Abstract
Background
Current treatment of chronic hepatitis C virus (HCV) infection has limited efficacy −especially among genotype 1 infected patients−, is costly, and involves severe side effects. Thus, predicting non-response is of major interest for both patient wellbeing and health care expense. At present, treatment cannot be individualized on the basis of any baseline predictor of response. We aimed to identify pre-treatment clinical and virological parameters associated with treatment failure, as well as to assess whether therapy outcome could be predicted at baseline.
Methodology
Forty-three HCV subtype 1b (HCV-1b) chronically infected patients treated with pegylated-interferon alpha plus ribavirin were retrospectively studied (21 responders and 22 non-responders). Host (gender, age, weight, transaminase levels, fibrosis stage, and source of infection) and viral-related factors (viral load, and genetic variability in the E1–E2 and Core regions) were assessed. Logistic regression and discriminant analyses were used to develop predictive models. A “leave-one-out” cross-validation method was used to assess the reliability of the discriminant models.
Principal Findings
Lower alanine transaminase levels (ALT, p = 0.009), a higher number of quasispecies variants in the E1–E2 region (number of haplotypes, nHap_E1–E2) (p = 0.003), and the absence of both amino acid arginine at position 70 and leucine at position 91 in the Core region (p = 0.039) were significantly associated with treatment failure. Therapy outcome was most accurately predicted by discriminant analysis (90.5% sensitivity and 95.5% specificity, 85.7% sensitivity and 81.8% specificity after cross-validation); the most significant variables included in the predictive model were the Core amino acid pattern, the nHap_E1–E2, and gamma-glutamyl transferase and ALT levels.
Conclusions and Significance
Discriminant analysis has been shown as a useful tool to predict treatment outcome using baseline HCV genetic variability and host characteristics. The discriminant models obtained in this study led to accurate predictions in our population of Spanish HCV-1b treatment naïve patients.
Introduction
Hepatitis C virus (HCV), with an estimated 170 million people infected worldwide, is the major causative agent of chronic liver disease, cirrhosis and hepatocellular carcinoma [1]. HCV is an enveloped positive single-stranded RNA virus and its genome exhibits significant genetic variability, which has been used to classify the virus into six major genotypes and a number of subtypes [2]. Furthermore, a high replication rate and the lack of proofreading activity of the viral RNA-dependent RNA polymerase generate a dynamic mosaic of closely related variants, usually referred to as quasispecies, within an infected individual. This phenomenon allows chronic infection establishment and may also have important implications in pathogenicity and resistance to antiviral drugs [3].
Pegylated-interferon alpha (PegIFN-α) and ribavirin (RBV) combination therapy constitutes the current standard of care for chronic hepatitis C treatment [4]. Despite recent advances in the development of “specifically targeted antiviral therapy for hepatitis C” (STAT-C) compounds, with protease inhibitors in phase III studies, possible future treatment regimens are likely to continue including these drugs in order to prevent HCV resistance [5]. Combination treatment is costly, requires long-term follow-up, and involves severe side effects. Furthermore, HCV genotype 1 infected patients fail to achieve a sustained virological response (SVR) in about 40–50% of the cases [6], [7]. Genotype 1 is the most common genotype worldwide; HCV subtype 1b (HCV-1b) is the most prevalent in Southern and Eastern Europe, Japan and other countries [8], [9] and is associated with a higher risk for hepatocellular carcinoma development [10].
A number of host-related factors have been associated with a lower likelihood of response to treatment, such as African-American ancestry, advanced liver fibrosis or cirrhosis, older age, male gender, obesity, transaminase levels, and host genetic polymorphisms [6], [7], [11]–[18]. Among the later, the rs12979860 polymorphism near the IL28B gene is the strongest predictive factor of SVR identified so far [14]; however, European-American patients not having the most favourable genotype (C/C) still have approximately 40% chance of responding to therapy (negative predictive value (NPV) around 60%). With regards to baseline virological factors, high viral loads, high levels of genetic variability within the E1–E2 and NS5A regions, as well as mutations in the so-called interferon sensitivity determining region (ISDR) and Core regions, have been related to therapeutic failure. Nevertheless, such findings have not been found in other studies and remain controversial [11], [19].
As predicting non-response prior to treatment is of major interest for both patient wellbeing and health care expense, several predictive models with variable accuracy have been proposed for HCV-1, such as those based in clinical variables in combination with viral load [20] or the ISDR mutant [21], as well as amino acid covariance in the full viral coding region [22]. However, according to present guidelines for patient management, individual treatment outcomes can only be precisely predicted once treatment is initiated on the basis of viral kinetics; a ≥2-Log(HCV-RNA) decline at week 12 (early virological response) is the most robust approach for identifying non-responder patients (NPV, 97–100%) and thus constitutes the earliest treatment-stopping rule [4].
The goal of this study was to identify pre-treatment clinical and virological parameters associated with treatment failure, as well as to assess whether therapy outcome could be predicted at baseline by means of comprehensive statistical methods in HCV-1b treatment naïve patients. Our results show that discriminant analysis could be a useful tool to predict treatment outcome using both baseline HCV genetic variability and host characteristics. The discriminant models obtained in this study lead to accurate predictions in our population of Spanish HCV-1b patients.
Results
Treatment response groups and adherence
Forty-three white Spanish patients met the inclusion criteria, 21 being responders and 22 non-responders. All patients were on treatment for the complete expected time and adherence to both drugs was overall >80%. No significant differences were observed between groups: 20 (95.2%) and 22 (100%) responders and non-responders had a good adherence to PegIFN-α, respectively (p = 0.488), and these proportions were 17 (80.9%) and 20 (90.9%) for RBV (p = 0.412).
Baseline clinical variables associated with treatment outcome
Baseline clinical characteristics of patients according to treatment outcome and bivariate analyses results are shown in Table 1 . Responder and non-responder groups were comparable in terms of gender, age, source of infection, and liver fibrosis stage (liver biopsy was not performed in 37.2% of the patients). Regarding body weight, one outlier was identified corresponding to a responder patient with 101.40 Kg, and differences between groups became significant when this patient was excluded (70.79±8.35 vs. 78.51±14.96 Kg in responder and non-responder groups, respectively p = 0.048). The alanine transaminase (ALT) quotient was significantly higher in responders than in non-responders (p = 0.009). Conversely, the gamma-glutamyl transferase (GGT) quotient tended to be higher in the non-responder group; two outliers were identified, which corresponded to two responder patients, and the GGT quotient was significantly higher in the non-responder group when these outliers were excluded (median, 0.58 and 1.07 in responders and non-responders, respectively, p = 0.033). The aspartate transaminase (AST) quotient was similar in both groups.
Table 1. Baseline clinical features of study patients according to treatment response group.
| Patient characteristic | Responders (n = 21) | Non-responders (n = 22) | p-value | |
| Male gender, n (%) | 9 (42.9) | 14 (63.6) | 0.172 | |
| Age a | 47.52±9.66 | 48.55±12.39 | 0.764 | |
| Weight (Kg) a | 72.24±10.53 | 78.51±14.96 | 0.122 | |
| Source of infection, n (%) | Blood transfusion | 6 (28.6) | 10 (45.5) | 1.000 |
| Non blood transfusion | 2 (9.5) | 2 (9.1) | ||
| Unknown | 13 (61.9) | 10 (45.5) | ||
| Liver fibrosis stage, n (%) | F0-2 | 11 (52.3) | 10 (45.5) | 0.648 |
| F3-4 | 2 (9.5) | 4 (18.2) | ||
| Unknown | 8 (38.1) | 8 (36.4) | ||
| ALT quotient (×ULN) b | 2.51 (1.32–4.15) | 1.53 (0.15–4.90) | 0.009 | |
| AST quotient (×ULN) a | 1.74±0.50 | 1.54±0.74 | 0.328 | |
| GGT quotient (×ULN) b | 0.58 (0.22–1.80) | 1.12 (0.18–2.50) | 0.111 | |
ALT, alanine transaminase; AST, aspartate transaminase; GGT, gamma-glutamyl transferase; ×ULN, factor times upper limit of normal used in our center for males and females: 41 and 31 U/L for ALT, 37 and 31 for AST, and 85 and 50 for GGT, respectively;
Data presented as mean ± SD, Student's t test;
Data presented as median (range), Mann-Whitney U test.
Baseline virological variables associated with treatment outcome
HCV viral load
Viral load did not differ significantly between groups (p = 0.210), with a mean value of 5.75±0.86 Log(IU/ml) in responders, and 6.03±0.58 Log(IU/ml) in non-responders.
E1–E2 genetic variability estimates
The median number of clones sequenced per patient was 22 (range, 20–33) in responders and 23 (range, 20–27) in non-responders (p = 0.291), yielding a total of 993 sequences. Genetic variability estimates according to treatment outcome and genomic region are shown in Table 2 . Although non-responder patients tended to have higher values than those with SVR for most E1–E2 genetic variability estimates, the number of quasispecies variants (number of haplotypes, nHap) was the only factor that significantly differed between groups (p = 0.003). Regarding the hypervariable regions (HVR), the HVR-1 showed the highest values for all parameters; the nHap and the number of synonymous substitutions per synonymous site (Ks) in this region were marginally significant, both being higher in non-responders.
Table 2. Summary of viral genetic variability estimates according to genomic region.* .
| E1–E2 region | HVR-1 subregion | HVR-2 subregion | HVR-3 subregion | |||||||||
| Estimator | Responders (n = 21) | Non-responders (n = 22) | p-value | Responders (n = 21) | Non-responders (n = 22) | p-value | Responders (n = 21) | Non-responders (n = 22) | p-value | Responders (n = 21) | Non-responders (n = 22) | p-value |
| S | 60.9±42.9 | 68.0±26.7 | 0.525 | 16.0 (1–48) | 17.0 (0–46) | 0.319 | 3.0 (0–13) | 4.0 (1–10) | 0.366 | 13.7±10.2 | 13.1±6.9 | 0.817 |
| η | 48.0 (9–154) | 63.5 (29–142) | 0.290 | 17.0 (1–64) | 18.0 (0–59) | 0.458 | 3.0 (0–13) | 4.0 (1–11) | 0.282 | 11.0 (2–42) | 12.0 (5–34) | 0.981 |
| nHap | 17 (5–25) | 22 (11–27) | 0.003 | 11 (2–17) | 12 (1–18) | 0.090 | 4 (1–12) | 5 (2–10) | 0.281 | 9.5±5.4 | 10.8±3.4 | 0.384 |
| π | 0.019 (0.002–0.089) | 0.024 (0.005–0.077) | 0.496 | 0.043 (0.001–0.261) | 0.063 (0.000–0.256) | 0.716 | 0.035 (0.000–0.186) | 0.032 (0.003–0.176) | 0.734 | 0.019 (0.001–0.132) | 0.021 (0.005–0.099) | 0.923 |
| Ka | 0.013 (0.000–0.063) | 0.014 (0.001–0.060) | 0.827 | 0.052 (0.000–0.294) | 0.057 (0.000–0.279) | 0.903 | 0.030 (0.000–0.195) | 0.028 (0.000–0.221) | 0.961 | 0.016 (0.000–0.084) | 0.009 (0.001–0.084) | 0.536 |
| Ks | 0.057±0.050 | 0.064±0.035 | 0.609 | 0.070±0.060 | 0.112±0.079 | 0.064 | 0.041 (0.000–0.591) | 0.048 (0.000–0.190) | 0.864 | 0.040 (0.004–0.322) | 0.056 (0.014–0.162) | 0.610 |
*Nucleotide positions corresponding to the H77 reference sequence (GenBank accession number AF009606): E1–E2 region, 1322–1853; HVR-1, 1491–1571; HVR-2, 1761–1787; HVR-3, 1632–1739.
S, total number of polymorphic sites; η, total number of mutations; nHap, number of haplotypes; π, nucleotide diversity corrected by Jukes-Cantor method; Ka, number of nonsynonymous substitutions per nonsynonymous site; Ks, number of synonymous substitutions per synonymous site; Data are expressed as mean ± SD, Student's t test or median (range), Mann-Whitney U test.
Phylogenetic analysis of the E1–E2 region
Differentiated clusters corresponding to responder and non-responder patients were not observed (Figure S1). Patients 1746 and 3468 appeared to be closely epidemiologically related since they shared a monophyletic clade with a 100% bootstrap support. In this clade, sequences from patient 1746 were a subgroup of those obtained from patient 3468, thus pointing to a source-recipient relationship. Patients 1634 and 3030, and 587 and 1313 might also be epidemiologically related, as inferred from the highly supported clade encompassing sequences from both patients in each group (100 and 90% bootstrap values, respectively), but no source-recipient relationship could be inferred.
Analysis of amino acid composition of the E1–E2 region
None of the nine amino acid positions initially identified by VESPA analysis showed a significantly different composition between responders and non-responders after the false discovery rate correction was applied (data not shown).
Analysis of amino acid composition of the Core region
VESPA analysis did not identify any amino acid position that differed between groups, although a polymorphism at position 70 was detected. On the other hand, when pairs of observed polymorphisms were subjected to bivariate analysis, the absence of both amino acids arginine (R) at position 70 and leucine (L) at position 91 was observed in 5 of 21 responder patients (23.8%) and in 12 of 22 non-responders (54.5%), (p = 0.039). R70 was substituted either by glutamine (Q) or histidine (H), and L91 mostly by methionine (M) and by cysteine (C) in one case. Since phylogenetic analysis showed that patients with this amino acid pattern did not group within the same cluster, the observed association was not attributed to sharing a common ancestry. This phylogenetic analysis provided similar evidence regarding to epidemiological relationships described for the E1–E2 region (data not shown).
Prediction of the treatment outcome according to baseline host and virological variables
Logistic regression analysis
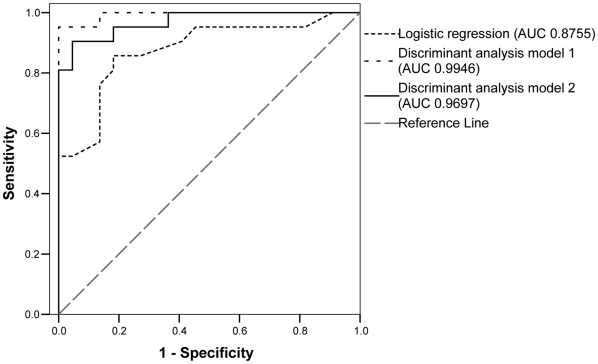
Variables showing a p-value <0.2 in the bivariate analyses (gender, Sqrt(ALT quotient), Sqrt(GGT quotient), weight, Core amino acid pattern, nHap_E1–E2, Log(Ks_E1–E2), nHap_HVR-1, and Sqrt(Ks_HVR-1)) were initially considered; the nHap_E1–E2 and the Core amino acid pattern persisted in the final model (Text S1), with an odds ratio (OR) of 1.47 (95% confidence interval, CI95% = [1.16–1.87]) and 25.47 (CI95% = [2.52–257.74]), respectively. Thus, the absence of amino acids R70 and L91 and a higher nHap_E1–E2 significantly increased the risk for treatment failure. An area under the curve (AUC) of 0.8755 was obtained in the receiver operating characteristic (ROC) curve ( Figure 1 ), and selecting a 0.500 cut-off yielded a sensitivity and positive predictive value (PPV) of 81.0%, and a specificity and NPV of 81.8%.
Figure 1. Receiver operating characteristic (ROC) curves for the multivariate logistic regression analysis and discriminant analysis models.
AUC, area under the ROC curve; Sensitivity, proportion of responders which are correctly identified; Specificity, proportion of non-responders which are correctly identified. Variables included in the models in decreasing order of significance: logistic regression model, Core amino acid pattern and nHap_E1–E2; discriminant analysis model 1, Core amino acid pattern, nHap_E1–E2, Sqrt(GGT quotient), Sqrt(ALT quotient), Log(viral load), Sqrt(S_HVR-2), body weight, and Log(Ks_E1–E2); discriminant analysis model 2, Core amino acid pattern, nHap_E1–E2, Sqrt(GGT quotient), Sqrt(ALT quotient), nHap_HVR-1, and Sqrt(Ks_HVR-1), and body weight.
Discriminant analysis
Two discriminant functions were obtained (Text S1) and cross-validated to assess how the results obtained would generalize to an independent but similar data set. Variables that persisted in Model 1 were: Core amino acid pattern, nHap_E1–E2, Sqrt(GGT quotient), Sqrt(ALT quotient), Log(viral load), Sqrt(total number of polymorphic sites in the HVR-2, S_HVR-2), body weight, and Log(Ks_E1–E2), in decreasing order of significance. The ROC curve obtained had an AUC of 0.9946 ( Figure 1 ). This model yielded a 95.2% sensitivity and a 100% specificity ( Table 3 ); however, sensitivity decreased to 76.2% and specificity to 72.7% after cross-validation. Therefore, we developed model 2 including the Core amino acid pattern, nHap_E1–E2, Sqrt(GGT quotient), Sqrt(ALT quotient), nHap_HVR-1, and Sqrt(Ks_HVR-1), and body weight, in decreasing order of significance. The AUC of the corresponding ROC curve was 0.9697 ( Figure 1 ). Treatment outcome was predicted with 90.5% sensitivity and 95.5% specificity (cut-off, 0.550), and these values remained high after the cross-validation (85.7% and 81.8%, respectively). Besides, the model could be optimized to correctly identify most responder patients by choosing a cut-off of 0.900, so that treatment is not denied to individuals that are likely to respond (NPV, 93.3% after cross-validation). Sensitivity, specificity, NPV and PPV for different cut-offs are shown in Table 3 . According to cross-validation, in an independent but similar data set, treatment could be omitted in 63.6 to 81.8% of the non-responder patients while most patients likely to respond would be identified and treated.
Table 3. Sensitivity, specificity, and predictive values for the discriminant models obtained.
| AUC | Cut-off | Sensitivity, % (cross-validated) | Specificity, % (cross-validated) | NPV, % (cross-validated) | PPV, % (cross-validated) | |
| Model 1 | 0.9946 | 0.500 | 95.2 (76.2) | 100 (72.7) | 95.7 (76.2) | 100 (72.7) |
| Model 2 | 0.9697 | 0.550 | 90.5 (85.7) | 95.5 (81.8) | 95.0 (81.8) | 91.3 (85.7) |
| 0.900 | 95.2 (95.2) | 68.2 (63.6) | 93.8 (93.3) | 74.1 (71.4) |
AUC, area under the receiver operating characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
Discussion
As combination treatment failure occurs in about half of all patients with chronic hepatitis C infected by genotype 1 [6], [7], prediction of treatment outcome at baseline would be highly beneficial. Although several factors have been identified as predictors of treatment outcome, none of them can provide a reliable individualized prediction when used independently. Based on our results in Spanish patients infected with HCV-1b, we propose the use of discriminant statistical models based on host and viral characteristics to provide an aggregate prediction of the treatment outcome at baseline.
Among the host-related factors studied baseline ALT levels, which are an indicator of liver damage, were significantly higher in responder patients than in non-responders (p = 0.009), as previously reported [11], [12]. Conversely, the GGT quotient tended to be higher in the non-responder group in agreement with other studies [12], [23]; higher GGT levels have been related to advanced fibrosis, steatosis and insuline resistance, which are more common among non-responders [24]. The body weight tended to be higher in non-responder patients; in fact, it has been suggested that obese subjects have an increased expression of the IFN-α signalling inhibitor factor SOCS-3 [25]. Some of the host factors that have previously been associated with treatment failure, such as male gender, advanced age, advanced liver fibrosis stage and cirrhosis [6], [7], [13] did not reach statistical significance in our study probably due to a limited sample size, especially regarding the liver biopsy, which was not performed in 37.2% of patients.
In relation to virus-related factors, HCV baseline viral load has been suggested as a predictor of SVR, but several cut-offs have been proposed [24]. In our study, average viral loads were higher in non-responders but differences were not significant. Additionally, several studies have reported an association between the level of variability in the HCV genome at baseline and treatment outcome. Envelope glycoprotein coding regions are highly variable; the HVR-1, which is the most variable region in the whole genome, is targeted by host neutralizing antibodies and plays a role in immune escape [26]. While the variability in this region has also been associated with treatment outcome [27]–[32], discrepancies on this matter have been noted probably due to the different treatment regimens, the different genetic variability estimates employed, and limitations in statistical analyses [33]–[35]. While our results show that treatment outcome was not related to the presence of a common evolutionary origin, in general terms, the E1–E2 genetic variability estimators suggested that a high heterogeneity in the baseline viral population could be involved in combination therapy failure, either through the pre-existence or the generation of drug-resistant viral variants. A higher number of quasispecies variants in the E1–E2 region (nHap_E1–E2) was significantly associated with treatment failure (p = 0.003). Additionally, when the analysis focussed on the HVR-1 subregion, nHap and Ks were marginally significant with higher values in the non-responder group. Although significant differences between groups at the amino acid level were not found, synonymous substitutions may have an effect on the secondary structure of the genomic RNA, which is an important selection target [36].
Pre-treatment Core amino acid substitutions at positions 70 (R by Q) and/or 91 (L by M) have been described as useful independent predictors of treatment failure in Japanese HCV-1b infected patients [37]. Similarly, our results show an association between the absence of both R70 and L91 amino acids and treatment failure (p = 0.039). Although it has been suggested that the Core protein may inhibit the transcription of antiviral genes induced by IFN-α [38], further studies are needed to clarify the role of the observed amino acid substitutions in treatment failure.
Since factors that significantly differed between groups in the bivariate analyses were not completely reliable in predicting treatment outcome when used independently, we developed predictive models that included a combination of variables. The logistic regression analysis identified the nHap_E1–E2 (OR = 1.47) and the Core amino acid pattern (OR = 25.47) as independent risk factors for treatment failure. However, predictive models obtained by discriminant analysis including additional variables showed better AUC values and more accurate predictions in our study population (90.5–95.2% sensitivity and 95.5–100% specificity). The most significant variables in both discriminant models were the Core amino acid pattern, nHap_E1–E2, and GGT and ALT quotients. Although prediction accuracy may deteriorate in an independent sample, the internal cross-validation pointed to a better reproducibility for model 2 in a comparable population (identifying 85.7% and 81.8% of the responder and non-responder patients, respectively), despite the fact that model 1 best predicted treatment outcome in our population. Besides, using model 2 the detection of those patients likely to respond to therapy could be maximized by adjusting the cut-off, leading to a higher NPV at the cost of a lower specificity (93.3% and 63.6%, respectively, after cross-validation). Thus, the results suggest that non-response could be predicted at baseline with high accuracy (NPV after cross-validation of 81.8% to 93.3% depending on the cut-off) in patient groups comparable to ours in terms of ethnicity, clinical background, and HCV subtype.
To our knowledge, this is the first study that describes a model for predicting individual combination therapy outcomes on the basis of baseline host and viral characteristics using a discriminant multivariate analysis. This comprehensive statistical method integrates the information of all variables included in the model thus improving the prediction with respect to more commonly used statistical approaches. Additionally, discriminant models may be adjusted to include the most significant predictors of treatment outcome in each population. However, our study has several limitations: i) other viral genome regions not included in the study might also be involved in resistance to therapy, such as the ISDR. Nevertheless, a meta-analysis suggested that the association between the number of mutations in this region and SVR achievement was more pronounced in Japanese than in European patients [39]. As most European HCV-1b strains present less than 3 mutations, large sample sizes would be required to find significant associations; ii) recent studies have suggested that single nucleotide polymorphisms in several human genes involved in the IFN mediated response are associated to treatment outcome in HCV-1 infected patients, especially the IL28B gene polymorphisms [14]–[18]. Since our study was retrospective, whole-blood samples were not available to assess host genetic polymorphisms; iii) the sample size was limited to 43 patients. However, a similar number of patients were included in each group, accounting for the fact that about 50% of patients infected by HCV-1b achieve an SVR. Although an independent but similar population was not available, we performed an internal cross-validation. This method is commonly used to reduce classification bias and estimate future model performance [40].
Our results show that both host and viral factors are involved in treatment failure, although the exact mechanisms should be further characterized. The host-related variables included in the prediction models are routinely used for patient management and relatively easy to obtain, while viral variability estimates are obtained through laborious methods. Even so, and if confirmed in further studies, the information obtained may help physicians to restrict treatment to those patients that are likely to benefit from it, thus reducing overall treatment costs. Those patients that are unlikely to respond could avoid current therapy and related side effects, and wait for more effective treatment regimens.
In conclusion, discriminant analysis using both baseline HCV genetic variability and host characteristics has been shown as a useful statistical tool allowing us to accurately predict combination treatment outcome in a high proportion of Spanish HCV-1b infected patients. Further studies including host genetic polymorphisms and larger numbers of patients are under way, and similarly generated models will probably have an increased predictive power.
Materials and Methods
Ethics statement
This study was approved by the Clinical Research Ethics Committee at our institution (“Comité Ético de Investigación Clínica”, CEIC). As this was a retrospective study, and data were analyzed anonymously, informed consent was specifically waived.
Patients and specimens
Patients with chronic hepatitis C by HCV-1b, treated with combination therapy at “Hospital Universitari Germans Trias i Pujol”, were retrospectively selected. Exclusion criteria were: previous IFN-based treatment, HIV or HBV coinfection, and having other causes of liver disease or alcohol abuse. Infection with HCV-1b was confirmed through NS5B sequencing followed by phylogenetic analysis, as previously described [41]. The patients had started antiviral therapy with PegIFN-α2a (180 µg/week) plus weight-based doses of RBV (1000–1200 mg/day) for 48 weeks between 2003 and 2008. The patients were classified into responders (patients with SVR, defined as undetectable HCV-RNA in serum 24 weeks after treatment cessation) and non-responders. Non-response was defined as continued presence of HCV-RNA during therapy (null response), rebound of HCV-RNA while on therapy (breakthrough) or 24 weeks after the end of treatment (relapse). All virological analyses were performed using serum specimens obtained before patients initiated treatment and conserved at −80°C until testing.
Baseline clinical and epidemiological host parameters
Variables considered were gender, age, weight, source of infection, stage of fibrosis according to the Scheuer scoring system [42], and serum levels of ALT, AST, and GGT. Liver enzyme levels were transformed into a quotient expressing the factor times upper limit of normal (ULN) according to gender. We defined good treatment adherence as having received ≥80% of total maximum dose prescribed of both drugs for ≥80% of the expected duration of therapy [43].
Baseline virological parameters
Serum viral load
HCV-RNA had been quantified by RT-PCR (Cobas® Amplicor HCV Monitor test, Roche Molecular Systems, Pleasanton, CA, USA) or by real-time RT-PCR (Abbott RealTime HCV assay, Abbott Molecular Inc., Des Plaines, IL, USA), according to manufacturer's instructions.
RNA extraction and reverse transcription (RT)
Total RNA was extracted from 220 µl of serum, using the QIAamp® viral RNA kit (QIAGEN® GmbH, Hilden, Germany) according to the manufacturer's protocol. RT was performed using random hexamers in order to prevent any bias during the reaction, as previously described [44].
PCR-cloning and sequencing of the E1–E2 region
A 532-bp sequence encompassing the E1 C-terminal and the E2 N-terminal regions (including the HVR-1, HVR-2 and HVR-3) was obtained and referred to as E1–E2 region (nucleotides 1322–1853 in the H77 reference sequence, GenBank accession number AF009606). PCR products were cloned and sequenced as previously described [44]. Briefly, a hemi-nested PCR was carried out with the proofreading Pfu DNA polymerase (Promega, Mannheim, Germany), and HCV-1b specific degenerated primers (2-Eg1 and 2-Ea, and 2-Eg2 and 2-Ea primers for the first and second rounds of PCR, respectively) [45]. Amplified DNA products were purified and cloned into EcoRV-digested pBluescript II SK(+) phagemid (Stratagene, La Jolla, CA, USA). Plasmids were transformed into Escherichia coli XL-1 blue MRF' competent cells (Stratagene). Between 25 and 35 colonies were selected and subjected to PCR followed by purification and sequencing of both strands using vector-based primers and the BigDyeTM Terminator v3.1 Ready Reaction Cycle Sequencing Kit on ABI Prism 3730 or 3100-Avant Genetic Analyzers (Applied Biosystems Foster City, CA, USA). Readings were assembled and edited with the STADEN package v1.6. [46].
PCR and direct sequencing of the Core region
The whole Core region (573 bp, H77 positions 342–914) was amplified using forward primer Cg1 (5′ GCCATRGTGGTCTGCGGAAC 3′, H77 positions 137–156), which was slightly modified from primer CC11 [37], and reverse primer Ca (5′ GTTGGAGCAGTCGTTCGTRA 3′, H77 positions 949–968). PCR was performed in 50 µl containing 5 µl of cDNA, 0.2 mM of each dNTP, 0.4 µM of each primer, Pfu buffer and 0.6 U of Pfu DNA polymerase (Promega). Thermocycler conditions were: 1 cycle at 94°C for 2 min, 35 cycles at 94°C for 1 min, 55°C for 2 min and 72°C for 3 min, and 1 cycle at 72°C for 7 min. PCR products were directly sequenced with the Cg2 primer (5′ GGGAGGTCTCGTAGACCGTGCAYCATG 3′, H77 positions 318–344), which was slightly modified from the Core-A1g primer [47], and the Ca primer.
Phylogenetic analysis of the E1–E2 region
The complete E1–E2 cloned region was subjected to phylogenetic analysis in order to rule out potential contamination between specimens and assess clustering of patients according to treatment outcome. Sequences were aligned by ClustalW implemented in MEGA 4 [48]. jModeltest [49] was used to obtain the evolutionary model that best fitted the data according to the Akaike Information Criterion. This model was employed to reconstruct a maximum-likelihood phylogenetic tree with PHYML [50]. RAxML software was used for evaluating tree reliability on the basis of branch support (1000 replicates) [51].
Genetic variability analysis of the E1–E2 region
Multiple alignments were generated for each patient for the complete E1–E2 region, and the HVR-1, HVR-2 and HVR-3 (H77 nucleotide positions 1491–1571, 1761–1787, and 1632–1739, respectively). The following genetic variability estimates were obtained for each multiple alignment with DnaSP v4.50 [52]: total number of polymorphic sites (S), total number of mutations (η), nucleotide diversity corrected by Jukes-Cantor method (π), and number of quasispecies variants (number of haplotypes, nHap). The number of nonsynonymous substitutions per nonsynonymous site (Ka) and Ks were obtained using the Nei-Gojobori method.
Amino acid composition analysis in the E1–E2 region
This analysis aimed to detect any amino acid position in the E1–E2 region that differed between groups but showed within-group homogeneity. Consensus sequences were compared between groups with the program VESPA [53] to obtain the predominant sequence for each group. The VESPA output file was employed to estimate the G-statistics in each amino acid position as previously described [31], where p-values ≤0.05 were considered significant. The false discovery rate procedure was used to correct for multiple comparisons.
Amino acid composition analysis of the Core region
Direct sequences obtained were analysed as described for the E1–E2 region. Sequences were also aligned to assess the presence of amino acid polymorphisms associated to treatment outcome.
Statistical analysis
Clinical and virological values were compared between responders and non-responders in bivariate analysis using Student's t test or Mann-Whitney U test for quantitative variables, and Chi-square or Fisher's exact tests for categorical variables. Data was expressed as mean ± standard deviation, median and range, or relative frequency. Values between 1.5 and 3 inter-quartile range above/below the upper/lower quartile of quantitative variables were identified as outliers.
Statistical models were developed to predict non-response. A multivariate logistic regression analysis was performed, where covariates included in the model were explanatory variables that achieved a p-value <0.20 on bivariate analyses. Variables which presented high correlations with other variables (Spearman's correlation >0.7) were also excluded to avoid colinearity problems. To obtain the final set of variables included in the model we used a backward stepwise selection procedure [54]. OR and CI95% were reported for significant variables. Two discriminant analyses were also carried out [55]. In model 1 all covariates analyzed but those which presented high correlations with other variables were considered. Variables with a skewed distribution were transformed using quadratic or Log transformations and multivariate normality was tested using Henze-Zirkler's test [56]. The final discriminant function was obtained using a backward stepwise variable selection procedure. To assess how the results obtained would generalize to an independent but similar data set, each case was classified by the functions from all cases other than that case (“leave-one-out” cross-validation); this validation was performed in the whole stepwise variable selection procedure. Chi-square test was used to test the equality of covariance structures across groups [57], considering a pooled covariance matrix when the value was not significant at the 0.1 level. Model 2 included covariates that achieved a p-value <0.15 on bivariate analyses with the goal to improve the cross-validation results. ROC curves were obtained and the following parameters were calculated to measure the effectiveness of prediction: AUC, sensitivity (proportion of responders which are correctly identified), specificity (proportion of non-responders which are correctly identified), NPV and PPV. These parameters were also computed after cross-validation taking into account all misclassified patients in any of the 43 replications. Cut-off values that yielded highest sensitivity and specificity were selected by ROC curve analysis for the three predictive models obtained. P-values <0.05 were considered significant. Statistical analyses were performed using the statistical software packages SPSS v15.0 and SAS v9.1 (SAS Institute Inc., Cary, NC, USA).
Accession numbers
All sequences obtained in this study were submitted to the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) under the following accession numbers: FN675941-FN675983, FN675984-FN676976, and FN676977-FN677019 for Core, E1–E2 and NS5B regions, respectively.
Supporting Information
All viral sequences obtained for each patient are identified with a vertical line, the patient identification number and the response group (R, responders; NR, non-responders). Substitution model: GTR+G+I (gamma shape parameter: 0.926, proportion of invariable sites: 0.271). All nodes corresponding to each individual patient were supported with bootstrap values >70%. The scale bar represents 0.05 substitutions per nucleotide position.
(0.02 MB PDF)
(0.03 MB DOC)
Acknowledgments
We would like to thank Elisabet Bascuñana and Victoria González for their help in sample processing and sequence edition, as well as Lurdes Matas, Sònia Molinos, Eva Erice and Anna Bargalló for their support in clinical records review.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by "Instituto de Salud Carlos III - Fondo de Investigaciones Sanitarias" (ISCIII-FIS) projects PI05/1131 (EM, RP, VA) and CP09/00044 (EM, MA, RP, VA, VS), grant numbers CD05/00258 ("Contratos Postdoctorales de Perfeccionamiento") and CP09/00044 ("Miguel Servet") from "Ministerio de Ciencia e Innovacion" (MICINN), within the "Plan Nacional de Investigación científica, Desarrollo e Innovación Tecnológica (I+D+I)" (EM); and with support from "Comissionat per a Universitats i Recerca del Departament d'Innovació, Universitats i Empresa de la Generalitat de Catalunya i del Fons Social Europeu" and "Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica", Spain (VS). This work was also partially supported by ISCIII-FIS projects CP07/00078 (MAB) and BFU2008-03000 (FG-C, MAB) from MICINN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Viral Cancers: Hepatitis C virus. 2010;28 Available: www.who.int/vaccine_research/diseases/viral_cancers/en/index2.html#disease%20burden. Accessed 2010 May. [Google Scholar]
- 2.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 3.Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- 4.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange CM, Sarrazin C, Zeuzem S. Review article: HCV - STAT-C era of therapy. Aliment Pharmacol Ther. 2010;32:14–28. doi: 10.1111/j.1365-2036.2010.04317.x. [DOI] [PubMed] [Google Scholar]
- 6.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 7.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 8.Esteban JI, Sauleda S, Quer J. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol. 2008;48:148–162. doi: 10.1016/j.jhep.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 9.Sy T, Jamal MM. Epidemiology of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:41–46. doi: 10.7150/ijms.3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142–1154. doi: 10.1016/j.jhep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 11.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 12.Berg T, Sarrazin C, Herrmann E, Hinrichsen H, Gerlach T, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37:600–609. doi: 10.1053/jhep.2003.50106. [DOI] [PubMed] [Google Scholar]
- 13.Hoofnagle JH, Wahed AS, Brown RS, Jr, Howell CD, Belle SH. Early changes in hepatitis C virus (HCV) levels in response to peginterferon and ribavirin treatment in patients with chronic HCV genotype 1 infection. J Infect Dis. 2009;199:1112–1120. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 14.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 15.Su X, Yee LJ, Im K, Rhodes SL, Tang Y, et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. J Hepatol. 2008;49:184–191. doi: 10.1016/j.jhep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukada H, Ochi H, Maekawa T, Abe H, Fujimoto Y, et al. A polymorphism in MAPKAPK3 affects response to interferon therapy for chronic hepatitis C. Gastroenterology. 2009;136:1796–1805. doi: 10.1053/j.gastro.2009.01.061. [DOI] [PubMed] [Google Scholar]
- 17.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 19.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20:23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Bauer E, Crespo J, Romero-Gómez M, Moreno-Otero R, Solá R, et al. Development and validation of two models for early prediction of response to therapy in genotype 1 chronic hepatitis C. Hepatology. 2006;43:72–80. doi: 10.1002/hep.21002. [DOI] [PubMed] [Google Scholar]
- 21.Shirakawa H, Matsumoto A, Joshita S, Komatsu M, Tanaka N, et al. Pretreatment prediction of virological response to peginterferon plus ribavirin therapy in chronic hepatitis C patients using viral and host factors. Hepatology. 2008;48:1753–1760. doi: 10.1002/hep.22543. [DOI] [PubMed] [Google Scholar]
- 22.Aurora R, Donlin MJ, Cannon NA, Tavis JE. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J Clin Invest. 2009;119:225–236. doi: 10.1172/JCI37085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kronenberger B, Herrmann E, Micol F, von Wagner M, Zeuzem S. Viral kinetics during antiviral therapy in patients with chronic hepatitis C and persistently normal ALT levels. Hepatology. 2004;40:1442–1449. doi: 10.1002/hep.20487. [DOI] [PubMed] [Google Scholar]
- 24.Kau A, Vermehren J, Sarrazin C. Treatment predictors of a sustained virologic response in hepatitis B and C. J Hepatol. 2008;49:634–651. doi: 10.1016/j.jhep.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Walsh MJ, Jonsson JR, Richardson MM, Lipka GM, Purdie DM, et al. Non-response to antiviral therapy is associated with obesity and increased hepatic expression of suppressor of cytokine signalling 3 (SOCS-3) in patients with chronic hepatitis C, viral genotype 1. Gut. 2006;55:529–535. doi: 10.1136/gut.2005.069674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farci P, Shimoda A, Wong D, Cabezon T, De GD, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh BI, Han KH, Lee HW, Sohn JH, Ryu WS, et al. Factors predictive of response to interferon-alpha therapy in hepatitis C virus type 1b infection. J Med Virol. 2002;66:481–487. doi: 10.1002/jmv.2169. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann WP, Sarrazin C, Kronenberger B, Schonberger B, Bruch K, et al. Mutations within the CD81-binding sites and hypervariable region 2 of the envelope 2 protein: correlation with treatment response in hepatitis C virus-infected patients. J Infect Dis. 2003;187:982–987. doi: 10.1086/368221. [DOI] [PubMed] [Google Scholar]
- 29.Ueda E, Enomoto N, Sakamoto N, Hamano K, Sato C, et al. Changes of HCV quasispecies during combination therapy with interferon and ribavirin. Hepatol Res. 2004;29:89–96. doi: 10.1016/j.hepres.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Morishima C, Polyak SJ, Ray R, Doherty MC, Di Bisceglie AM, et al. Hepatitis C virus-specific immune responses and quasi-species variability at baseline are associated with nonresponse to antiviral therapy during advanced hepatitis C. J Infect Dis. 2006;193:931–940. doi: 10.1086/500952. [DOI] [PubMed] [Google Scholar]
- 31.Torres-Puente M, Cuevas JM, Jiménez-Hernández N, Bracho MA, García-Robles I, et al. Genetic variability in hepatitis C virus and its role in antiviral treatment response. J Viral Hepat. 2008;15:188–199. doi: 10.1111/j.1365-2893.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 32.Cuevas JM, Torres-Puente M, Jiménez-Hernández N, Bracho MA, García-Robles I, et al. Refined analysis of genetic variability parameters in hepatitis C virus and the ability to predict antiviral treatment response. J Viral Hepat. 2008;In press doi: 10.1111/j.1365-2893.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 33.Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, et al. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99:3081–3086. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbate I, Lo IO, Di SR, Cappiello G, Girardi E, et al. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J Hepatol. 2004;40:831–836. doi: 10.1016/j.jhep.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Chambers TJ, Fan X, Droll DA, Hembrador E, Slater T, et al. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79:3071–3083. doi: 10.1128/JVI.79.5.3071-3083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Guillou-Guillemette H, Vallet S, Gaudy-Graffin C, Payan C, Pivert A, et al. Genetic diversity of the hepatitis C virus: impact and issues in the antiviral therapy. World J Gastroenterol. 2007;13:2416–2426. doi: 10.3748/wjg.v13.i17.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–380. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 38.de Lucas S, Bartolomé J, Carreno V. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J Infect Dis. 2005;191:93–99. doi: 10.1086/426509. [DOI] [PubMed] [Google Scholar]
- 39.Pascu M, Martus P, Höhne M, Wiedenmann B, Hopf U, et al. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53:1345–1351. doi: 10.1136/gut.2003.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukasiewicz E, Gorfine M, Freedman LS, Pawlotsky JM, Schalm SW, et al. Prediction of nonSVR to therapy with pegylated interferon-α2a and ribavirin in chronic hepatitis C genotype 1 patients after 4, 8 and 12 weeks of treatment. J Viral Hepat. 2010;17:345–351. doi: 10.1111/j.1365-2893.2009.01183.x. [DOI] [PubMed] [Google Scholar]
- 41.Martró E, González V, Buckton AJ, Saludes V, Fernández G, et al. Evaluation of a new assay in comparison with reverse hybridization and sequencing methods for hepatitis C virus genotyping targeting both 5′ noncoding and nonstructural 5b genomic regions. J Clin Microbiol. 2008;46:192–197. doi: 10.1128/JCM.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheuer PJ. Classification of chronic viral hepatitis: a need for reassessment. J Hepatol. 1991;13:372–374. doi: 10.1016/0168-8278(91)90084-o. [DOI] [PubMed] [Google Scholar]
- 43.McHutchison JG, Manns M, Patel K, Poynard T, Lindsay KL, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 44.Jiménez-Hernández N, Torres-Puente M, Bracho MA, García-Robles I, Ortega E, et al. Epidemic dynamics of two coexisting hepatitis C virus subtypes. J Gen Virol. 2007;88:123–133. doi: 10.1099/vir.0.82277-0. [DOI] [PubMed] [Google Scholar]
- 45.Bracho MA, García-Robles I, Jiménez N, Torres-Puente M, Moya A, et al. Effect of oligonucleotide primers in determining viral variability within hosts. Virol J. 2004;1:13. doi: 10.1186/1743-422X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 47.López-Labrador FX, Bracho MA, Berenguer M, Coscolla M, Rayon JM, et al. Genetic similarity of hepatitis C virus and fibrosis progression in chronic and recurrent infection after liver transplantation. J Viral Hepat. 2006;13:104–115. doi: 10.1111/j.1365-2893.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 49.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 52.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 53.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 54.Hosmer DW, Lemeshow S. New York: Wiley; 2000. Applied logistic regression.375 [Google Scholar]
- 55.McLachan GJ. New York: Wiley; 1992. Discriminant analysis and statistical pattern recognition.526 [Google Scholar]
- 56.Henze N, Zirkler B. A class of invariant consistent tests for multivariate normality. Communications in Statistics - Theory and Methods. 1990;19:3595–3618. [Google Scholar]
- 57.Morrison DF. New York: McGraw-Hill; 1967. Multivariate Statistical Methods.338 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All viral sequences obtained for each patient are identified with a vertical line, the patient identification number and the response group (R, responders; NR, non-responders). Substitution model: GTR+G+I (gamma shape parameter: 0.926, proportion of invariable sites: 0.271). All nodes corresponding to each individual patient were supported with bootstrap values >70%. The scale bar represents 0.05 substitutions per nucleotide position.
(0.02 MB PDF)
(0.03 MB DOC)