The double-antibody sandwich assay has become a mainstay of clinical diagnostics.1-3 In a typical assay, a specific monoclonal antibody is used to capture a target molecule on the walls of a plastic microtiter plate. A second antibody directed against a second epitope on the same target is then added, forming an antibody–antigen–antibody “sandwich.” If the second antibody is linked to an enzyme (ELISA) or radiolabeled (radioimmunoassay), the presence of the target is then readily determined. Because double-antibody sandwich assays require the simultaneous binding of two antibodies, they are extremely specific, rendering them suitable for use directly in complex materials, such as blood serum. Likewise, because signaling is typically coupled to a catalytic (enzymatic) signaling mechanism, the approach often achieves impressive, subnanomolar detection limits for protein targets.4,5
The specificity and low detection limits of double-antibody sandwich assays have ensured their widespread use. Despite these attributes, however, the approach suffers from several drawbacks stemming from the requirement that the target exposes two distinct epitopes (i.e., is capable of simultaneously binding two different antibodies). First, because steric hindrance renders it unlikely that a small molecule will simultaneously bind to two antibodies, the approach is poorly suited to the detection of low molecular weight targets. Second, the requirement for dual binding specificity has also largely precluded the use of aptamer recognition elements in the sandwich assay format; whereas, as aptamers, DNA or RNA molecules selected in vitro for their ability to bind specific targets offer potentially important gains in shelf life and ease of synthesis,6,7 the number of targets for which multiple, distinct aptamers have been generated to date is small8 and thus rather few electrochemical aptamer-based sandwich assays have been reported for proteins, much less small molecules.9-11 Here, however, we demonstrate a selective, sensitive sandwich assay requiring only a single aptamer sequence and which is suitable for the detection of small molecule targets.
A single aptamer, split into two fragments, will equilibrate between its two dissociated parts and a folded, associated complex. If the target binds the complex with high affinity, the presence of the target will drive the equilibrium toward the formation of this complex (Figure 1), creating a “sandwich assay” from the two halves of a single aptamer. We have exploited this mechanism to create an electrochemical sandwich assay by splitting aptamers into two fragments, one of which is attached to a gold electrode via alkane thiol self-assembled monolayer chemistry and the second of which is modified with the redox moiety methylene blue. Target-induced association of the two fragments thus increases the concentration of methylene blue at the electrode surface, which can be readily monitored via voltammetry.
Figure 1.
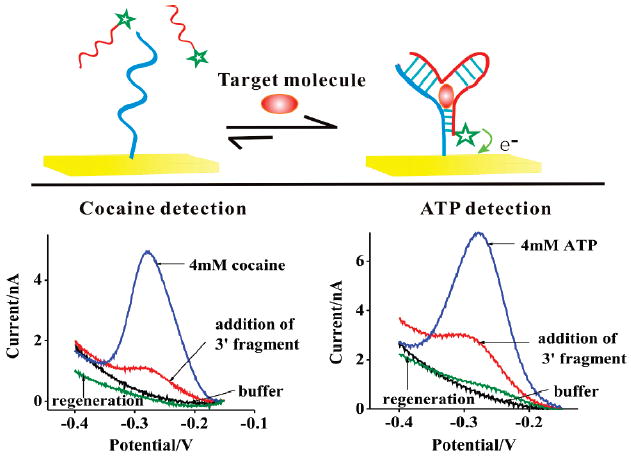
Here we demonstrate a new, electrochemical sandwich assay approach based on single aptamer sequences and capable of detecting small molecule targets. (Top) The sensor is predicated on the observation that, when split into two fragments most, if not all, aptamers will dissociate. Target binding, however, will stabilize their associated, native conformation, leading to a large increase in faradaic current when target is present (Bottom). To demonstrate this approach, we have fabricated sensors against the targets cocaine and ATP. In the absence of the methylene blue modified 3′ fragment, neither sensor exhibits any significant faradaic current. After addition of the relevant methylene blue modified 3′ fragment a small faradaic current is observed, presumably due to interactions between the two fragments that occur even in the absence of the target. After addition of the specific target molecule, however, we observe an ~600% increase in faradaic current for the cocaine sensor and an ~400% increase for the ATP sensor (both targets at 4 mM). As shown, both sensors are readily regenerated via a 30 s rinse with distilled, room-temperature water.
As test beds for the development of this new class of sandwich assays, we have fabricated sensors against the small molecules cocaine and ATP. To fabricate the cocaine sensor we synthesized 12-base, 5′ and 20-base, 3′ fragments corresponding to the 32-base cocaine binding aptamer of Stojanovich.12 The cut site was selected by inspection of the aptamer’s predicted secondary structure and via circular permutation studies that indicate that a cut at the selected site does not impede target binding (data not shown). We attached the 5′ fragment to an interrogating gold electrode via a hexane-thiol at its 5′ terminus and modified the 3′ fragment by adding a methylene blue to its 3′ terminus. The ATP sensor was similarly fabricated by cutting the 27-base aptamer of Szostak13 into 13-base, 3′ and 14-base, 5′ fragments and modifying them as in our cocaine sensor. In the absence of targets we observe small reduction peaks at the −0.27 V potential expected for methylene blue (Figure 1). This background peak presumably arises due to interactions between the two fragments that occur even in the absence of the target. The addition of the target molecules stabilizes the associated complexes, leading to large increases in faradaic current for both sensors. We then rinsed the sensor with room temperature distilled water for 30 s which regenerated the sensor, allowing us to reuse it many times before significant degradation is observed (see Supporting Information Figure S1).
The performance of a sensor such as this represents a trade-off: higher concentrations of the methylene blue modified 3′ fragment increase the population of the associated complex observed in the absence of target, thus increasing the sensor’s background current and degrading sensitivity. Lower concentrations of this fragment, however, destabilize the complex, reducing its affinity for the target. We find that a 3′ fragment concentration of 4 μM produces optimal signal gain for our cocaine sensor (Figure S2). Using this concentration we achieve statistically significant detection of cocaine at concentrations as low as 1 μM (Figure 2, left panel inset). Above this concentration we observe a monotonically increasing current with increasing cocaine concentration until a gain of 1200% is achieved at concentrations above a few millimolar (Figure 2, left panel). The ATP sensor likewise readily detects ATP at concentrations as low as 1 μM (Figure 2, right panel inset), above which the observed redox current increases monotonically until saturating at 400% above 4 mM target (Figure 2, right panel).
Figure 2.
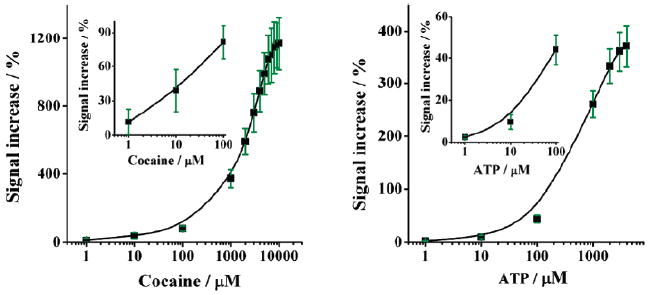
Both sensors are sensitive and cover a broad dynamic range. Shown are average values and standard deviations obtained from at least three independently fabricated electrodes; the error bars illustrate sensor-to-sensor reproducibility and are dominated by the reproducibility of sensor fabrication.
The new sensor architecture is specific, selective, and rapid. For example, neither sensor responds significantly (signal change < 2 ± 1 %) when challenged with the other’s target at 1 mM (Figure S3). Likewise, our cocaine sensor performs well when challenged directly in whole blood or any of a number of cutting and masking agents commonly found as contaminants in street cocaine, and our ATP sensor readily detects its target in crude, ATP-doped cell lysates (Figure S3). Finally, both sensors respond rapidly to their target molecules, exhibiting saturated or near saturated signaling within 1 min of the addition of their targets (Figure S4).
Our new detection scheme is intellectually related to the design of a prior electrochemical sensor based on the binding-induced folding of partially folded aptamers.14-17 These so-called E-AB (electrochemical, aptamer-based) sensors are, like the assay described here, specific and selective enough to deploy directly in complex sample matrices.18,19 Moreover, unlike the approach described here E-AB sensors are comprised of a single, continuous, electrode-bound aptamer chain and thus are reagentless. The gain of the E-AB platform, however, is relatively small (typically 30% to 200%14,15); because the signal-generating redox moiety is held near the electrode even in the absence of target, this approach suffers from high background currents. Thus, at the cost of requiring a reagent (the methylene-blue modified fragment), the sandwich assay described here improves on the gain of this earlier platform by at least 6-fold leading to significantly improved detection limits.14
Our new detection scheme is likewise intellectually related to a previously described strand displacement strategy for the detection of small molecule targets using aptamer recognition elements.20-23 In this approach, partial or complete hybridization of the aptamer with a cDNA strand prevents the redox moiety from approaching the electrode surface. The target, however, stabilizes the binding conformation of the aptamer, leading to dissociation of the blocking, complementary strand and a concomitant increase (if the redox is placed on the aptamer22) or decrease (if the redox moiety is on the complementary strand24) in faradaic current. The sandwich assay described here, however, exhibits a significantly larger signal gain and significantly improved response times when compared with these earlier approaches. The signal-on strand-displacement ATP sensor, 20,22 for example, exhibits an equilibration time constant of more than 1 h, a sharp contrast from the <1 min response time of the sensor described here.
Our approach is less sensitive than some established methods for detecting small molecules, such as chromatography (for the detection of cocaine) or colorimetric enzymatic assays (for the detection of ATP). These existing assays, however, require slow, multistep processes and expensive instrumentation, and thus our approach appears much more convenient. Indeed, our sandwich assay detects low micromolar concentrations in seconds, directly in complex samples such as blood and cellular lysates, and in a convenient, readily, reusable electrochemical format. Perhaps equally importantly, because our approach does not require the binding of multiple aptamers and the recognition aptamer to undergo a target-induced conformation change, it appears generalizable to most any target against which a suitable aptamer can be generated.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant No. 2R01EB002046 (to K.W.P).
Footnotes
Supporting Information Available: Detailed description of the experimental procedures and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Engvall E, Perlmann P. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 2.Leech D. Chem Soc Rev. 1994;23:205–213. [Google Scholar]
- 3.Yalow RS, Berson SA. Nature. 1959;184:1648–1649. doi: 10.1038/1841648b0. [DOI] [PubMed] [Google Scholar]
- 4.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DS, Nock S. Curr Opin Chem Biol. 2002;6:81–85. doi: 10.1016/s1367-5931(01)00281-2. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Willner I, Zayats M. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y, Plaxco KW. Electrochemical aptamer sensors. In: Lu Y, Li Y, editors. Functional nucleic acids for sensing and other analytical applications. Kluwer/Springer; New York: 2009. pp. 179–198. [Google Scholar]
- 9.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 10.Ikebukuro K, Kiyohara C, Sode K. Biosens Bioelectron. 2005;20:2168–2172. doi: 10.1016/j.bios.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Mir M, Vreeke M, Katakis L. Electrochem Commun. 2006;8:505–511. [Google Scholar]
- 12.Stojanovic MN, de Prada P, Landry DW. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 13.Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 14.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J Am Chem Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 15.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, Lai RY, Plaxco KW. Nat Protocols. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 17.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem Int Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 18.Lai RY, Plaxco KW, Heeger A. J Anal Chem. 2007;79:229–33. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Y, Rowe AA, Plaxco KW. J Am Chem Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 21.Zayats M, Huang Y, Gill R, Ma CA, Willner I. J Am Chem Soc. 2006;128:13666–13667. doi: 10.1021/ja0651456. [DOI] [PubMed] [Google Scholar]
- 22.Zuo XL, Song SP, Zhang J, Pan D, Wang LH, Fan CH. J Am Chem Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Shlyahovsky B, Elbaz J, Willner I. J Am Chem Soc. 2007;129:5804–5805. doi: 10.1021/ja070180d. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Zhu N, Yu P, Mao L. Analyst. 2008;133:1256–1260. doi: 10.1039/b807913g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


