Abstract
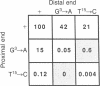
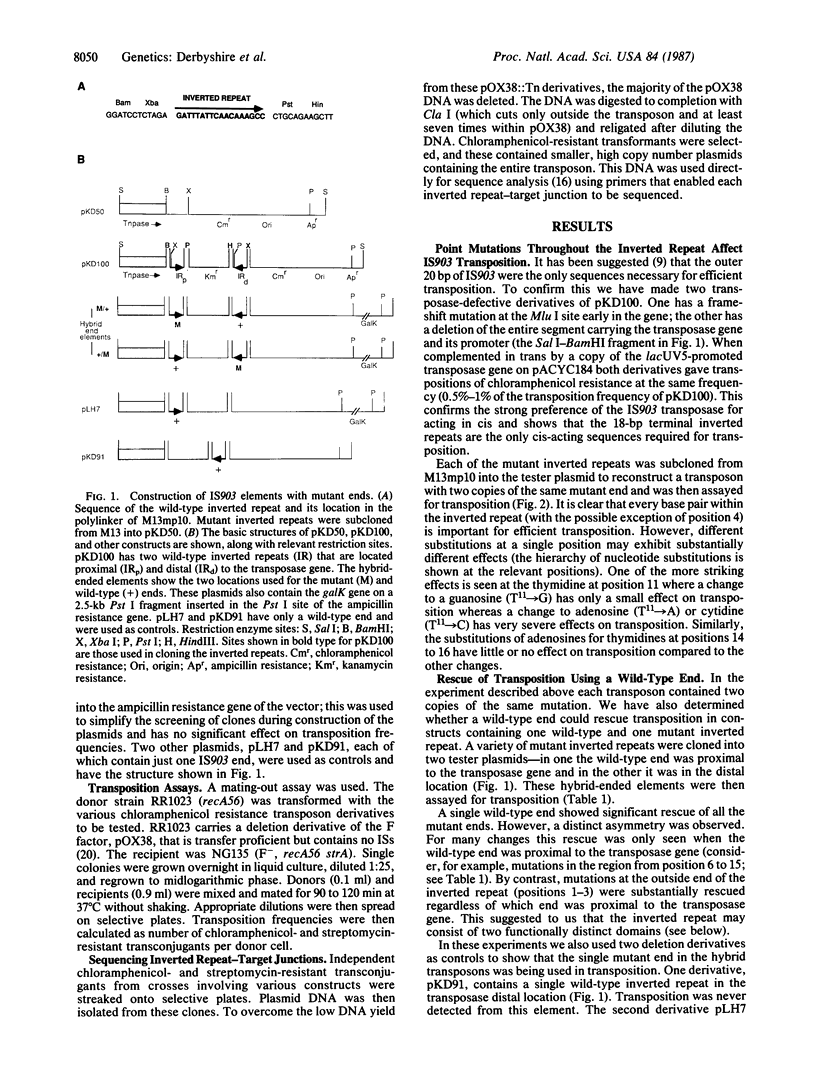
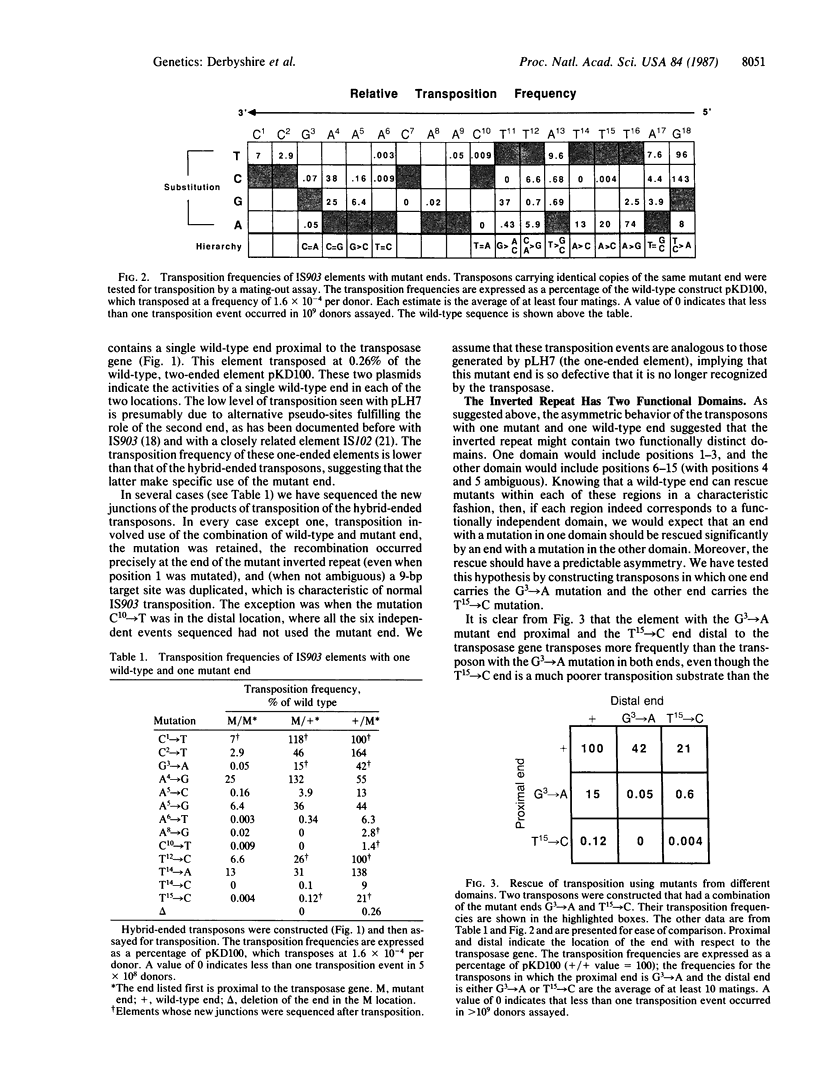
The insertion sequence IS903 has perfect, 18-base-pair terminal repeats that are the presumed binding sites of its transposase. We have isolated mutations throughout this inverted repeat and analyzed their effect on transposition. We show that every position in the inverted repeat (with the possible exception of position 4) is important for efficient transposition. Furthermore, various substitutions at a single position can have a wide range of effects. Analysis of these hierarchical effects suggests that transposase contacts the minor groove in the region from position 13 to position 16 but makes major groove (or more complex) interactions with the outer portion of the inverted repeat. Our data indicate that the transposase exhibits relaxed specificity for the "second" end of a transposed segment; the defect in transposition of virtually all mutant inverted repeats can be rescued by a wild-type end. However, this rescue exhibits a pronounced position effect; in most cases, it is efficient only when the wild-type end is close to the 3' end of the transposase gene. This confirms the cis-acting nature of the transposase protein and suggests the initial transposase-inverted repeat interaction is the rate-limiting step in transposition. From the behavior of transposons with one mutant and one wild-type end, we infer that the inverted repeat contains two functional domains--one for initial complex formation with transposase and the other for effective completion of transpositional recombination. To support this hypothesis we show that an end with a mutation in one domain can significantly rescue an end with a mutation in the other domain.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Salvo J. J., Grindley N. D. A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene. 1986;46(2-3):145–152. doi: 10.1016/0378-1119(86)90398-7. [DOI] [PubMed] [Google Scholar]
- Diaz-Aroca E., Mendiola M. V., Zabala J. C., de la Cruz F. Transposition of IS91 does not generate a target duplication. J Bacteriol. 1987 Jan;169(1):442–443. doi: 10.1128/jb.169.1.442-443.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Replicon fusions promoted by the inverted repeats of Tn5. The right repeat is an insertion sequence. J Mol Biol. 1981 Jul 25;150(1):15–32. doi: 10.1016/0022-2836(81)90322-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. A novel type of transposon generated by insertion element IS102 present in a pSC101 derivative. Cell. 1982 Aug;30(1):29–36. doi: 10.1016/0092-8674(82)90008-3. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo H., Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci U S A. 1982 Jan;79(2):277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Transposase promotes double strand breaks and single strand joints at Tn10 termini in vivo. Cell. 1984 Nov;39(1):181–190. doi: 10.1016/0092-8674(84)90204-6. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Greene P. Eco RI* specificity and hydrogen bonding. DNA. 1982;1(2):117–124. doi: 10.1089/dna.1.1982.1.117. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Berg D. E. IS50-mediated inverse transposition. Discrimination between the two ends of an IS element. J Mol Biol. 1982 Aug 5;159(2):257–271. doi: 10.1016/0022-2836(82)90495-8. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Carle G. F., Berg D. E. Sequences essential for transposition at the termini of IS50. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7293–7297. doi: 10.1073/pnas.80.23.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C., Phadnis S. H., Carle G. F., Berg D. E. Sequences essential for IS50 transposition. The first base-pair. J Mol Biol. 1985 Apr 20;182(4):487–493. doi: 10.1016/0022-2836(85)90235-9. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Kleckner N. Essential sites at transposon Tn 10 termini. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3452–3456. doi: 10.1073/pnas.81.11.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Derbyshire K. M., Hughson F. M., Grindley N. D. Replicative and conservative transpositional recombination of insertion sequences. Cold Spring Harb Symp Quant Biol. 1984;49:251–260. doi: 10.1101/sqb.1984.049.01.029. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Schaus N. A., Grindley N. D. Insertion sequence duplication in transpositional recombination. Science. 1983 Nov 18;222(4625):755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]