region DNA recombination, in centroblasts/centrocytes, abundance of mature VHDJH -Cγ transcripts in centrocytes and, finally, the expression of surface IgG, IgA or IgE in centrocytes62–64 (Figure 2). In the mouse, aicda expression and Iγ1-Cγ1 germline transcription, two early and essential events for CSR to IgG1, as triggered by immunization with hapten 4-hydroxyl-3-nitrophenylacetyl conjugated to chicken γglobulin (NP-CGG), begin in early centroblasts, while Iγ1-Cμ circular transcripts, as expressed from the excised Sγ1-Sμ circle, appear later in the centroblasts/centrocytes65.
FIGURE 2.
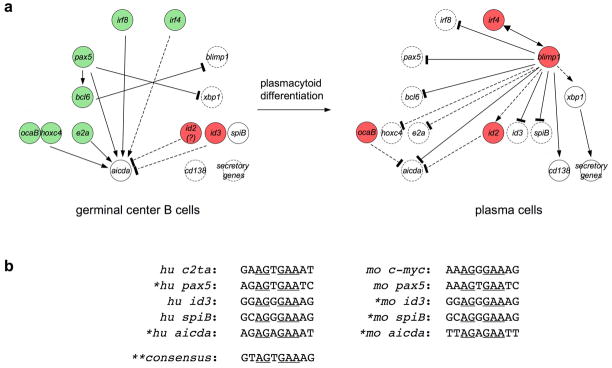
Germinal center B cell differentiation, including SHM and CSR, stage-specific expression of AID, and positive regulators (depicted in blue) and negative modulators (depicted in gray) of aicda gene transcription. AID expression initiates in early centroblasts, is maximal in full-blown centroblasts, significantly decreases in centrocytes and extinguishes in plasma cells. The expression profile of AID and transcription factors in memory B cells is not shown here. The dual role of OcaB and Irf4 is dependent on different germinal center reaction stages. Most data are derived from studies in human tonsil or mouse spleen B cells and plasma cells. The expression profile of Id2 was not available and is deduced from those of Irf4 and Bcl6, two putative id2 transcription regulators.
Consistent with the unfolding of SHM and CSR processes, expression of AID in germinal centers initiates in early centroblasts, is maximal in full-blown centroblasts, significantly decreases in centrocytes and extinguishes in plasma cells (Figure 2). In early centroblasts, CD154:CD40 engagement activates a B cell transcription program, which involves paired box protein 5 (Pax5, also termed B cell-lineage specific activation protein, BSAP), interferon (IFN) regulatory factor 4 (Irf4) and Irf8, to initiate aicda expression. This transcription program likely requires de novo protein synthesis of those transcription factors, as suggested by the suppression of optimal aicda induction in CH12F3-2 B cells by cycloheximide, an inhibitor of the mRNA translation9. In the dark zone, few if any CD154-positive CD4+ T cells can be detected66, suggesting that the aicda regulation program, as triggered in early centroblasts, continues to be active and drives B cell division and differentiation (Figure 2). In human tonsil germinal centers, AID expression is predominant in the dark zone and limited in the light zone, which displays a rich follicular dendritic cell (FDC) network67. In mouse spleen germinal centers induced by NP-CGG, AID is also preferentially expressed in the dark zone 14 days after immunization (our unpublished data). Downregulation of AID as centroblasts transition to centrocytes is likely mediated by changes in the expression level of transcription factors, i.e., decreased expression of positive aicda transcription regulators and increased expression of aicda negative modulators, in light zone B cells (Figure 2). The abated expression of AID, as well as its positive transcriptional regulators, likely results from insufficient signals B cells receive from CD4+ T cells or FDCs, as localized in the light zone, to overcome the attenuation of the aicda transcription program (Figure 2). Insufficient T cell help could be caused by more transient T:B cell synapses, as compared to the T:B cell engagements involving early centroblasts59,68, or a possible downregulation of CD154 expression in CD4+ T cells, or both. Finally, the differential expression of AID in centroblasts and centrocytes, which display different proliferation rates, is consistent with the cell cycle-dependent regulation of aicda expression69, i.e., a higher steady-state aicda mRNA level in B cells undergoing a higher than a lower rate of proliferation70. The extinction of aicda expression during plasmacytoid differentiation is discussed later (see below).
The intracellular carboxyl-terminal tail of CD40 contains binding sites for different TNFα receptor-associated factors (Trafs). Traf6 binds to the KxxPxE motif in the human CD40 and to the RQDPQE motif in the mouse CD40; Traf2 and Traf3 bind to the PxQxT motif in both human and mouse CD40. Depending on the Traf adaptor proteins that are recruited to clustered CD40 trimers, different signal transduction pathways are activated, leading to the differential activation of trans-factors, such as NF-κB and p38. While a wildtype CD40 transgene fully rescued germinal center development, aicda expression and humoral immune responses in cd40−/− mice, cd40−/− mice carrying a CD40 transgene with mutations at both the Traf2/Traf3- and Traf6-binding sites (cd40Δtraf2/traf3Δtraf6) displayed severely impaired germinal center development, mimicking the effect of complete CD40 deficiency35,71. However, the Traf2/Traf3- and Traf6-binding sites of CD40 were individually dispensable for B cell proliferation or germinal center formation, as shown by similar trans-complementation studies in cd40Δtraf2/traf3 and cd40Δtraf6 mice71. In vitro, cd40Δtraf6 B cells displayed a similar level of aicda induction by an anti-CD40 monoclonal antibody (mAb) as wildtype B cells. In contrast, cd40Δtraf2/traf3 B cells displayed significantly impaired aicda induction and CSR efficiency upon stimulation by an anti-CD40 mAb, while aicda and CSR induction by LPS is apparently normal, suggesting that the CD40-Traf2/Traf3, but not the CD40-Traf6, signal transduction pathway plays a major role in CD40-mediated induction of aicda expression. The impaired aicda expression in anti-CD40 mAb-stimulated cd40Δtraf2/3 B cells may indirectly result from the low proliferation rate of these B cells71, as suggested by the cell cycle-dependent induction of aicda expression70.
In human B cells, the recruitment of Traf2 to the intracellular tail of CD40 and NF-κB activation are hampered upon engagement of CD30, a TNFR family protein expressed on the B cell surface, by its ligand CD153, which is expressed on the T cell surface31,32,72 (Figure 1). Expression of CD30 in human B cells is upregulated by CD154:CD40 engagement. In human tonsil germinal centers, CD30 and AID expression in the dark zone were mutually exclusive and, in CD30+ and AID+ B cells, expression of germinal center reaction-related transcription factors displayed a reciprocal profile67. For instance, AID+ B cells were positive for the E-box protein E47 and negative for inhibitor of differentiation (Id2) and IRF4, while CD30+ B cells were positive for Id2 and IRF4 and negative for E47 (see below for the role of E47, Id2 and IRF4 in regulating aicda expression), suggesting that CD30 negatively modulates AID expression, likely through its repression of CD40-Traf2 signaling. CD154:CD40 engagement-mediated CD30 induction could be inhibited by BCR crosslinking72, suggesting that the BCR signaling plays an important role in relieving CD30-mediated suppression of aicda expression. Thus, the crosstalk between CD40− and BCR-activated signal transduction pathways is critical to for AID induction in germinal center B cells (Figure 2).
CD40-Traf signaling-mediated NF-κB activation depends on the phosphorylation and degradation of NF-κB inhibitor IκB. Upon recruitment to clustered CD40 tails, multimerized Traf adaptor proteins polyubiquitinate and activate the NF-κB essential modulator (NEMO) (also termed IκB kinase γ, IKKγ), a scaffolding molecule for the two other subunits of the IKK complex, IKKα and IKKβ. Missense mutations at Cys417 in the zinc finger domain of NEMO lead to the X-linked hyper-IgM syndrome with ectodermal dysplasia (XHM-ED) (Table 2)37,73,74. In B cells from XHM-ED patients, the CD40-dependent NF-κB activation was abolished and CSR was impaired. However, induction of aicda was seemingly normal upon B cell stimulation by CD154 and IL-437, suggesting that NEMO activity is not essential for AID expression. Instead, an alternative NF-κB activation pathway, as mediated by NF-κB-inducing kinase (NIK) and IKKα, or a compensating Traf-dependent signal transduction pathway involving p38 likely leads to aicda induction in XHM-ED B cells. Among the five mammalian NF-κB proteins, p50 (NF-κB1), p52 (NF-κB2), Rel A (p65), Rel B and c-Rel, p50 seems to play a role in regulating aicda expression, as suggested by the partial impairment of aicda induction in p50−/− mouse B cells stimulated with an anti-CD40 mAb and IL-475. Upon activation by an anti-CD40 mAb, p50 and Rel A, but not p52, c-Rel or Rel B, bound to a double-stranded oligodeoxyribonucleotide containing the two putative NF-κB sites, as identified in the upstream regulatory region of the aicda gene75 (Table 3), suggesting that NF-κB directly binds to the aicda promoter and induces a basal transcription level that can be further raised by other B cell stimuli.
Table 3.
Transcription factors and their putative binding sites in the murine aicda promotera and intronic enhancer regionb
| Transcription factor | Inducing stimuli | Binding sites | Sequence | Reference |
|---|---|---|---|---|
| NF-κB | CD154; LPS/IL-4; BCR crosslinking | −1203 ~ −1193 | 5′ GGGATTTCCC 3′ | 75 |
| −1267 ~ −1258 | 5′ GGGTCTTTCC 3′ | |||
| Stat6 | IL-4 | −1327 ~ −1319 | 5′ TTCCCACAA 3′ | 75 |
| Pax5 | ND | −97 ~ −85 | 5′ GGATACCTGGTGG 3′ | 143 |
| −68 ~ −57 | 5′ GGGGAGGAGCCC 3′ | |||
| E47 | LPS; CD154/IL-4; BCR crosslinking/IL-4 | −105 ~ −100 | 5′ CAGATG 3′ | 143,147 |
| +1642 ~ +1647 | 5′ CAGCTG 3′ | |||
| +1654 ~ +1659 | 5′ CAGCTG 3′ | |||
| HoxC4/Oct HoxC4c |
CD154/IL-4; LPS/IL-4; CD154/IL-4/TGF-β | −20 ~ −13 | 5′ ATTTGAAT 3′ | 144 |
| −169 ~ −166 | 5′ ATTT 3′ | |||
| −615 ~ −612 | 5′ ATTT 3′ | |||
| −600 ~ −597 | 5′ AAAT 3′ | |||
| Irf8 Blimp1 |
Not determined | −607 ~ −600 | 5′ AAAAGAAA 3′ | 163 |
| LPS; IL-2/IL-5; IL-6; BCR crosslinking/CD154/IL-4/IL-5 | −593 ~ −584 | 5′ TTAGAGAATT 3′ | 136 |
The promoter region is defined as the 1500 bp-DNA upstream the putative transcription initiation site9.
The two putative enhancer regions within the first intron were previously143,147; the cr1 encompasses about 360 bp DNA (+300 ~ +660) and the cr2 encompasses about 230 bp DNA (+1570 ~ +1800).
There are six more putative HoxC4-binding site at the −900 ~ −400 region, ten more in the cr1 and two more in the cr2 region, respectively. The three HoxC4 sites as shown are close to the binding site of E47 or Irf8 and likely play a role in the assembly of the aicda enhanceosome.
3.1.2. IL-4R
IL-4 is a pleiotropic cytokine produced by activated type 2 CD4+ T cells. By inducing both aicda expression and germline Iγ-Cγ transcription, IL-4 plays an essential role in regulating CSR to IgG4 and IgE in the human and CSR to IgG1 and IgE in the mouse76. Murine B cells stimulated in vitro by CD154 or LPS proliferate at a high rate, but switch to IgG2b and IgG3 at an efficiency at least two to five-fold lower than the switching efficiency to IgG1, as induced by CD154 or LPS together with IL-4, possibly reflecting a sub-optimal induction of AID in the absence of IL-49,35,38. The high-affinity binding of IL-4 to the 140-kDa α chain subunit of its receptor IL-4R (IL-4Rα) causes heterodimerization of IL-4Rα with the IL-4R γ common chain (γc) subunit (Figure 1), an element shared by receptors for other cytokine, including IL-2, IL-7, IL-9 and IL-1376. Upon IL-4Rα and γc heterodimerization, IL-4Rα-associated receptor tyrosine kinase Janus kinase-1 (Jak1) and γc-associated Jak3 autophosphorylate and/or reciprocally phosphorylate each other, leading to their own activation. Activated Jak1 and Jak3 in turn phosphorylate tyrosine residues in the cytoplasmic tail of IL-Rα and γc, respectively. Upon phosphorylation, the five conserved tyrosine residues in IL-4Rα, Tyr497 (Y1), Tyr575 (Y2), Tyr603 (Y3), Tyr631 (Y4) and Tyr713 (Y5), become docking sites for downstream signaling molecules, which are then phophorylated by Jaks and become activated76. Phosphorylated Y1 recruits insulin receptor substrate (IRS) and Shc adapter proteins and plays a critical role in IL-4-stimulated B cell proliferation77; phosphorylated Y2, Y3 and Y4 interact with the Src homology 2 (SH2) domain of the signal transducer and activator of transcription 6 (Stat6), which plays a major role in activating IL-4-induced gene expression, including aicda expression (see below); and finally, phosphorylated Y5 recruits Src homology-containing phosphatase 1 (Shp1), which dephosphorylates Stat6, thereby negatively regulating Stat6 signaling78. How individual Tyr residues in IL-4Rα contribute to AID induction and CSR to IgG and IgE remains to be determined. An alternative IL-4R (Type II), consisting of IL-13Rα1 and γc, is also the receptor for IL-13, which controls the TH2 response together with IL-479. The dual role of the IL-13Rα1/γc heterodimer as the type II IL-4R and IL-13R likely explain why IL-13, together with an anti-CD40 mAb, induced aicda expression80, and why IL-13 induced CSR to IgG4 and IgE in primary human B cells co-cultured with CD4+ T cell membrane in an IL-4-independent fashion81.
Stat6 is one of the seven mammalian Stat proteins, all of which are latent in the cytoplasm before their phosphorylation at Tyr residues by Jak family kinases. Different Jaks and Stats are activated in response to different extracellular stimuli, such as IFNs, growth factors and ILs. Phosphorylated Stat6 disengages from IL-4Rα and forms dimers that translocate to the nucleus, where they bind to DNA and direct gene transcription82,83. In the mouse, Stat6 is critical for IL-4-induced B cell activation and CSR to IgG1 and IgE, but negatively regulates CSR to other IgG isotypes84–86. Stat6-deficiency leads to a severely impaired aicda induction in murine B cells stimulated with an anti-CD40 mAb and IL-475. Stat6 signaling is negatively regulated by Shp1 phosphatase, which is recruited by IL-4Rα Tyr713. In addition, the CD45 phosphatase, when induced by an anti-CD45 mAb, negatively regulates IL-4-induced aicda expression in human B cells, possibly through dephosphorylation of Jak1, Jak3 and Stat687. A gain-of-function polymorphic allele IL-4Rα in the human, IL-4Rα(Glu576Arg), is associated with hyper-IgE syndrome. This can be explained by the accelerated recruitment/activation or dimerization of Stat6 near the IL-4Rα Tyr575 and/or suppressed dephosphorylation of Stat676, and would call for an analysis of AID induction in human B cells carrying this mutation. Similar to NF-κB p50, Stat6 binds to a double-stranded oligodeoxyribonucleotide containing a canonical Stat-binding motif 5′-TTCnnn(n)GAA-3′, as identified upstream of the aicda promoter (Table 3). However, whether NF-κB and Stat6 directly bind to the aicda promoter region in B cells in unclear. NF-κB and Stat6 seem to directly interact with each other and synergize in promoting transcription of IL-4-inducible reporter genes in human B lymphoma cells88, suggesting NF-κB and Stat6 synergistically bind to the upstream regulatory region (−1400 to −1200) of the aicda promoter (Table 3), thereby mediating the converging CD40− and IL-4R-activated signal transduction pathways for efficient aicda induction.
3.1.3. BCR
The BCR consists of IgH, IgL and the two intracytoplasmic signaling components, Igα (CD79a) and Igβ (CD79b) (Figure 1). BCR signaling is required for early-stage B cell development and mature B cell activation and differentiation when antibody affinity maturation occurs89,90. Upon crosslinking by antigen, most BCR molecules remain non-phosphorylated and are internalized by endocytosis, eventually resulting in the localization of the BCR-antigen complex into late endosomes for processing and antigen presentation91. A minor population of BCR molecules are phosphorylated at Tyr in Igα and Igβ and retained by the lipid raft at the B cell surface, thereby initiating the assembly of the BCR signalosome, a macromolecular protein complex containing enzymes and adaptor/scaffolding proteins involved in pathways leading to Ca2+ response and NF-κB activation91–93. In vitro, BCR crosslinking by an anti-IgM F(ab′)2 fragment or an anti-IgD antibody conjugated to dextran (anti-δ dextran) mimics antigen-mediated BCR crosslinking. In human B cells, aicda expression is efficiently induced by co-stimulation of CD154 and anti-IgM F(ab′)2, two of the signals that are required for induction of germinal center-like differentiation, including SHM and CSR31,32,36. Anti-δ dextran does not induce appreciable AID expression if used alone, but potentiates LPS-induced AID expression in primary murine B cells38. In vivo, BCR signaling is essential for the T-dependent germinal center reaction in the spleen, but dispensable for aicda induction in gut-associated lymphoid tissues (GALTs), as suggested by studies in the BCR-deficient mouse strain DHLMP2A90. In DHLMP2A mice, IgH expression was ablated and BCR signaling was mimicked by the Epstein-Barr virus (EBV) latent membrane protein 2A (LMP2A), which contains multiple domains that are functionally homologous to those in Igα and Igβ90. LMP2A co-opted the lipid raft and functioned as a BCR surrogate to promote survival, development and maintenance in DHLMP2A mouse B cells90,94. DHLMP2A mice displayed no germinal center formation in response to T-dependent antigens, likely due to the lack of antigen presentation and cognate T:B cell interactions, but displayed a number of germinal centers in GALTs, including Peyer’s patches, comparable to their congenic Balb/C counterparts. In DHLMP2A GALTs, aicda expression, SHM and CSR were seemingly normal, possibly as a result of chronic exposure of B cells to gut microbial flora that activate TLR signal transduction pathways90. Thus, BCR signaling likely regulates aicda induction by enhancing the CD40-activated aicda transcription program and/or by relieving the inhibitory effect of CD30 (as discussed above) during the initiation of aicda induction (Figure 2).
During B cell activation, the threshold of BCR signal transduction is critically regulated by the BCR co-receptor CD19/CD21/CD81 complex (Figure 1), which is recruited by antigen-associated complement cleavage products C3d/C3dg and co-aggregates with crosslinked BCR95. BCR crosslinking, together with co-engagement of CD21 and CD19, led to ready induction of aicda expression and SHM in the human Burkitt’s lymphoma B cell line BL1296. Translocation of CD19, as co-aggregated with crosslinked BCR, to the lipid raft activates PI 3-K signal transduction pathways by recruiting the p85α regulatory subunit of PI 3-K through two CD19 Tyr residues, Y482 and Y51358,97,98. In cd19−/− mice expressing a transgenic CD19 with mutations in Y482 and Y513 (cd19−/−TgY482F/Y513F), immunization with 2-phenyloxazolone coupled to chicken serum albumin (phOx-CSA) induced a seemingly normal germinal center development, centroblast proliferation and a significant recruitment of the VκOx1 gene, a Vκ4/5 family member used by 129/Sv mice for the phOx-specific antibody response, into the BCR repertoire. However, SHM frequency in the VκOx1 DNA in cd19−/−TgY482F/Y513F mice is about half that in control wildtype mice58, suggesting that AID might be expressed at a suboptimal level in the CD19 mutant mice. PI 3-K and its antagonistic enzyme PTEN likely play multiple roles in regulating aicda expression, CSR and plasma cell differentiation, as further discussed in the next section.
CD81, or its palmitoylated form, is necessary for the partitioning of co-aggregated BCR and CD19-CD21 into the lipid raft99–102. CD81 is also a co-receptor for the hepatitis C virus (HCV) in both human hepatocytes and B cells. In the human B cell line Raji, binding of recombinant HCV E2 protein to CD81 led to Ig SHM, as underpinned by aicda induction, double-strand DNA breaks (DSBs) in VHDJH DNA and induction of error-prone DNA polymerases103, three hallmark events of SHM21,32,36. Thus, E2 protein-activated CD81 induces aicda expression autonomously.
3.1.4. TLR
TLRs are pattern recognition receptors (PRRs) that recognize and bind microbial components, including LPS, flagellin, double-stranded RNA (dsRNA), CpG DNA and certain glycoproteins and lipoproteins104–106. TLRs can be divided into two groups based on their subcellular localization, i.e., the transmembrane TLR1, TLR2, TLR4, TLR5, and TLR6, which belong to the IL-1R/TLR protein superfamily, and the intracellular TLR3, TLR7 and TLR9, which likely initiate signal transduction from acidic endosomes107,108. All TLRs contain a Toll-IL-1R (TIR) domain, a protein-protein interaction module that recruits TIR-containing adaptor proteins to activated TLRs105. In mammalian cells, there are at least four TIR adaptor proteins, including myeloid differentiation factor 88 (MyD88), TIR adapter protein/MyD88 adapter like (Tirap/Mal), TIR domain-containing adapter inducing IFN-β/TIR domain-containing adapter molecule-1 (Trif/Ticam1) and Trif-related molecule/TIR domain-containing adapter molecule-2 (Tram/Ticam2). TIR-containing adaptor proteins, upon recruitment to TLRs, activate signal transduction pathways, thereby resulting in the activation of transcriptions factors, such as NF-κB, p38 and IRFs109. For instance, MyD88, possibly the best characterized TIR-containing adaptor proteins, activates the serine/threonine kinases IL-1R-associated kinase-1 (Irak1) and Irak4, which, in turn, recruit and activate Traf6. A Traf6-mediated activation of the IKK complex would lead to NF-κB activation. A MyD88-independent TLR4 signal transduction pathway, as mediated by Trif, intersects with the MyD88-dependent pathway at the level of Traf6 and leads to the activation of the downstream NF-κB and other transcription factors.
TLRs play an important role in directing efficient T-dependent adaptive immunity in the human and the mouse110,111. TLR4, which is expressed in the mouse, but not in the human, together with membrane-anchored CD14 and extracellular MD-2 proteins, form the receptor for E. coli LPS112 (Figure 1). When used together with TH cytokines, such as IL-4, IL-5, IFN-γ and/or TGF-β, E. coli LPS stimulate primary murine B cells to undergo proliferation and efficient CSR in vitro. While LPS alone induces weak AID expression, it synergizes with IL-4 and/or BCR signals for effective AID induction9,38. In vivo, LPS induce polyclonal B cell activation as a type 2 T-independent antigen. In addition, LPS, which are likely present in the form of the DC-captured immune complex, would bind to TLR4 expressed on mouse germinal center B cells, thereby enhancing DC-mediated BCR signaling events. It is conceivable that CD40, BCR and TLRs all contribute to the optimal AID induction in immune responses against bacterial infection (Figure 1), as suggested by the intrinsic defect of myD88−/− B cells in generating effective antibody response to LPS-conjugated ovalbumin (LPS-OVA)110. However, the role of TLRs in the antibody response during natural bacterial infection and the relative contribution of MyD88-dependent and MyD88-independent pathways in LPS-induced AID expression remain to be determined.
TLR4-MyD88 pathway may also play a role in viral-induced AID expression. Human papillomavirus type 16 (HPV16) L1 virus-like particles (VLP) induce aicda expression and CSR in B cells through TLR4, as demonstrated by the lack of aicda induction in C3H/HeJ mice B cells, which carry a mutation in the TIR domain of TLR4. In addition, aicda transcription and secreted IgG levels were greatly decreased in HPV16-simulated myD88−/− B cells, as compared to wildtype B cells113. HPV16 also synergizes with the CD40 signaling to further upregulate AID expression113, possibly reflecting the non-overlapping CD40-Traf2/3 and TLR4-MyD88-Traf6 signal transduction pathways and their synergistic activation of NF-κB. However, primary T-dependent responses in vivo and aicda expression in germinal center B cells were normal in MyD88-deficient mice upon infection with mouse polyoma virus114. Upon infection with Abelson murine leukemia retrovirus, in bone marrow B cells isolated from mice deficient in MyD88 or doubly-deficient in MyD88 and Trif, aicda was induced to a level comparable to that in wildtype bone marrow B cells115, suggesting a complex role of TLR-MyD88 in virus-induced AID expression in vivo.
Receptor-interacting protein (Rip) is a death domain kinase that is associated with Tnfr1 death receptor. Rip-deficiency in the mouse causes embryonic lethality that can be partially rescued by combined Tnfr1 deficiency. The interaction of Rip with Traf2 and Traf6 points to a possible role of Rip in CD40− and/or TLR4-mediated AID induction. Indeed, rip−/−tnfr1−/− murine B cells displayed more than 75% reduction of aicda expression and CSR to IgG and IgE upon stimulation with LPS and IL-4, as compared to rip+/+tnfr1−/− B cells. The comparable aicda induction in rip−/−tnfr1−/− and rip+/+tnfr1−/− B cells stimulated with anti-CD40 mAb and IL-4 suggests that Rip functions through Traf6, but not Traf2, in these B cells to activate the NF-κB signaling116. The exact role of different NF-κB proteins in LPS-mediated AID induction remains to be investigated in such knockout B cells75.
Induction of aicda expression is also mediated by the intracellular TLR9, which is expressed in both the human and the mouse, in B cells stimulated with CpG DNA. CpG DNA is prevalent in bacterial, but not mammalian, genome117 and is a potent AID-inducing agent118. TLR9 is localized in the endoplasmic reticulum (ER) (Figure 1) and is recruited to lysosomes after acidification of endosomes and lysosomes, as agents that block endosomal acidification, such as chloroquine and bafilomycin A1, completely abrogate CpG DNA-induced signaling. TLR9 intracellular localization is critical for it to distinguish foreign DNA from self DNA, which can be released to the extracellular space from necrotic cells107. TLR9 expression was upregulated in B cells stimulated by an anti-CD40 mAb or BCR crosslinking agents119. CD40 expression was upregulated in a time- and dose-dependent fashion in human B cells stimulated by CpG DNA, but not by control GpC DNA120, suggesting that a reciprocal activation of CD40 and TLR9 exists to establish a positive feedback loop in regulating their expression. CpG DNA induced expression of TLR9 in B cells, thereby possibly further enhancing signaling119. Further, primary human B cells stimulated by CD154, BCR crosslinking agents and CpG DNA responded with a higher proliferation rate, a stronger aicda induction and a higher CSR efficiency than their counterparts stimulated by CD154 and BCR crosslinking only, thereby suggesting that TLR9 plays an important role in enhancing the CD40 and BCR signals that lead to AID induction and CSR111. Finally, in human B cells, the TLR9 TIR domain, MyD88, Irak1 and Traf6, but not Traf2, were critical for CpG DNA and IL-10-induced aicda expression and CSR121.
In contrast to its enhancement of CD40− and BCR-induced CSR, CpG DNA suppressed IL-4-induced CSR to IgG1 and IgE in a TLR9- and MyD88-dependent fashion120,122,123, possibly in a fashion that is independent of downregulation of AID expression. More likely, this specific CpG DNA-induced suppression of CSR is mediated by T-bet, a T-box family transcription factor that selectively downregulates germline Iγ1-Cγ1 and Iε-Cε transcription, and upregulates germline Iγ2a-Cγ2a and/or Iγ2b-Cγ2b transcription120,122,123. Finally, that CpG modulates aicda expression through pathways independent of Id2, an E-box transcriptional suppressor that participates in the negative regulation of CD40− and IL-4 receptor-mediated aicda induction (see below), is suggested by the demonstration that the suppression of CSR to IgE by CpG DNA does not involve Id2124.
3.2. TRANSCRIPTION PROGRAMS REGULATING aicda GENE EXPRESSION
In germinal center B cells, a characteristic gene expression profile arises that is distinct from that in naïve B cells, post-switched plasma cells or memory B cells. Although proteomic approaches are starting to be used to identify signature gene expression profiles, the prevalent approach in analyzing such profiles in germinal center B cells is quantitative RT-PCR and/or DNA microarray. These two approaches allow only for analyses of gene transcripts that show changes in steady-state level, while excluding analyses of post-translational modification. Due to this limitation, here we will discuss mainly the modulation of aicda expression by transcriptional regulators, whose own expression is regulated during the germinal center reaction.
A typical gene expression profile in cells undergoing differentiation consists of at least three hierarchical levels: (i) genes whose expression directly specifies major cellular functions, hence the term “terminal node” genes; (ii) genes that encode transcription factors which directly regulate terminal node gene expression; hence the term “proximal node” genes; and (iii) genes that encode “master switch” transcription factors that control expression of proximal node genes; hence the term “master switch node”. Intermediate level nodes between master switch and proximal nodes may exist (Figure 3).
FIGURE 3.
Downregulation of aicda expression by Blimp1 and B cell plasmacytoid differentiation. (a) Expression profiles of genes that regulate aicda expression in germinal center B cells (left panel) and plasmacytes (right panel). Positive regulators and negative regulators of aicda transcription at each stage of the germinal center reaction are depicted in solid green and red circles, respectively. Broken circles depict genes with a low steady-state expression level. Solid and broken arrows/lines indicate direct and indirect regulation of gene expression, respectively. In germinal center B cells, multiple transcription factors synergize to induce aicda expression. In plasmacytes, the transcription suppressor Blimp1 downregulates expression of both aicda and the positive regulators of aicda expression, such as pax5, e2a and irf8 and upregulates negative regulators of aicda expression, such as id2. Irf4 switches from a positive to a negative regulator of aicda expression during plasmacytoid differentiation and its gradient-like expression pattern in light zone B cells is depicted as a light green circle. c-myc is one of the germinal center B cells-specific genes and cd138 (syndecan1); other genes involved in protein folding and secretion are highly expressed in plasmacytes. (b) Blimp1-binding site in the promoter of its direct target genes: “*” denotes predicted Blimp1-binding sites and “**” denotes the consensus Blimp1-binding motif135. Underlined nucleotides are present in all Blimp1-binding sites and likely contribute to Blimp1-DNA interaction that is essential for the transcription regulation activity of Blimp1.
In germinal center B cells where SHM and CSR unfold, aicda is a terminal node gene. In plasma cells that produce and secrete thousands of antibody molecules per second, terminal node genes include secretory pathway genes encoding factors involved in protein translocation across the ER membrane, protein folding and glycosylation in the ER and vesicle trafficking125 (Figure 3). For some terminal node genes, such as aicda, the expression is highly specific; many other terminal node genes are expressed in a broad range of cell types/tissues, such as secretory pathway genes of plasma cells.
Proximal nodes genes are often expressed in multiple cell types or at different differentiation stages of the same cell lineage at fluctuating rates. For instance, e2a, an aicda proximal node gene in germinal center B cells (see below), is also preferentially expressed in B cell and T cell progenitors where e2a-encoded E-box protein E12 and/or E47 are essential for Rag1- and Rag2-mediated gene recombination126. Proximal node genes function as signal “integrators”, i.e. their expression is induced by multiple signal transduction pathways, as activated by a combination of same stimuli that synergize to induce terminal node gene expression. In addition, several proximal node factors could be assembled into an “enhanceosome”127,128, which determines the cell type- or cell stage-specificity in terminal node gene induction. For instance, CD154, IL-4 or BCR crosslinking each induces aicda expression at a basal level, but they synergize to potentiate aicda induction through upregulation of several proximal node transcription factors (see below) (Table 3), thereby restricting aicda expression primarily in germinal center B cells. Negative proximal node genes contribute to the specificity of terminal node gene expression in sequential differentiation stages. For instance, downregulation of AID in plasma cells, possibly effected by Id2 and B lymphocyte-induced maturation protein 1 (Blimp1) (Figure 3), would ensure the preservation of Ig sequences that are fixed to encode high affinity and class switched mature antibodies and prevent potential genotoxic effects of AID in plasma cells.
Master switch node genes regulate the expression of genes at different levels, including terminal node, proximal node and intermediate node gene. In germinal center B cells, the master node gene pax5 directly regulates the expression of the terminal node gene aicda and an intermediate node gene B cell lymphoma 6 (bcl6). In fact, in addition to maintaining the B cell identity129, Pax5 is critical for the germinal center reaction, which is, at least partially, due to its specific induction of bcl6 and aicda in these cells. The linear cause-effect relationship of pax5 and bcl6130,131 suggests that bcl6 functions as a de facto master node gene, although whether bcl6 is a proximal node gene for aicda is unclear. An analogous master-intermediate node gene pair in plasma cells involves prdm1 and xbp1, which encode Blimp1 (also termed positive regulatory domain containing protein 1, Prdm1) and X-box binding protein 1 (Xbp1), respectively (Figure 3). A unique feature of most master switch node genes is their dual functions as both transcription activators and suppressors, as exemplified by the intertwined gene expression signature files in germinal center B cells and plasma cells (Figure 3). In germinal center B cells, Pax5 directly induces c-myc and aicda expression, but suppresses xbp1 expression132; Bcl6 forms a co-repressor complex with MTA3 and Mi-2/NuRD to directly suppress prdm1 expression133. Conversely, in plasma cells, Blimp1 positively regulates irf4 expression134, but directly suppresses pax5 expression, thereby relieving the inhibition of xbp1 expression135; Xbp1 regulates gene expression only positively, suggesting that it functions as an intermediate and/or proximal node gene, but not a de facto master node gene.
The reciprocal suppression between pax5-bcl6 and prdm1-xbp1 suggests the existence of a transitional state during which the pax5-bcl6 axis is gradually downregulated while the prdm1-xbp1 axis is upregulated136. Recent evidence suggests that the downregulation of pax5 precedes the initiation of prdm1 expression137, although the nature of events leading to the pax5 downregulation is still unclear. In addition, Irf4 controls prdm1 expression138,139, suggesting that Irf4 plays an important role during the transition to plasma cells. A further, and possibly more drastic, dowregulation of AID expression in this transitional state, as compared to the already decreased AID expression level in centrocytes (Figure 2), is possibly mediated by both Pax5 downregulation and expression of Blimp1. Here, we will review different levels of regulation, both positive and negative, of aicda expression, either direct or indirect, by Pax5, E47, HoxC4, Irf4, Irf8 and Blimp1, along the B cell germinal center reaction and plasmacytoid differentiation pathway (Figure 2).
3.2.1. Pax5
Pax5 is expressed from the pro-B cell to the mature B cell stage, being required to maintain B cell identity throughout the cell ontogeny129. In addition to a highly conserved paired DNA-binding domain (PRD) and a partial homeodomain, Pax5 contains a transcription-activation domain and a transcription-repression domain, which mediate its dual transcriptional regulation activity. Pax5 controls the expression of many B lineage-specific genes, including cd19, cd79a and the B cell linker protein-encoding gene blnk, through binding to their promoters and/or other regulatory elements140. In human and mouse germinal centers, the pax5 transcription level is significantly upregulated in centroblasts141 (Figure 2), possibly as a result of activated CD40 and/or IL-4 receptor signal transduction pathways, as suggested by the efficient Pax5 induction in murine spleen B cells stimulated with an agonistic anti-CD40 mAb142. In mouse spleen B cells stimulated with LPS, pax5 mRNA level increases by day 2, coincident with the initiation of aicda expression, and then decreases by day 4, with concomitant decrease of aicda expression38,141. Enforced overexpression of Pax5 in a mouse pro-B cell line induces expression of the otherwise silent aicda gene, suggesting that Pax5 indeed regulates AID expression, possibly through its recruitment to Pax5-binding sites in the aicda promoter143 (Table 3). The concomitant expression of Pax5 and AID is also kinetically associated with the expression of another transcription factor HoxC4 in mouse germinal center B cells as well as in in vitro-stimulated human and mouse B cells144 (see below) (Figure 2). Indeed, optimal induction of the aicda promoter requires both Pax5 and HoxC4144, suggesting that their simultaneous expression constitutes one of checkpoints that restrict AID expression to germinal center B cells, thereby avoiding its genotoxic effect in other cell types. Downregulation of expression of pax5, and, possibly, hoxc4 by Blimp1 during the plasmacytoid differentiation would contribute to the downregulation of AID and termination of CSR, as suggested by the abrogation of CSR in B cells in which pax5 expression is repressed by enforced Blimp1 expression. While stimulation of a subset of human chronic lymphocytic leukemia (CLL) B cells by CD154 plus IL-4 induced expression of the full length pax5 transcript and aicda, the expression of an alternative spliced form pax5/ΔEx, which encodes a Pax5 isoform without the activation domain but with a preserved suppression domain, was dampened145. Both full length Pax5 and Pax5/ΔEx8 bound to the aicda promoter, suggesting that a balance between two isoforms plays a role in regulating aicda expression. Such a balance could be further compounded if Pax5/ΔEx8 precludes other trans-factors, such as E47 and HoxC4, from accessing the aicda promoter.
3.2.2. E47
The e2a gene, through differential splicing, encodes two transcription factor E12 and E47. Both E12 and E47 belong to a family of basic helix-loop-helix (bHLH) proteins known as E-proteins, which bind DNA at E-box sites. Similar to that of pax5, expression of e2a is essential for B cell development, maturation and activation. In mouse germinal centers, E47 expression level is the highest in dark zone B cells, reduced in the light zone and drastically decreased during plasmacytoid differentiation. This expression pattern is similar to that of Pax5, HoxC4 and AID, suggesting that E47, Pax5 and HoxC4 form the aicda enhanceosome that co-integrates signal transduction pathways to activate aicda expression (Figure 2). In vitro, the DNA-binding activity of E47 is induced in B cells activated by CD40, LPS, IL-4 and BCR crosslinking146, the same signals that also induce aicda expression. E47 is recruited to the conserved region 2 (cr2) DNA in the first intron of the aicda gene through high-affinity binding to two E-box sites and transactivate the aicda promoter147. E47 may also directly bind the aicda promoter at another E-box site, which is in proximity of both Pax5- and HoxC4-binding sites (Table 3), and synergize with Pax5 and HoxC4 in aicda induction.
The transactivation activity of E12 and E47 is antagonized by the Id family of transcriptional inhibitor proteins, including Id1, Id2, Id3 and Id4, all of which lack the basic domain essential for DNA binding. E-proteins that heterodimerize with Id proteins cannot bind to E-box sites, thereby failing to transactivate their target genes126. In mature B cells, Id2 and Id3 are the two dominantly expressed Id proteins. Overexpression of Id3 in mouse splenic B cells results in a defective CSR, which can be partially rescued by enforced AID expression147, suggesting that E47 mediates CSR by activating AID expression. Paradoxically, Id3 expression is induced by BCR crosslinking and Id3 is critical for BCR signaling-activated, but not CD40, IL-4 or LPS-activated, B cell proliferation148. Thus, the Id3-mediated AID inhibition is independent of the B cell proliferation rate, suggesting that the downregulation of AID expression in the light zone is, at least partially, due to the Id3 expression in this area (Figure 2).
E47 expression is inhibited in plasma cells, possibly a result of decreased e2a gene expression, as effected by Blimp1134. Blimp1 downregulates the expression of Id3 in plasma cells134, suggesting that Id3 is not essential for the negative regulation of AID expression. In contrast, expression of another Id protein, Id2, may be indirectly upregulated by Blimp1 through the inhibition of Bcl6, a transcriptional suppressor of the id2 gene149. Id2 plays a role in the negative regulation of aicda expression, as suggested by the five-fold increase of the aicda transcript level in id2−/− B cells stimulated with LPS plus IL-4, as compared to that in wildtype B cells143. Conversely, overexpression of Id2 suppresses aicda induction, likely by antagonizing the transactivation activities of both E-proteins and Pax5143. The inhibitory effect of Id2 can be overcome by enforced overexpression of Pax5, suggesting that in dark zone B cells, where Pax5 and E2A are highly expressed, the potential inhibition of AID expression by Id2 is suppressed.
3.2.3. HoxC4
Hox proteins are a family of transcription factors that specify anterior-posterior axis differentiation and regulate cellular differentiation and organogenesis during embryogenesis150. All Hox transcription factors contain a helix-loop-helix homeodomain, as encoded by a highly conserved 180-bp homeobox DNA sequence. Hox genes within each family are chromosomally clustered151 and their expression is temporally and spatially regulated150,152. Among Hox genes, hoxc4 is preferentially expressed in B cells in the human and the mouse and expression and activities of HoxC4 are regulated in different stages of B cell development and activation144,153,154. In vitro, hoxc4 expression is induced by the same stimuli that induce efficient aicda expression: (i) CD154 plus IL-4 in human B cells; (ii) CD154, IL-4 plus TGF-β in the mouse B cell line CH12F3-2; and (iii) CD154 or LPS plus IL-4 in primary mouse spleen cells144 (Table 3). Conversely, in HoxC4-deficient B cells stimulated with CD154 and IL-4, aicda induction is significantly impaired, resulting, as compared to that in wildtype B cells144, strongly suggesting that HoxC4 regulates aicda gene expression. The impact of HoxC4 on aicda expression is further emphasized by the severe impairment of SHM in HoxC4-deficient mice155 and the significantly reduced CSR in hoxc4−/− B cells, which can be rescued by enforced expression of AID144.
The binding of Hox proteins to DNA through their homeodomain is inherently weak, but can be greatly enhanced by their interaction with transcription factors that are recruited to the same region in target genes, thereby effectively transactivating gene expression. For instance, HoxC4, together with Oct1 and/or Oct2 homeodomain transcription factors and OcaB co-activator, play a critical role in the induction of the IgH 3′ hs1,2 enhancer in human B cells by binding to the evolutionarily conserved HoxC4 5′ ATTT 3′ site and composite HoxC4/Oct 5′ ATTTGCAT 3′ site154. Likewise, an embedded HoxC4/Oct binding site 5′ ATTTGAAT 3′ in the aicda promoter is essential for aicda promoter induction by HoxC4144 (Table 3), through the assembly of the HoxC4/Oct1/2/OcaB complex, as part of the aicda enhanceosome (Figure 1). While Oct1/2 are highly expressed throughout the germinal center reaction, expression of OcaB is highest in dark zone B cells, at least partly due to its upregulation by the CD40, IL-4R and BCR signaling156, further emphasizing the synergistic aicda induction by the enhanceosome involving the HoxC4/Oct1/2/OcaB complex, Pax5 and E47. OcaB deficiency leads to impaired induction of prdm1 transcription and concomitant de-repression of expression of the pax5, bcl6 and aicda genes in cultured antibody-secreting cells157, suggesting that OcaB may switch from a positive to a negative regulator of aicda expression during plasmacytoid differentiation.
3.2.4. Irf4 and Irf8
Irf4 (also termed Pip, ICSAT and LSIRF) and Irf8 (also termed IFN consensus sequence-binding protein, ICSBP) are two closely related members of the Irf transcription factor family. They bind to elements in promoters of IFN-inducible genes through a DNA-binding domain that contains five conserved Trp repeats. Irf4 and Irf8 interact with each other as well as other transcription factors, including SpiB, Pu.1, E47 and Stat6 to regulate cell proliferation, apoptosis and differentiation158. Like HoxC4, Irf4 and Irf8 possess weak intrinsic DNA-binding activity and are only recruited to DNA as components of protein complexes, such as Pu.1-Irf4 and SpiB-Irf8, that bind to the Ets-Irf composite elements (EICE)159.
Expression of Irf4 and Irf8 is restricted to B and T lymphocytes and other immune cells. In the mouse, Irf4 expression is high in bone marrow immature B cells, decreases in mature B cells, further decreases in centroblasts in germinal centers, and then increases again to become maximal in plasma cells160,161 (Figure 2). This expression pattern of Irf4 is consistent with an important role of Irf4 in initiating aicda expression in early centroblasts and CSR, as suggested by the impaired aicda induction and CSR in irf4−/− B cells stimulated with CD154 or LPS plus IL-4, and impaired plasmacytoid differentiation in mice deficient in Irf4 expression in germinal centers138,139. Irf4 likely regulates AID expression through other trans-factors, such as Pax5, which are induced in germinal center B cells but downregulated in plasma cells by Blimp1139. Alternatively, Irf4 may suppress the expression of a putative transcription repressor of aicda, i.e. Id2, in germinal center B cells, as suggested by the increased id2 gene expression level observed in vitro stimulated irf4−/− B cells138. Accordingly, Irf4 may regulate the expression and activity HoxC4 or other components of the aicda enhanceosome.
Irf4 and Irf8 signal transduction pathways are partially overlapping, as suggested by the complete block of the pre-B to B cell transition in the bone marrow of irf4−/− irf8−/− doubly-deficient mice but not in irf4−/− or irf8−/− mice162. Irf8 displays an expression pattern similar to that of Irf4, albeit with some difference, as it is expressed in bone marrow immature B cells, downregulated in peripheral mature B cells, maximally expressed in dark zone B cells and then downregulated in plasma cells163 (Figure 2). Consistent with the expression and function of Irf8 in centroblasts, Irf8 likely binds to the bcl6 or aicda promoter and possibly regulates their expression, as suggested by the significant reduction of bcl6 or aicda transcripts in irf8−/− B cells, as compared to wildtype B cells. Irf8 expression is negatively regulated by Blimp1 during plasmacytoid differentiation134, either directly or through negatively regulated expression of bcl6, thereby contributing to the overall downregulation of aicda expression. A short stretch of the aicda promoter sequence 5′-ATTTTAAAAA[AAAAGAAA]AAT-3′ (Table 3) contains two HoxC4-bindings sites, which are in addition to the embedded HoxC4/Oct site discussed above, and one consensus Irf-binding site [AAxxGAAA]. HoxC4 and Irf8 may co-occupy this site to regulate aicda expression, in a fashion similar to the regulation of the Igλ enhancer by Pu.1/Irf4 complex through a composite EICE 5′-AAAAGG[AAGTGAAA]CCA-3′159.
3.2.5. Blimp1
As the master node gene controlling plasmacytoid differentiation, Blimp1 effectively inhibits gene expression of an array of transcription factors, including Pax5, E2A, Irf8, Pu.1, SpiB and Stat6134, all involved in the positive regulation of aicda expression. Downregulation of these factors is partially responsible for the Blimp1-mediated aicda downregulation. Like the direct downregulation of Blimp1 target genes bcl6, spiB and id3, the downregulation of aicda expression cannot be rescued by the re-expression of Pax5, suggesting a direct suppression of the aicda promoter by Blimp1 through a putative Blimp1-binding site134,144 (our unpublished data, Figure 3). In addition, Blimp1 positively regulates expression of Id2 and Irf4, albeit indirectly, during plasmacytoid differentiation. As discussed above, an increased level of Id2 inhibits Pax5 and E2A activities. Although Irf4 upregulates aicda expression in early centroblasts, it does so indirectly, either through upregulating a positive intermediate trans-factor, such as Pax5, or through suppression of a negative intermediate factor, such as Id2. Both Irf4 activities can be overridden by the powerful transcriptional suppression effects of Blimp1. Finally, Blimp1 expression is also upregulated by Irf4, thereby turning Irf4 from a positive to a negative aicda regulator138,139. In summary, Blimp1 may inhibit AID expression through at least three different mechanisms, by: (i) direct suppression; (ii) suppression of positive regulators, such as Pax5, E2A, Irf8 and possibly HoxC4; and, finally, (iii) induction of negative regulators, such as Id2. Blimp1-mediated negative regulation of AID expression is consistent with the increased CSR in prdm1−/− cells stimulated with LPS or LPS plus IL-4164.
During the germinal center reaction, Bach2, a transcriptional repressor, represses Blimp1 expression165, thereby relieving Blimp1-mediated AID downregulation, as suggested by the striking reduction of aicda transcription and CSR in bach2−/− B cells stimulated with LPS166. The impaired CSR in stimulated bach2−/− B cells could not be rescued by AID re-expression166, suggesting that Bach2 also regulates expression of other genes critical for the germinal center reaction, SHM and CSR166. How Bach2 is downregulated during plasmacytoid differentiation to allow Blimp1 expression is unclear.
The recent finding that pax5 downregulation precedes the initiation of prdm1 induction suggests that downregulation of bcl6 is critical in this transitional stage, as Bcl6 is a direct negative regulator of the prdm1 gene. In addition, expression of the Bcl6 protein is regulated at a post-transcriptional/post-translational level, as implied by the ubiquitination and subsequent proteasome-dependent degradation of phosphorylated Bcl6, as induced by BCR crosslinking in spontaneously hypermutating Ramos B cells167, suggesting that engagement of a somatically hypermutated and high-affinity BCR by antigen leads to decreased Bcl6 protein level. Finally, the balance between the Irf4 and Bcl6, the positive and negative regulator of the prdm1 gene, respectively, likely determines how Blimp1 expression is initiated during plasmacytoid differentiation, as both Irf4 and Bcl6 bind to a conserved non-coding region of the intron 5 of the prdm1 gene.
Impaired SHM results in failure to generate high-affinity BCR molecules, which, in turn, may result in a defective plasmacytoid differentiation. That Blimp1 expression is indeed driven by a high-affinity antigen-BCR interaction is demonstrated by studies using transgenic HyHEL10 mice, which express a high-affinity BCR specific for hen egg lysozyme (HEL)168. Immunization with HEL led to normal germinal center formation and plasmacytoid differentiation of adoptively transferred HyHEL10 B cells, in the absence of mutations in the HyHEL10 transgene in plasma cells. In contrast, immunization with a mutated HEL (HEL3X), which displays a 10,000-fold lower affinity for HyHEL10, resulted in an Tyr53 to Asp53 amino acid change in HyHEL10, which increases the affinity of HyHEL10 for HEL3X, but not to HEL. Further, in B cells double-transgenic for HyHEL10 and prdm1+/GFP, GFP, whose expression is driven by the prdm1 promoter, was expressed almost exclusively in B cells that acquired a high affinity anti-HEL3X HyHEL10, suggesting a critical role for BCR signaling in Blimp1 expression and AID downregulation.
One of the major pathways activated by BCR signaling is the PI 3-K pathway98. In B cells deficient in Pten, reduced AID expression and impaired CSR could be partially rescued by a putative PI 3-Kδ inhibitor, IC87114164,169, suggesting a role of PI 3-K in downregulating AID expression/activity. The Pten deficiency-mediated CSR impairment could also be partially restored by Blimp1 deficiency164, suggesting that prdm1 expression is downstream of PI 3-K pathways. Increased CSR in prdm1−/− B cells could be further enhanced by the PI 3-K inhibitor IC87114, suggesting a Blimp1-independent negative regulation of CSR by PI 3-K. In summary, PI 3-K signaling likely plays a double role in the germinal center reaction and plasmacytoid differentiation by: (i) enhancing AID induction, as triggered by BCR crosslinking in early centroblasts to initiate SHM and CSR; and (ii) activating Blimp1 and/or other factors to downregulate AID expression and extinguish CSR upon strong antigen-BCR interaction, i.e., crosslinking of hypermutated and high affinity BCRs, in the post-SHM stage.
3.3. POST-TRANSLATIONAL REGULATION OF AID ACTIVITY
AID undergoes a series of post-translational modification, such as dimerization/oligomerization, nuclear/cytoplasmic translocation and phosphorylation, all of which play important roles in regulating AID activity and function in SHM and CSR. The importance of AID post-translational modifications is further emphasized by the failure of enforced AID expression to induce SHM or CSR in transgenic mice170. Here, we will discuss these AID post-translational modifications with an attempt to identify the physiologically active form of AID and the regulation of such specific AID form. Not only expression of the aicda gene is restricted, as effected by the assembly of the aicda enhanceosome, in germinal center B cells, and activity of AID is tightly controlled, as effected by its post-translational modification, an additional level of AID regulation is the alternative splicing of the primary aicda transcript(s). Two major aicda mRNA transcripts in activated mouse B cells result from differential polyadenylation9; and only one major aicda mRNA transcript is expressed in human B cell lines of germinal center origin, such as Ramos and Raji171. However, up to five alternative aicda mRNA molecules were identified in B cells from patients with CLL172,173. While putative AID mutants encoded by those transcripts await biochemical and functional characterization, it is clear that some of alternative aicda mRNA transcripts are implicated in the prognosis of CLL; and readers are referred to the original papers172,173.
3.3.1. Dimerization/Oligomerization
In activated B cell nuclei, the single-stranded DNA deamination activity of AID existed in a broad fraction containing proteins or protein complexes of 20 to 70 kDa in size174, suggesting a heterogeneity of AID oligomer and/or AID/co-factor complex. Indeed, recombinant AID expressed in E. coli or non-B mammalian cells formed dimers and/or oligomers175. Exogenous AID expressed in HEK293 cells existed as an oligomer, or within a small protein complex, about 30–60 kDa in size, at a high salt concentration, i.e. 750 mM NaCl; at a low salt concentration, i.e. 100 mM NaCl, AID exists as a multimer, or within a large protein complex, about more than 500 kDa in size176. Both the 500 kDa and the 30–60 kDa protein complexes displayed the dC deaminase activity176. It is conceivable that the AID monomer, which is 20 kDa in size, forms a stable dimer/trimer or strongly interacts with a putative binding partner (or a complex of several smaller proteins) about 40 kDa in size. The 32 kDa subunit of the replication protein A (RPA) complex, which specifically binds to phosphorylated AID in B cells, but not to non-phosphorylated AID expressed in HEK293 cells174 (see below), does not seem to be the putative partner. Recombinant AID with a streptavidin tag (StrepAID) expressed in E. coli migrated at about 80 kDa on an SDS-PAGE, reflecting the formation of a tetramer that is resistant to the denaturation by EDTA or β-mercaptoethanol. Such StrepAID tetramer could be disassociated into a monomer and a stable trimer upon treatment with EDTA together with strong denaturants, such as urea or guanidine, further emphasizing the ability of AID to form complex oligomers177.
It is suggested that AID dimerization/oligomerization is essential for the processive deamination of cytidines in single-stranded DNA by AID178,179. Structural analyses of human AID, as modeled using the solved crystal structure of human Apobec2, which share 33% identity and 44% homology overall and 75% identity and 96% homology in buried residues with AID, suggest that a β-strand (β-2 strand) encompassing amino acid residues from Ser41 to Asn51 in each AID monomer provides the major interface of the two monomers180 (Figure 4). Two AID dimers may further form head-to-head tetramers, as suggested by the structure of Apobec2180. AID with the double mutation Phe46Ala/Tyr48Ala in the β-2 strand displayed a 4-fold decrease in deaminase activity, likely due to the weakened, but not abolished, dimerization of this AID mutant180. The extensive interface between the two AID β-2 strands suggests that a single amino acid mutation within the β-2 strand may result in only a partial loss of the deaminase activity and the residual AID activity may still be sufficient to mediate SHM and CSR. Amino acid residues between Gly47 and Gly54 have been shown to be important for AID dimerization and CSR175, which is largely consistent with the modeled AID structure.
FIGURE 4.
Predicted three-dimensional structure of the human AID dimer (Egest, Xu and Casali, unpublished data). The structure of amino acid residues 11 to 180 of AID is modeled, as based on the solved crystal structure of the human Apobec2 (PDB No: 2NYT) (33.3% identity and 44% homology). The structure of the carboxyl-terminal NES (amino acid residues 184–198) is modeled, as based on the solved crystal structure of the human p53 tetramerization domain (PDB No: 1C26) (33% homology), which contains a helical NES. The Pro 183 of AID likely hampers the formation of a long helix spanning carboxyl-terminal amino acid residues 168–198. Shown are the side chains of His56, Glu58, Cys87 and Cys90, which forms the active site in the AID cytidine deamination domain. His56, Cys87 and Cys90 coordinate with a Zn2+ ion, which is essential for the AID activity. The conformational change of the β1′-turn from a loop to a hairpin, as shown in purple, likely controls the access of the AID active site to DNA/RNA substrates. The β2 strand (amino acid residues Ser 41 to Asn 51) of each AID monomer mediates the dimerization. Shown is the side chain of Ser38, the PKA phosphorylation site.
Several carboxyl-terminal truncated AID mutants isolated from HIGM2 patients displayed normal cytidine deaminase activity and effectively drive SHM in fibroblasts181, suggesting that defective CSR results from the inability of these AID mutants to interact with a critical CSR-specific cofactor(s). A likely role of the AID dimerization/oligomerization in mediating CSR is to interact with a dimer/multimer of the elusive CSR co-factor, as the interaction between monomeric AID and this putative CSR co-factor might be insufficient to target AID to the IgH locus. As such, the eight carboxyl-terminal amino acid resides would be essential for this protein-protein interaction, as suggested by the dominant negative activity of an AID mutant truncated at Arg190181. The formation of a dimer/oligomer consisting of both wildtype and this truncated AID would best explain the dominant negative activity of the AID mutant, as suggested by the intermediate HIGM2 phenotypes in patients that are heterozygotic for the mutated allele181.
3.3.2. Nuclear/Cytoplasmic Translocation
AID is specifically expressed in germinal center B cells. Subcellular fractionation analyses of tonsil germinal center B cells demonstrated that AID is predominant in the cytoplasm182. As a small size protein of 20 kDa, which would otherwise freely diffuse through nuclear membrane pores, AID is either retained in the cytoplasm after translation or actively transported outside the nucleus. An efficient nuclear export would explain the low steady-state level of AID protein level in the nucleus, although this can also be explained by a non-mutually exclusive possibility that AID is subjected to a faster degradation in the nucleus. In cells treated with leptomycin B, which blocks nuclear transport by specifically inhibiting the ubiquitous soluble shuttle receptor chromosome region maintenance/exportin 1 (CRM1), AID localized in both nucleus and cytoplasm, suggesting that AID is actively transported out of the nucleus in a CRM1-dependent fashion183,184. The AID carboxyl-terminal 189LRDAF(R/K)(T/M)LGL/F198 sequence is rich in Leu and hydrophobic Phe residues, thereby providing a putative nuclear export signal (NES) (Figure 4). Indeed, truncation of the carboxyl-terminal sequence or mutation of Phe198 to Ala led to nuclear localization of GFP-tagged AID, suggesting that this NES mediates the cytoplasmic localization of GFP-tagged AID and, likely, endogenous AID.
As a DNA or pre-RNA deaminase, AID would perform its function in the nucleus. It is possible that the low AID level in the nucleus is sufficient to exert its deamination activity in germinal center B cells. However, it is unlikely that the larger amount of AID localizing in the cytoplasm is turned over without expressing its activity, thereby raising the question of when and how AID is translocated from the cytoplasm into the nucleus. It is suggested that a bipartite nuclear localization signal (NLS) at the amino-terminus of human and mouse AID, together with the carboxyl-terminal NES, effectively regulates the dynamic shuttling of AID between the cytoplasm and the nucleus183. However, the existence of such NLS has not yet been independently verified184,185.
Although the molecular mechanism that governs the balance of AID nuclear import/export is unclear, it is conceivable that such a molecular mechanism provides B cells with two important functional features to: (i) hamper the highly active and mutagenic AID from accessing its substrate, DNA or pre-RNA, thereby preventing indiscriminate cytidine deamination, DNA damage and/or insertion of deleterious mutations, as suggested by the higher SHM rate brought by an AID mutant lacking the NES on an artificial DNA substrate183,184; and (ii) generate a cytoplasmic AID reservoir, in which AID could be assembled with either SHM or CSR co-factors into a protein complex that, upon delivery of appropriate stimuli, can be promptly translocated into the nucleus and preferentially targeted to the Ig locus (Figure 5). Such a fast-kinetic nuclear localization of AID is suggested by the rapid nuclear accumulation of red fluorescent protein (RFP)-tagged AID when cells are treated with agents causing DSBs185. It would be important to identify the stimulus that directs the nuclear localization of AID and analyze its role in regulating SHM and CSR during the germinal center reaction.
FIGURE 5.
An integrated model of AID post-translational modifications. AID forms dimers/oligomers spontaneously. Most AID molecules localize in the cytoplasm, likely through active nuclear export, as effected by CRM1. Phosphorylation of cytoplasmic AID dimers by PKA would result in efficient nuclear import. After localizing to the nucleus, phosphorylated AID dimers binds to RPA and a putative AID cofactor, depicted as a dimer, to effect its DNA deaminase functions. Alternatively, PKA-mediated phosphorylation of cytoplasmic AID dimers increases AID binding to the putative cofactor before translocation into the nucleus, where the AID-cofactor complex binds to RPA and then deaminates dC in DNA. After deamination, AID is released, possibly dephosphorylated by a phosphatase and then exported out of the nucleus to replenish the cytoplasmic AID reservoir.
3.3.3. Phosphorylation
Recombinant AID displays deaminase activity on single-stranded, but not double-stranded, DNA substrates176,178. When actively transcribed, double-stranded DNA can be effectively deaminated by phosphorylated AID, but not by non-phosphorylated AID174,176, suggesting that phosphorylation plays an important role in targeting AID to its natural substrates, i.e., functionally single-stranded DNA, as occurring in actively transcribed Ig V(D)J and S regions. When expressed in HEK293 cells, AID is phosphorylated at a much lower degree, as compared to AID protein purified from in vitro activated mouse spleen B cells118,174,186 (Figure 4), suggesting a B cell-specific phosphorylation of AID. AID phosphorylation may explain the apparent size heterogeneity of the AID-containing complexes, ranging from 20 kDa to 70 kDa, in the B cell nucleus174.
In activated B cells, AID can be phosphorylated at Ser38 and Tyr184118,174,186. The functional meaning of AID Tyr184 phosphorylation is unclear. PKA-mediated phosphorylation of Ser38, which is located within a consensus PKA phosphorylation site RRxS118,186,187, is critical for AID to interact with the RPA complex, which potentiates AID deamination of transcribed double-stranded DNA174,186,188–190. Impaired CSR in in vitro stimulated aicda−/− B cells can be rescued by wildtype AID, but not by an AID molecule carrying a Ser38Ala mutation. An AID Ser38Asp mutant, which likely mimics the constitutively phosphorylated AID at Ser38, is defective in rescuing CSR in aicda−/− B cells118,174,186, suggesting that a dynamic phosphorylation/dephosphorylation process is critical in regulating AID activity. Further, in CH12F3 mouse B cells treated with PKA inhibitors, CSR to IgA is reduced by five folds, as compared to similar cells untreated or treated with inhibitors of other kinases, such as PI 3-K. Conversely, in in vitro stimulated B cells that are deficient in PKA R1α, one of the two inhibitory subunits of the PKA holoenzyme, and displaying a PKA kinase activity increased by four folds, CSR is significantly upregulated, further underlying the important role of PKA in regulating AID activity and CSR187. Both Ser38Ala and Ser38Asp AID mutants display a reduced activity in mediating SHM of an artificial substrate, suggesting that AID phosphorylation at Ser38 is also critical in regulating the SHM process118.
Only a fraction of all AID molecules is phosphorylated at Ser38118. However, phosphorylated AID is enriched in the chromatin-associated nuclear fraction, as compared to the cytoplasmic and soluble nuclear fractions118, suggesting that phosphorylated AID is the active form of AID that exerts its DNA deaminase function in the nucleus. This selective enrichment of phosphorylated AID to chromatin likely results from either AID phosphorylation by PKA localizing in the nucleus or selective import of phosphorylated AID from the cytoplasm to the nucleus. Transiently expressed Flag-tagged AID specifically bound to S region DNA and to RNA polymerase II, thereby linking germline IH-CH transcription with AID targeting to S region DNA191. It is possible that phosphorylated endogenous AID preferentially targets V(D)J or S region DNA, as compared to non-phosphorylated AID. If AID phosphorylation and its nuclear localization are indeed linked, there may exist a phosphatase that dephosphorylates AID shortly after the dissociation of AID from V(D)J DNA and/or export of AID out of the nucleus (Figure 5). How PKA and, possibly, the putative AID phosphatase, in B cells are activated by CD40, IL-4R, BCR and/or TLR signals remains to be determined. Finally, if phosphorylation precedes AID dimerization/oligomerization, both phosphorylated and non-phosphorylated forms of AID would be present in the AID dimer/oligomer, which would be less active than an AID dimer/oligomer containing only the phosphorylated form. It is likely that AID dimerization/oligomerization occurs before phosphorylation and only a selective fraction of AID dimer/oligomers are phosphorylated (Figure 5).
4. Conclusions
The highly active nature and tightly regulated levels of AID in B cells account for the significant, albeit transient, burst of events leading to SHM and CSR in germinal centers. Restricted aicda expression in germinal center B cells is induced by specific stimuli from CD4+ T cells and/or invading microbial pathogens and is effectively downregulated by Blimp1 during plasmacytoid differentiation. AID induction is likely mediated by the formation of an enhanceosome consisting of specific transcription factors upregulated in centroblasts, including Pax5, HoxC4/Oct1/2/OcaB, E47 and Irf8. Blimp1 likely exerts its transcription repressor functions on multiple genes, thereby effectively shutting down aicda expression. A major fraction of AID is non-phosphorylated and confined to the cytoplasm, a compartment where AID cannot access DNA or pre-RNA, thereby avoiding its potential genotoxic effects. In spite of our knowledge about regulation of aicda expression and AID activities, several important issues remain to be addressed, including: (i) identifying the dynamic composition of the aicda enhanceosome. To address this issue, a high-resolution proteomic approach would be required to analyze how expression of transcription factors, such as Pax5, HoxC4, E47, Irf4, Irf8 and Blimp1, changes from centroblasts in the dark zone to centrocytes in the light zone and then to plasmablasts, and systematically analyzing the role of these transcription factors in SHM and CSR; (ii) addressing how PKA and, possibly, a putative AID phosphatase, are regulated to modulate AID activity, SHM and CSR; (iii) defining the modulation of AID protein level and activity170; and, finally, (iv) unraveling the mechanism(s) mediating the specific targeting of AID to the Ig locus, possibly as a result of multiple interactions between Ig cis-elements and germinal center-specific trans-factors.
Acknowledgments
Owing to space limitations, we could cite only a fraction of the papers relevant to the topic discussed in this review article. We apologize to the authors of the publications that are not cited here. This work was supported by NIH grants AR 40908, AI 45011 and AI 60573 to P.C.
References
- 1.Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med. 1993;178:1293–307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nat Rev Immunol. 2002;2:605–14. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–52. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 4.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 5.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–83. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 6.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–45. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 8.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–6. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 10.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–77. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 11.Schlissel MS, Kuo TC. AID for innate immunity to retroviral transformation. Immunity. 2006;24:671–2. doi: 10.1016/j.immuni.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 13.Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–61. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Fulop Z, Zhong Y, Evinger AJ, III, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann NY Acad Sci. 2005;1050:146–62. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenter AL. Class switch recombination: an emerging mechanism. Curr Top Microbiol Immunol. 2005;290:171–99. doi: 10.1007/3-540-26363-2_8. [DOI] [PubMed] [Google Scholar]
- 16.Martomo SA, Gearhart PJ. Somatic hypermutation: subverted DNA repair. Curr Opin Immunol. 2006;18:243–8. doi: 10.1016/j.coi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–8. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–21. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–71. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase h is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 21.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–53. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase ζ. J Immunol. 2001;167:327–35. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 23.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA, Weill JC. DNA polymerase η is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–70. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase η- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–61. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–6. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci USA. 2005;102:13986–91. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–8. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–23. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z, Zan H, Pal Z, Casali P. DNA replication to aid somatic hypermutation. Adv Exp Med Biol. 2007;596:111–27. doi: 10.1007/0-387-46530-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zan H, Cerutti A, Dramitinos P, Schaffer A, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β: evidence for TGF-β but not IL-10-dependent direct Sμ→Sα and sequential Sμ→Sγ, Sγ→Sα DNA recombination. J Immunol. 1998;161:5217–25. [PMC free article] [PubMed] [Google Scholar]
- 32.Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: definition of the requirements and modalities of hypermutation. J Immunol. 1999;162:3437–47. [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–5. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 34.Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–6. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 35.Jabara H, Laouini D, Tsitsikov E, Mizoguchi E, Bhan A, Castigli E, Dedeoglu F, Pivniouk V, Brodeur S, Geha R. The binding site for TRAF2 and TRAF3 but not for TRAF6 is essential for CD40-mediated immunoglobulin class switching. Immunity. 2002;17:265–76. doi: 10.1016/s1074-7613(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 36.Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–38. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain A, Ma CA, Lopez-Granados E, Means G, Brady W, Orange JS, Liu S, Holland S, Derry JM. Specific NEMO mutations impair CD40-mediated c-Rel activation and B cell terminal differentiation. J Clin Invest. 2004;114:1593–602. doi: 10.1172/JCI21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–8. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Ginkel FW, Wahl SM, Kearney JF, Kweon MN, Fujihashi K, Burrows PD, Kiyono H, McGhee JR. Partial IgA-deficiency with increased Th2-type cytokines in TGF-β 1 knockout mice. J Immunol. 1999;163:1951–7. [PubMed] [Google Scholar]
- 40.Cazac BB, Roes J. TGF-β receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–51. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 41.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 42.Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–9. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castigli E, Scott S, Dedeoglu F, Bryce P, Jabara H, Bhan AK, Mizoguchi E, Geha RS. Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci USA. 2004;101:3903–8. doi: 10.1073/pnas.0307348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, Geha RS. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–34. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 45.Castigli E, Wilson SA, Scott S, Dedeoglu F, Xu S, Lam KP, Bram RJ, Jabara H, Geha RS. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med. 2005;201:35–9. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagarasan S, Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol. 2003;3:63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 47.Mackay F, Schneider P, Rennert P, Browning J. BAFF and APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–64. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 48.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol. 2005;17:282–9. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Bossen C, Schneider P. BAFF, APRIL and their receptors: structure, function and signaling. Semin Immunol. 2006;18:263–75. doi: 10.1016/j.smim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Cancro MP. The BLyS/BAFF family of ligands and receptors: key targets in the therapy and understanding of autoimmunity. Ann Rheum Dis. 2006;65(Suppl 3):iii34–6. doi: 10.1136/ard.2006.058412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dillon SR, Gross JA, Ansell SM, Novak AJ. An APRIL to remember: novel TNF ligands as therapeutic targets. Nat Rev Drug Discov. 2006;5:235–46. doi: 10.1038/nrd1982. [DOI] [PubMed] [Google Scholar]
- 52.Quezada SA, Jarvinen LZ, Lind EF, Noelle RJ. CD40/CD154 interactions at the interface of tolerance and immunity. Annu Rev Immunol. 2004;22:307–28. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- 53.Armitage RJ, Fanslow WC, Strockbine L, Sato TA, Clifford KN, Macduff BM, Anderson DM, Gimpel SD, Davis-Smith T, Maliszewski CR, et al. Molecular and biological characterization of a murine ligand for CD40. Nature. 1992;357:80–2. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 54.Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, Bajorath J, Grosmaire LS, Stenkamp R, Neubauer M, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 55.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–78. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–31. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 57.Durandy A, Peron S, Fischer A. Hyper-IgM syndromes. Curr Opin Rheumatol. 2006;18:369–76. doi: 10.1097/01.bor.0000231905.12172.b5. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Carter RH. CD19 regulates B cell maturation, proliferation, and positive selection in the FDC zone of murine splenic germinal centers. Immunity. 2005;22:749–61. doi: 10.1016/j.immuni.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 59.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–31. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 60.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180:329–39. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu YJ, Malisan F, de Bouteiller O, Guret C, Lebecque S, Banchereau J, Mills FC, Max EE, Martinez-Valdez H. Within germinal centers, isotype switching of immunoglobulin genes occurs after the onset of somatic mutation. Immunity. 1996;4:241–50. doi: 10.1016/s1074-7613(00)80432-x. [DOI] [PubMed] [Google Scholar]
- 62.Cerutti A, Schaffer A, Goodwin RG, Shah S, Zan H, Ely S, Casali P. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J Immunol. 2000;165:786–94. doi: 10.4049/jimmunol.165.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaffer A, Kim EC, Wu X, Zan H, Testoni L, Salamon S, Cerutti A, Casali P. Selective inhibition of class switching to IgG and IgE by recruitment of the HoxC4 and Oct-1 homeodomain proteins and Ku70/Ku86 to newly identified ATTT cis-elements. J Biol Chem. 2003;278:23141–50. doi: 10.1074/jbc.M212952200. [DOI] [PubMed] [Google Scholar]
- 64.Zan H, Pal Z, Yel L, Casali P. AID- and Ung-dependent generation of resected double-strand DNA breaks in immunoglobulin class switch DNA recombination. 2007 doi: 10.1016/j.molimm.2008.07.003. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103:7396–401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, van Kooten C, Liu YJ, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 67.Cattoretti G, Buttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 2006;107:3967–75. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 68.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–73. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 69.Yang G, Obiakor H, Sinha RK, Newman BA, Hood BL, Conrads TP, Veenstra TD, Mage RG. Activation-induced deaminase cloning, localization, and protein extraction from young VH-mutant rabbit appendix. Proc Natl Acad Sci USA. 2005;102:17083–8. doi: 10.1073/pnas.0501338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc Natl Acad Sci USA. 2005;102:13242–7. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahonen C, Manning E, Erickson LD, O’Connor B, Lind EF, Pullen SS, Kehry MR, Noelle RJ. The CD40-TRAF6 axis controls affinity maturation and the generation of long-lived plasma cells. Nat Immunol. 2002;3:451–6. doi: 10.1038/ni792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerutti A, Schaffer A, Shah S, Zan H, Liou HC, Goodwin RG, Casali P. CD30 is a CD40-inducible molecule that negatively regulates CD40-mediated immunoglobulin class switching in non-antigen-selected human B cells. Immunity. 1998;9:247–56. doi: 10.1016/s1074-7613(00)80607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, Wood P, Rabia SH, Headon DJ, Overbeek PA, Le Deist F, Holland SM, Belani K, Kumararatne DS, Fischer A, Shapiro R, Conley ME, Reimund E, Kalhoff H, Abinun M, Munnich A, Israel A, Courtois G, Casanova JL. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat Genet. 2001;27:277–85. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 74.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2:223–8. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 75.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFκB. Int Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 76.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–38. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 77.Keegan AD, Nelms K, White M, Wang LM, Pierce JH, Paul WE. An IL-4 receptor region containing an insulin receptor motif is important for IL-4-mediated IRS-1 phosphorylation and cell growth. Cell. 1994;76:811–20. doi: 10.1016/0092-8674(94)90356-5. [DOI] [PubMed] [Google Scholar]
- 78.Hanson EM, Dickensheets H, Qu CK, Donnelly RP, Keegan AD. Regulation of the dephosphorylation of Stat6. Participation of Tyr-713 in the interleukin-4 receptor alpha, the tyrosine phosphatase SHP-1, and the proteasome. J Biol Chem. 2003;278:3903–11. doi: 10.1074/jbc.M211747200. [DOI] [PubMed] [Google Scholar]
- 79.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300:1527–8. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 80.Kajiwara K, Shinazawa M, Morishima H, Yanagihara Y. Differential effect of IL-4 and IL-13 on the expression of recombination-activating genes in mature B cells from human peripheral blood. Cell Immunol. 2004;227:121–8. doi: 10.1016/j.cellimm.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Punnonen J, Aversa G, Cocks BG, McKenzie AN, Menon S, Zurawski G, de Waal Malefyt R, de Vries JE. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90:3730–4. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 83.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–12. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 84.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 85.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 86.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 87.Zhou C, Saxon A, Zhang K. Human activation-induced cytidine deaminase is induced by IL-4 and negatively regulated by CD45: implication of CD45 as a Janus kinase phosphatase in antibody diversification. J Immunol. 2003;170:1887–93. doi: 10.4049/jimmunol.170.4.1887. [DOI] [PubMed] [Google Scholar]
- 88.Shen CH, Stavnezer J. Interaction of stat6 and NF-κB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Niiro H, Clark EA. Regulation of B-cell fate by antigen-receptor signals. Nat Rev Immunol. 2002;2:945–56. doi: 10.1038/nri955. [DOI] [PubMed] [Google Scholar]
- 90.Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–27. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 91.Hou P, Araujo E, Zhao T, Zhang M, Massenburg D, Veselits M, Doyle C, Dinner AR, Clark MR. B cell antigen receptor signaling and internalization are mutually exclusive eventsg. PLoS Biol. 2006;4:e200. doi: 10.1371/journal.pbio.0040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fruman DA, Satterthwaite AB, Witte ON. Xid-like phenotypes: a B cell signalosome takes shape. Immunity. 2000;13:1–3. doi: 10.1016/s1074-7613(00)00002-9. [DOI] [PubMed] [Google Scholar]
- 93.Kurosaki T. Functional dissection of BCR signaling pathways. Curr Opin Immunol. 2000;12:276–81. doi: 10.1016/s0952-7915(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 94.Dykstra ML, Longnecker R, Pierce SK. Epstein-Barr virus coopts lipid rafts to block the signaling and antigen transport functions of the BCR. Immunity. 2001;14:57–67. doi: 10.1016/s1074-7613(01)00089-9. [DOI] [PubMed] [Google Scholar]
- 95.Tedder TF, Inaoki M, Sato S. The CD19-CD21 complex regulates signal transduction thresholds governing humoral immunity and autoimmunity. Immunity. 1997;6:107–18. doi: 10.1016/s1074-7613(00)80418-5. [DOI] [PubMed] [Google Scholar]
- 96.Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud CA, Weill JC. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 2002;419:944–7. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- 97.Deane JA, Fruman DA. Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–98. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 98.Fruman DA. Phosphoinositide 3-kinase and its targets in B-cell and T-cell signaling. Curr Opin Immunol. 2004;16:314–20. doi: 10.1016/j.coi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 99.Cherukuri A, Cheng PC, Pierce SK. The role of the CD19/CD21 complex in B cell processing and presentation of complement-tagged antigens. J Immunol. 2001;167:163–72. doi: 10.4049/jimmunol.167.1.163. [DOI] [PubMed] [Google Scholar]
- 100.Cherukuri A, Cheng PC, Sohn HW, Pierce SK. The CD19/CD21 complex functions to prolong B cell antigen receptor signaling from lipid rafts. Immunity. 2001;14:169–79. doi: 10.1016/s1074-7613(01)00098-x. [DOI] [PubMed] [Google Scholar]
- 101.Cherukuri A, Shoham T, Sohn HW, Levy S, Brooks S, Carter R, Pierce SK. The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts. J Immunol. 2004;172:370–80. doi: 10.4049/jimmunol.172.1.370. [DOI] [PubMed] [Google Scholar]
- 102.Cherukuri A, Carter RH, Brooks S, Bornmann W, Finn R, Dowd CS, Pierce SK. B cell signaling is regulated by induced palmitoylation of CD81. J Biol Chem. 2004;279:31973–82. doi: 10.1074/jbc.M404410200. [DOI] [PubMed] [Google Scholar]
- 103.Machida K, Cheng KT, Pavio N, Sung VM, Lai MM. Hepatitis C virus E2-CD81 interaction induces hypermutation of the immunoglobulin gene in B cells. J Virol. 2005;79:8079–89. doi: 10.1128/JVI.79.13.8079-8089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Netea MG, van Deuren M, Kullberg BJ, Cavaillon JM, Van der Meer JW. Does the shape of lipid A determine the interaction of LPS with Toll-like receptors? Trends Immunol. 2002;23:135–9. doi: 10.1016/s1471-4906(01)02169-x. [DOI] [PubMed] [Google Scholar]
- 105.Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 106.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–90. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 107.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 108.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–55. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 109.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 110.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–8. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 111.Ruprecht CR, Lanzavecchia A. Toll-like receptor stimulation as a third signal required for activation of human naive B cells. Eur J Immunol. 2006;36:810–6. doi: 10.1002/eji.200535744. [DOI] [PubMed] [Google Scholar]
- 112.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 113.Yang R, Murillo FM, Delannoy MJ, Blosser RL, Yutzy WHt, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. B lymphocyte activation by human papillomavirus-like particles directly induces Ig class switch recombination via TLR4-MyD88. J Immunol. 2005;174:7912–9. doi: 10.4049/jimmunol.174.12.7912. [DOI] [PubMed] [Google Scholar]
- 114.Guay HM, Andreyeva TA, Garcea RL, Welsh RM, Szomolanyi-Tsuda E. MyD88 is required for the formation of long-term humoral immunity to virus infection. J Immunol. 2007;178:5124–31. doi: 10.4049/jimmunol.178.8.5124. [DOI] [PubMed] [Google Scholar]
- 115.Gourzi P, Leonova T, Papavasiliou FN. Viral induction of AID is independent of the interferon and the Toll-like receptor signaling pathways but requires NF-κB. J Exp Med. 2007;204:259–65. doi: 10.1084/jem.20061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vivarelli MS, McDonald D, Miller M, Cusson N, Kelliher M, Geha RS. RIP links TLR4 to Akt and is essential for cell survival in response to LPS stimulation. J Exp Med. 2004;200:399–404. doi: 10.1084/jem.20040446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 118.McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci USA. 2006;103:8798–803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 120.Gantner F, Hermann P, Nakashima K, Matsukawa S, Sakai K, Bacon KB. CD40-dependent and -independent activation of human tonsil B cells by CpG oligodeoxynucleotides. Eur J Immunol. 2003;33:1576–85. doi: 10.1002/eji.200323444. [DOI] [PubMed] [Google Scholar]
- 121.He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J Immunol. 2004;173:4479–91. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- 122.Fujieda S, Iho S, Kimura Y, Yamamoto H, Igawa H, Saito H. Synthetic oligodeoxynucleotides inhibit IgE induction in human lymphocytes. Am J Respir Crit Care Med. 2000;162:232–9. doi: 10.1164/ajrccm.162.1.9906136. [DOI] [PubMed] [Google Scholar]
- 123.Liu N, Ohnishi N, Ni L, Akira S, Bacon KB. CpG directly induces T-bet expression and inhibits IgG1 and IgE switching in B cells. Nat Immunol. 2003;4:687–93. doi: 10.1038/ni941. [DOI] [PubMed] [Google Scholar]
- 124.Kusunoki T, Sugai M, Gonda H, Nambu Y, Nagata-Nakajima N, Katakai T, Kusunoki M, Sakamoto A, Tokuhisa T, Nakahata T, Yokota Y, Shimizu A. CpG inhibits IgE class switch recombination through suppression of NF κB activity, but not through Id2 or Bcl6. Biochem Biophys Res Commun. 2005;328:499–506. doi: 10.1016/j.bbrc.2004.12.192. [DOI] [PubMed] [Google Scholar]
- 125.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 126.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–86. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 127.Thanos D, Maniatis T. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 128.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 129.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 130.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–81. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 131.Nera KP, Kohonen P, Narvi E, Peippo A, Mustonen L, Terho P, Koskela K, Buerstedde JM, Lassila O. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24:283–93. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 132.Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 134.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 135.Calame KL, Lin KI, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol. 2003;21:205–30. doi: 10.1146/annurev.immunol.21.120601.141138. [DOI] [PubMed] [Google Scholar]
- 136.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–42. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 137.Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, Nutt SL. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–66. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 138.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–82. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 139.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 140.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 141.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–80. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Merluzzi S, Moretti M, Altamura S, Zwollo P, Sigvardsson M, Vitale G, Pucillo C. CD40 stimulation induces Pax5/BSAP and EBF activation through a APE/Ref-1-dependent redox mechanism. J Biol Chem. 2004;279:1777–86. doi: 10.1074/jbc.M305418200. [DOI] [PubMed] [Google Scholar]
- 143.Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori KJ, Yokota Y, Shimizu A. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–37. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pal Z, Park S-R, Al-Qahtani A, Xu Z, Zan H, Capecchi MR, Casali P. Critical role of the homeodomain HoxC4 protein in regulating aicda gene expression, immunoglobulin somatic hypermutation and class switch DNA recombination. 2007 In preparation. [Google Scholar]
- 145.Oppezzo P, Dumas G, Lalanne AI, Payelle-Brogard B, Magnac C, Pritsch O, Dighiero G, Vuillier F. Different isoforms of BSAP regulate expression of AID in normal and chronic lymphocytic leukemia B cells. Blood. 2005;105:2495–503. doi: 10.1182/blood-2004-09-3644. [DOI] [PubMed] [Google Scholar]
- 146.Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–18. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–93. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 148.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol. 1999;19:5969–80. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 150.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–3. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 151.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martinez P, Amemiya CT. Genomics of the HOX gene cluster. Comp Biochem Physiol B Biochem Mol Biol. 2002;133:571–80. doi: 10.1016/s1096-4959(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 153.Meazza R, Faiella A, Corsetti MT, Airoldi I, Ferrini S, Boncinelli E, Corte G. Expression of HOXC4 homeoprotein in the nucleus of activated human lymphocytes. Blood. 1995;85:2084–2090. [PubMed] [Google Scholar]
- 154.Kim EC, Edmonston CR, Wu X, Schaffer A, Casali P. The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3′ hs1,2 enhancer in human B cells. Relevance to class switch DNA recombination. J Biol Chem. 2004;279:42258–69. doi: 10.1074/jbc.M407496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Boulet AM, Capecchi MR. Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev Biol. 1996;177:232–49. doi: 10.1006/dbio.1996.0159. [DOI] [PubMed] [Google Scholar]
- 156.Qin XF, Reichlin A, Luo Y, Roeder RG, Nussenzweig MC. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 1998;17:5066–75. doi: 10.1093/emboj/17.17.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P, Hodgkin PD. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201:1385–96. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 159.Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell. 2002;10:1097–105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- 160.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–87. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 161.Cattoretti G, Shaknovich R, Smith PM, Jack HM, Murty VV, Alobeid B. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–9. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 162.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–8. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, van der Zouwen B, Zhou JX, Lougaris V, Lipsky PE, Grammer AC, Morse HC., 3rd Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–57. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 165.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–34. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 166.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–71. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 167.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–61. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Phan TG, Paus D, Chan TD, Turner ML, Nutt SL, Basten A, Brink R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;30:2419–24. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suzuki A, Kaisho T, Ohishi M, Tsukio-Yamaguchi M, Tsubata T, Koni PA, Sasaki T, Mak TW, Nakano T. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–67. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Muto T, Okazaki IM, Yamada S, Tanaka Y, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci USA. 2006;103:2752–7. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Muto T, Muramatsu M, Taniwaki M, Kinoshita K, Honjo T. Isolation, tissue distribution, and chromosomal localization of the human activation-induced cytidine deaminase (AID) gene. Genomics. 2000;68:85–8. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- 172.Albesiano E, Messmer BT, Damle RN, Allen SL, Rai KR, Chiorazzi N. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 2003;102:3333–9. doi: 10.1182/blood-2003-05-1585. [DOI] [PubMed] [Google Scholar]
- 173.McCarthy H, Wierda WG, Barron LL, Cromwell CC, Wang J, Coombes KR, Rangel R, Elenitoba-Johnson KS, Keating MJ, Abruzzo LV. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor-prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–8. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- 174.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–8. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 175.Wang J, Shinkura R, Muramatsu M, Nagaoka H, Kinoshita K, Honjo T. Identification of a specific domain required for dimerization of activation-induced cytidine deaminase. J Biol Chem. 2006;281:19115–23. doi: 10.1074/jbc.M601645200. [DOI] [PubMed] [Google Scholar]
- 176.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 177.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–6. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 179.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–21. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 180.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–51. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 181.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–8. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 182.Pasqualucci L, Guglielmino R, Houldsworth J, Mohr J, Aoufouchi S, Polakiewicz R, Chaganti RS, Dalla-Favera R. Expression of the AID protein in normal and neoplastic B cells. Blood. 2004;104:3318–25. doi: 10.1182/blood-2004-04-1558. [DOI] [PubMed] [Google Scholar]
- 183.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–80. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–44. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 186.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–11. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 187.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–6. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 189.Shinkura R, Tian M, Smith M, Chua K, Fujiwara Y, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–41. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 190.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 191.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–40. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 192.Anant S, Blanc V, Davidson NO. Molecular regulation, evolutionary, and functional adaptations associated with C to U editing of mammalian apolipoproteinB mRNA. Prog Nucleic Acid Res Mol Biol. 2003;75:1–41. doi: 10.1016/s0079-6603(03)75001-6. [DOI] [PubMed] [Google Scholar]
- 193.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–23. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Turelli P, Trono D. Editing at the crossroad of innate and adaptive immunity. Science. 2005;307:1061–5. doi: 10.1126/science.1105964. [DOI] [PubMed] [Google Scholar]
- 195.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–30. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 196.Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–86. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]