Abstract
Background
The novel influenza A pandemic virus (H1N1pdm) caused considerable morbidity and mortality worldwide in 2009. The aim of the present study was to evaluate the clinical course, duration of viral shedding, H1N1pdm evolution and emergence of antiviral resistance in hospitalized cancer patients with severe H1N1pdm infections during the winter of 2009 in Brazil.
Methods
We performed a prospective single-center cohort study in a cancer center in Rio de Janeiro, Brazil. Hospitalized patients with cancer and a confirmed diagnosis of influenza A H1N1pdm were evaluated. The main outcome measures in this study were in-hospital mortality, duration of viral shedding, viral persistence and both functional and molecular analyses of H1N1pdm susceptibility to oseltamivir.
Results
A total of 44 hospitalized patients with suspected influenza-like illness were screened. A total of 24 had diagnosed H1N1pdm infections. The overall hospital mortality in our cohort was 21%. Thirteen (54%) patients required intensive care. The median age of the studied cohort was 14.5 years (3–69 years). Eighteen (75%) patients had received chemotherapy in the previous month, and 14 were neutropenic at the onset of influenza. A total of 10 patients were evaluated for their duration of viral shedding, and 5 (50%) displayed prolonged viral shedding (median 23, range = 11–63 days); however, this was not associated with the emergence of a resistant H1N1pdm virus. Viral evolution was observed in sequentially collected samples.
Conclusions
Prolonged influenza A H1N1pdm shedding was observed in cancer patients. However, oseltamivir resistance was not detected. Taken together, our data suggest that severely ill cancer patients may constitute a pandemic virus reservoir with major implications for viral propagation.
Introduction
The emergence of the novel influenza A/H1N1 pandemic virus (H1N1pdm) significantly affected the utilization of healthcare resources and increased morbidity and mortality in children and young adults [1], [2]. From April through September 2009, during the fall/winter in the southern hemisphere, Brazil experienced the first wave of the H1N1pdm virus, and by the end of December 2009, over 1600 H1N1pdm-related deaths had been reported in Brazil [3].
Emerging data on the clinical course of severe H1N1pdm infection have allowed the identification of high-risk groups, which include pregnant women and patients with morbid obesity [4], [5]. However, an analysis of the impact of this novel virus in a highly susceptible population, such as cancer patients, through clinical and virological perspectives, needs to be highlighted [6], [7], [8], [9], [10], [11]. The atypical clinical presentation of influenza infections in cancer patients, which delays clinical suspicion, antiviral treatment and adequate prevention of viral transmission, is a major challenge for clinical management in this population [12]. Cancer patients are more likely to suffer from severe seasonal influenza infections [12], [13], [14] and prolonged viral shedding, as has been reported for an H3N2 seasonal virus [15]. Prolonged shedding and the development of oseltamivir resistance in cancer patients infected with the H1N1pdm virus have not been thoroughly evaluated. Data on these aspects could have major implications for the clinical management and infection control practices for H1N1pdm-infected cancer patients [16].
Because the analysis of this novel viral infection in cancer patients is an important component of the 2009 pandemics, we conducted a prospective cohort study aimed at evaluating the clinical course of influenza infection, the duration of viral shedding, H1N1pdm evolution and the emergence of antiviral resistance in hospitalized cancer patients with a severe H1N1pdm infection in a reference cancer center during the winter of 2009 in Brazil.
Results
Characteristics of the study population
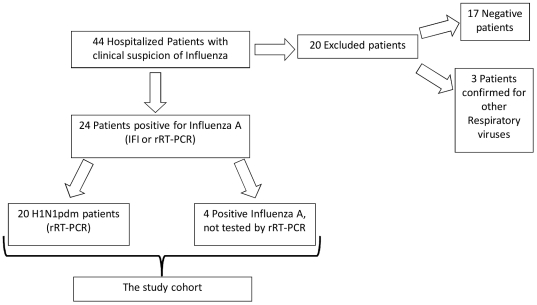
During the study period, 44 hospitalized cancer patients with a suspected influenza infection were screened, and 24 had a confirmed influenza A diagnosis using a rapid indirect immunofluorescence (IFI) test or World Health Organization (WHO)-recommended real-time RT-PCR (rRT-PCR) (Figure 1 and Table S1). Among these, 20 patients were confirmed to be positive for the H1N1pdm virus using rRT-PCR (Figure 1 and Table S1). The remaining four patients were positive for influenza A using IFI only. Considering the pandemic case definitions with reference to international guidelines [17], these last four cases were categorized as H1N1pdm-confirmed cases. Altogether, these 24 cases constituted the study population. All of the respiratory samples collected from the 20 rRT-PCR-confirmed patients were inoculated in cell cultures. We recovered the virus from 13 individuals after at least two passages in MDCKs, constituting 15 isolated samples. These isolates were also analyzed for oseltamivir resistance using a functional assay.
Figure 1. Study flow chart.
Patients diagnosed with H1N1pdm were young (median age = 14.5, range 3–69 years). In total, 14 (58.3%) were under 18 years old, and 17 (70.8%) were less than 50 years old. Hematologic cancer occurred in 75% (18) of the patients, whereas solid tumors occurred in 25% (6) patients (Tables S2 and S3). A total of 22 (∼92%) patients had received immunosuppressive therapy in the previous 30 days. Among these individuals, 18 patients (75%) were on chemotherapy, 14 (58.3%) received systemic corticosteroids and 1 (4%) received radiation therapy (Table 1 and S4). No patient received erythropoietin (EPO) or immunomodulatory agents. A total of 14 patients (58.3%) presented febrile neutropenia (<500 neutrophils/mm3) at the time H1N1pdm was diagnosed. The median duration of neutropenia after the onset of viral disease was two days (ranging from one to six days; Table S5). According to the Brazilian National Cancer Institute's protocol, all patients that presented neutropenia received G-CSF until normalization of neutrophil counts. The clinical characteristics and comparisons among groups are shown in Table 1 and Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, and S11. The overall mortality in our cohort was around 21% (n = 5), and four patients (n = 16.6%) had at least one comorbidity besides cancer. Of these patients with comorbidities, one died. No pregnant or morbidly obese patients were identified.
Table 1. Patient characteristics according to survival statusa.
| Variable | All patients (n = 24) | Survivors (n = 19–79.2%) | Non-survivors (n = 5–20.8%) | P value b |
| Age (years) | 14.5 (3–69) | 14 (3–69) | 17 (4–62) | 0.50 |
| Male gender | 12 (50%) | 7 (36.8%) | 5 (100%) | 0.03 |
| Type of cancer | ||||
| Solid tumor | 6 (25%) | 5 (26.3%) | 1 (20%) | 0.99 |
| Hematological malignancy | 18 (75%) | 14 (73.7%) | 4 (80%) | 0.99 |
| Cancer status | ||||
| Controlled/remission | 3 (12.5%) | 3 (15.8%) | 0 | 0.99 |
| Active - newly-diagnosed | 9 (37.5%) | 6 (31.6%) | 3 (60%) | 0.32 |
| Active – recurrence/progression | 12 (50%) | 10 (52.6%) | 2 (40%) | 0.99 |
| Performance status | ||||
| 0–1 | 6 (31.6%) | 6 (31.6%) | 0 | 0.28 |
| 2–4 | 18 (75%) | 13 (68.4%) | 5 (100%) | 0.28 |
| Previous chemotherapy | 18 (75%) | 13 (54.2%) | 5 (100%) | 0.28 |
| Previous use of corticosteroids | 14 (58.3%) | 10 (52.6%) | 4 (80%) | 0.36 |
| Neutropenia | 14 (58.3%) | 7 (36.8%) | 3 (60%) | 0.67 |
| Need for mechanical ventilation | 10 (41.6%) | 5 (26.3%) | 5 (100%) | 0.06 |
| Presence of extra-pulmonary organ dysfunction | 8 (33.3%) | 4 (21.1%) | 4 (80%) | 0.03 |
| Oseltamivir treatment duration (days) | 7 (0–19) | 9 (1–19) | 5 (1–18) | 0.38 |
Results are expressed as the mean ± standard deviation, median (range), n (%).
Reported P values refer to comparisons between survivors and non-survivors.
A total of 23 (95.8%) patients were treated with oseltamivir, and the median time from the initial symptoms to the initiation of therapy was three days (0–15 days; Tables 1, S2 and S6). One patient that died due to severe acute respiratory failure 24 h after clinical suspicion of H1N1pdm infection never received antiviral treatment. Oseltamivir was used for a median of seven days (0–19 days), and double doses (150 mg bid for adults and twice the recommended dose per kg for children) were administered for 11 (47.8%) patients (Table S6). A total of 11 (47.8%) patients received oseltamivir for more than seven days. Six patients (25%) received this antiviral within 48 h of clinical suspicion. All patients that died received oseltamivir more than 48 h after the onset of the illness. However, when we compared the mortality of patients that received oseltamivir either within or after 48 h of the onset illness, no significant difference was observed (0/6 vs. 5/18, p = 0.28). No differences in prolonged viral shedding were observed between these two groups.
At presentation, all patients were treated with broad-spectrum intravenous antimicrobial agents to combat community-acquired pneumonia and/or febrile neutropenia [18], and five (20.8%) had concomitant positive cultures (Tables S7 and S8). Hypoxemia was frequent, and the median PaO2/FiO2 on the first arterial blood gas evaluation was 192 mmHg (range: 64–367 mmHg).
Intensive care unit admission
Overall, 13 patients (five adults and eight children) were admitted to the ICU. Six patients were directly admitted from the emergency department, and the other seven patients were transferred from other hospital wards (Table S2). Ventilatory support was given to 12 patients (Table 1 and S9). Invasive mechanical ventilation was performed in 10 patients (76.9%), and non invasive ventilation (NIV) was performed in 3 patients (23.1%; Table 1). Among the NIV patients, one required subsequent endotracheal intubation and mechanical ventilation, and all three patients were discharged from the hospital. Extra-pulmonary organ failure occurred in eight patients (33.3%; Table 1 and S9).
Of the 13 critically ill patients, 12 were treated with oseltamivir, and treatment was initiated 48 h after the first signs/symptoms of viral infection in 5 of them. Adjunct or non-conventional supportive therapies for ARDS were performed for 12 of the 13 patients that entered the ICU (92.3%). A total of 10 patients (76.9%) received systemic corticosteroids (eight due to previous use and two for shock and persistent ARDS); five (38.5%) were ventilated in a prone position, and four (30.8%) required recruitment maneuvers. No patient received extra-corporeal membrane oxygenation.
Prolonged H1N1pdm shedding
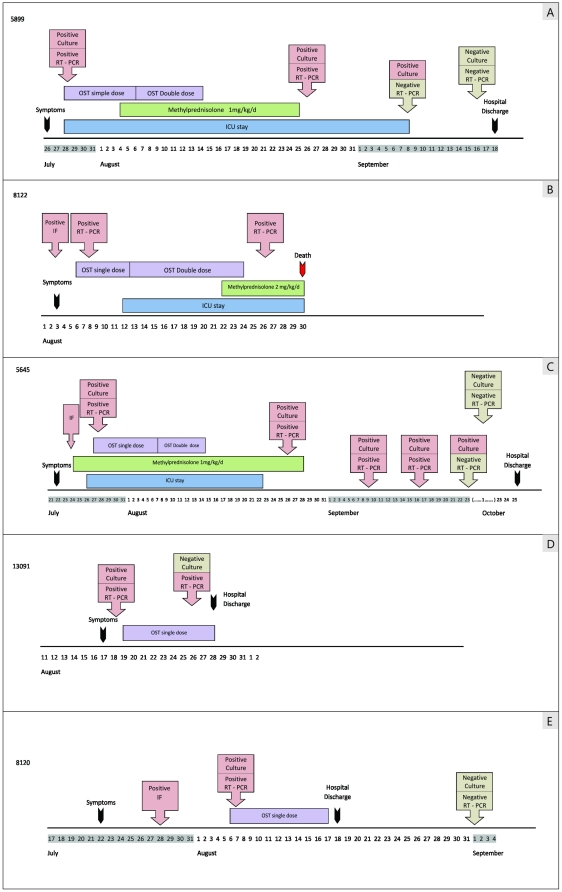
Although prolonged influenza A shedding has been observed for a cancer patient infected with the H3N2 seasonal virus [15], more detailed data on H1N1pdm secretion in severely ill cancer patients are required. We evaluated viral shedding in 10 mechanically ventilated patients by collecting sequential respiratory samples at different time-points after the onset of illness. The duration of viral shedding was considered to be the time frame from the initial symptoms to the last H1N1pdm-confirmed sample. Five (50%) patients in this group showed viral shedding for at least 11 days during oseltamivir treatment (Figure 2 and Table S12). The median duration of H1N1pdm shedding was 23 days (ranging from 11 to 63 days; Figure 2 and Table S12).
Figure 2. Time course of viral shedding in hospitalized cancer patients.
Most importantly, the maximum duration of H1N1pdm shedding in our investigation was 63 days, followed by 44 days for another patient. To our knowledge, these periods constitute the longest registered cases of H1N1pdm shedding described to date. All rRT-PCR-positive samples from these patients with the longest viral shedding durations (5645 and 5899) were culturable, meaning that these were infectious viruses (Figure 2 and Table S12). In addition, the last H1N1pdm-confirmed samples from these patients were only detected using cell culture assays, suggesting the presence of low viral loads in these specimens (Figure 2 and Table S12). These patients still shed the virus for an additional 25 to 40 days after cessation of the antiviral treatment (Figure 2).
H1N1pdm molecular evolution in clinical isolates
To date, no significant variation has been detected at the amino acid level in the hemagglutinin (HA) of the 2009 pandemic virus [19]. Therefore, we examined the genetic diversity of the H1N1pdm virus recovered from these severely ill patients by performing sequence analysis of the viral HA gene. No significant divergence was found in the H1N1pdm isolates collected during the onset of illness from the four patients with prolonged shedding (Figure S1).
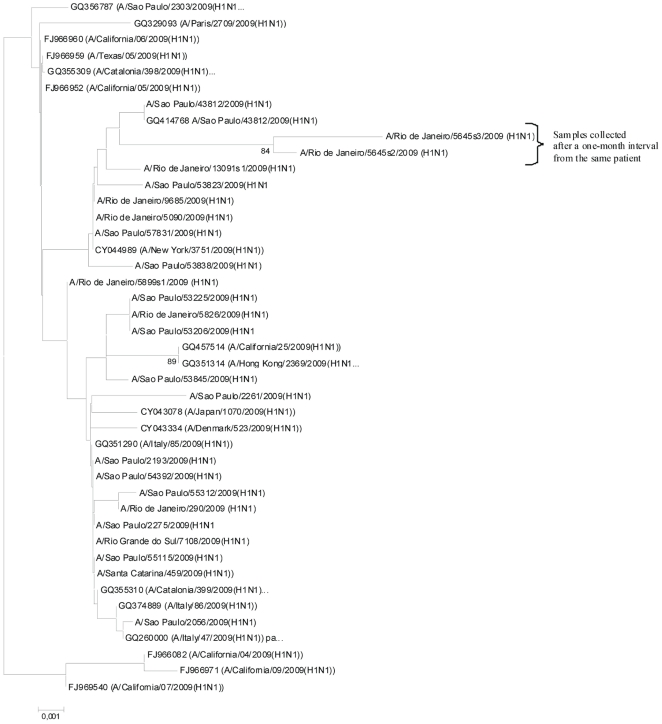
To evaluate the genetic characteristics of the isolates from patients with prolonged viral shedding, we sequenced two consecutive samples from a single individual (5645s2/09 and 5645s3/09) and compared their HA sequences to other H1N1pdm viruses from mild, severe and fatal cases from different countries. We observed that samples collected a month apart (5645s2/09 and 5645s3/09) clustered together and displayed a relatively large branch length from other H1N1pdm viruses (Figure 3). This result may have occurred because strain 5645s2/09 diverged from CA/04 in the amino acid residues L52S, L70P, P100S, C153L, T214A, Q293R and I321V (H1 numbering). Additionally, isolate 5645s3/09, which was sequenced from amino acid residues 167 to 413, also had the mutations T214A, Q293R and I321V fixed in the viral population a month after the initial sampling. In the isolate 5645s3/09, another mutation was also acquired, D238P, suggesting continuous viral evolution. Although we cannot determine the role of these mutations in viral pathogenesis by this result alone, the retention of the changed residues over time strongly suggests viral persistence rather than re-infection.
Figure 3. Phylogenetic tree of H1N1pdm strains from HCI-INCa and other Brazilian strains based on the HA gene.
The bootstrap probability is indicated for each interior branch, and values below 80% are hidden. The scale bar indicates the number of amino acid changes per site. The sampling number and the state of origin in Brazil are displayed. This tree is rooted by the California/04/2009 HA sequence.
H1N1pdm susceptibility to oseltamivir
Considering that influenza resistance to antivirals is likely to emerge in immunocompromised individuals and that, with respect to the H1N1pdm virus, only a few studies have detected oseltamivir resistance [20], we investigated the emergence of antiviral resistance in H1N1pdm samples isolated in cell culture. Thus, oseltamivir carboxylate IC50 values were measured for 15 H1N1pdm samples isolated from 13 patients. We found that 10 isolates were sensitive (0.85±0.27 nM), and 5 displayed high IC50 values for the antiviral used (165.13±42.26 nM). These samples were sent to the WHO collaborating center at the CDC in Atlanta for resistance confirmation. We found that the isolates with high IC50 values were endowed with a neuraminidase (NA) activity that was cross-resistant to oseltamivir, zanamivir and peramivir, suggesting the presence of a co-pathogen endowed with NA activity within these isolates. Co-infections with other respiratory viruses (coronavirus (229, 43 and 63), parainfluenza (1, 2, 3 and 4), human metapneumovirus, parechovirus, rhinovirus, RSV A/B, adenovirus and enterovirus) or atypical bacteria (Mycobacterium pneumonia) were not identified in these samples, suggesting that no other viral source of NA activity was responsible for these high IC50 values. However, we co-isolated a Streptococcus sp. from the samples with high IC50 values. The NA activity of this bacterial strain displayed a phenotype resistant to NAIs. In further testing with bacteria-free H1N1pdm isolates, the virus' IC50 were consistent with a sensitive isolate. Pyrosequencing analyses were also performed and revealed that the other high-IC50 samples had the WT H275 residue and thus did not contain this oseltamivir resistance marker. Viruses isolated from the initial onset of illness were A/California/07-like viruses (Figure S2).
Discussion
Prolonged influenza shedding in cancer patients has been observed for seasonal strains [15]. Regarding the H1N1pdm virus, it has been shown that prolonged virus shedding in cancer patients may occur, although such a phenomenon has only been documented through cases involving a single studied patient [21], [22], [23], [24], [25]. Here, we prospectively and systematically collected information from a cohort of hospitalized cancer patients with severe H1N1pdm infections. These patients presented high mortality, prolonged viral shedding and H1N1pdm evolution without the emergence of oseltamivir resistance. This is the first study to address viral shedding and resistance in cancer patients with H1N1pdm infections; thus, it may provide insight into the role of cancer patients as potential human reservoirs for this pandemic virus.
Unlike previous reports, our population was composed of hospitalized, severely immunocompromised cancer patients [26]. Most of them were young, had hematologic malignancies and received chemotherapy and systemic steroids in the weeks that preceded the H1N1pdm infection. The patients were treated with oseltamivir in the early course of the infection (the median time to antiviral initiation was three days). A total of 13 patients (54%) required intensive care and presented severe respiratory distress. In these patients, the mortality rates were higher (38%) than those observed for general ICU patients suffering from H1N1pdm infections [2], [27] as well as for non-critically ill cancer patients [26]. However, the outcomes were not different from those reported for cancer patients requiring mechanical ventilation [28], [29].
Interestingly, during the influenza season, 14 patients (58.3%) with febrile neutropenia were identified as H1N1pdm cases, a condition that is not usually investigated in this scenario. However, febrile neutropenic cancer patients have an increased risk of developing respiratory distress and multi-organ failure. Therefore, screening for respiratory viruses and prompt initiation of oseltamivir treatment should be considered in these patients. Febrile neutropenia indicates a poor prognostic with respect to a patient's outcome, but neutropenia duration in our cohort of patients was less than seven days. Thus, prolonged viral shedding might not have a correlation with neutropenia.
We observed the persistence of H1N1pdm in 5 out of 10 patients studied for this purpose. In these individuals, viral shedding continued for at least 11 days, despite the use of oseltamivir. The median duration of viral shedding in our population was 23 days, and two pediatric patients with acute lymphoblastic leukemia showed even longer virus secretions (44 and 63 days; Figure 2 and Table S10), although it is difficult to determine whether viral persistence was due to cancer per se or to acute lung injury and mechanical ventilation.
Influenza shedding is not considered to last long, and it disappears seven days after the onset (2.4 and 4.5 days for oseltamivir- and placebo-treated groups, respectively) [30]. Studies aimed at monitoring 2009 H1N1pdm virus shedding using randomized trials with appropriate controls, such as outpatients and hospitalized or immunocompromised individuals, have not yet been conducted. Because we were also unable to establish age-matched controls with or without immunosuppression, since this study was conducted during the peak of the first wave of the 2009 pandemics in Brazil, we compared our work to other studies on H1N1pdm shedding in general, hospitalized or immunocompromised populations [21], [22], [23], [24], [25], [31], [32], [33], [34], [35], [36]. In Table 2, we summarize the cohort used in each study, whether or not they were immunocompromised, their underlying diseases and the number of patients analyzed for viral shedding in each of these studies. We compared the maximum periods of shedding among these different populations and the number of patients that secreted the virus for more than seven days. These data would be more informative and relevant from a public health point of view because the duration of the quarantine for H1N1pdm was approximately that long [21]. We found (Table 2) that in households in Hong Kong [31] and Canada [34], the maximum periods of H1N1pdm shedding ranged from 8 to 11 days. These periods were not different from what was observed with military cadets [22] and during the containment phase of the pandemics in Vietnam [23] (Table 2). Regarding H1N1pdm shedding among infants, Hien et al. showed that children five to nine years old could secrete the H1N1pdm virus for five to six days, which is markedly lower than what is observed for the seasonal influenza virus [30], [33]. Compared to our results, we observed higher periods of viral shedding in two seven-year-old patients with acute lymphoblastic leukemia (Table 2 and Figure 2). However, a single report on two travelers in France showed that these apparently immunocompetent individuals secreted the H1N1pdm virus for 14 and 28 days [25] (Table 2). Although these periods of time are comparable to the time frame of virus secretion in hospitalized patients in China [23] and our work, the study from Felury et al. might be as biased as ours by the small size of the cohort (Table 2). Influenza-infected immunocompromised individuals may have prolonged influenza shedding [15], [21], [32], [36]; however, more insights are necessary to better comprehend the dynamics of the H1N1pdm virus in these individuals. Mora et al. showed that in HIV-1-infected individuals, co-infection with the H1N1pdm virus might lead to an outcome not different from the one expected for immunocompetent subjects, although no systematic analysis of viral shedding was performed [36]. A similar conclusion was also drawn for transplant recipient individuals, whose longest periods of viral shedding did not exceed 11 days [32] (Table 2). In our study, we found periods of H1N1pdm shedding similar to what the CDC observed for leukemia patients [21] (Table 2). Although the small size of these cohorts of immunocompromised individuals [21], [32], including ours, may require a more conclusive and mechanistic analysis, these observations may stimulate further systematic studies to understand or gain insight into factors associated with prolonged H1N1pdm shedding. In addition, it might give insights on basic studies on influenza pathogenesis.
Table 2. Comparisons of the period of viral shedding in the general, hospitalized and immunocompromised populations.
| General Population | Hospitalized | Immunocompromised | |||||||
| Authors | Cowling et al. | Hien et al. | De Serres et al. | Fleury et al. | Witkop et al. | Cao et al. | Seville et al. | CDC | Ours |
| Reference | [31] | [33] | [34] | [25] | [22] | [23] | [32] | [21] | |
| General population | Households, Hong Kong | Containment of the pandemics in Ho Chi Minh City (HCMC), Vietnam | Households, Canada | Travelers, France | Military cadets | First 426 patients hospitalized in China | No | No | No |
| Immunocompromised | ND | ND | ND | No | ND | ND | Yes | Yes | Yes |
| Underlying disease | ND | ND | ND | No | ND | Various | Transplant recipients | Leukemia | Cancer |
| Number of patients evaluated for virus shedding | 54 | 932 | 43 | 2 | 29 | 350 | 6 | 2 | 10 |
| Number of patients with virus shedding ≥7 days | 2 children | ∼80 | 8–14b | 2 | 7 | 238 | 1 | 2 | 5 |
| Highest period of shedding (days) | 8 | 11–12a | 8–11b | 14–28c | 9 | 17 | 11 | 37–44c | 63 |
– Five to six days for children under nine years old.
– Eight patients shed the virus for at least 8 days, while 14 shed for at least 11 days.
– Only two patients were evaluated.
ND – Not determined.
Our results highlight the need for closer surveillance of cancer patients with H1N1pdm infections until the detection of the first negative sample. We hypothesize that follow-up protocols aimed at monitoring the persistence of viral shedding in cancer patients may be relevant if patients are submitted to immunosuppressive therapies in the days or weeks prior to or following an H1N1pdm infection.
Next, we sought to determine viral evolution during prolonged shedding. We found that some amino acid changes persisted from the initial symptoms until 30 days thereafter, suggesting that these patients had viral persistence rather than re-infection. In addition, an extra amino acid change (D238P) was found in the viral HA sequenced a month after the onset of illness, suggesting continuing viral evolution. Although some of the mutations that we found (L52S, L70P, P100S, C153L, T214A, D238P, Q293R and I321V) in strains 5645s2/09 and 5645s3/09 have not been previously described, other amino acid residue changes that were detected in our study (P100S and T214A) have been found in H1N1pdm viruses throughout the world without a significant link to viral pathogenesis or antigenic variation [37], [38], [39].
Influenza viruses resistant to antiviral drugs have been reported in immunocompromised patients, [21], [40] and this resistance might be associated with prolonged viral shedding [41], [42]. Notably, five isolates from two patients had high IC50 values to neuraminidase inhibitors (NAIs). An NA activity that was multi-resistant to NAIs was identified and could be due to the presence of a Streptococcus strain found in throat swabs and tracheal aspirates. Pyrosequencing analyses of samples with high IC50 values revealed that these specimens were H275 wild-type sensitive viruses. These results reinforce the need for additional genotyping assays to confirm the identification of putative resistant strains identified using functional assays. In addition, our findings show the need for investigating other sources of NA activity in virus isolates with odd IC50 values.
The apparent paradox of prolonged viral shedding without antiviral resistance could be explained by either the inability of the immunocompromised host to effectively clear the H1N1pdm virus [12], [43] or inefficient absorption of the drug [44]. Because we combined both clinical and molecular virology data, our results might contribute to the discussion on the adequate duration and type of anti-H1N1pdm treatment in immunocompromised patients with a protracted course. In these patients, the use of parenteral systemic or inhaled antivirals should also be investigated.
Although our work further investigates the unique dynamics of H1N1pdm virus infection in immunocompromised hosts, some caveats must be noted. Because our investigation started during a new pandemic, the clinical evaluation and management protocols changed during the course of the study as new data emerged from the literature and from updated recommendations [45]. As the pandemic reached its peak in South America, the establishment of a larger and more diverse cohort with age-matched controls with or without immunosuppression became complex. Thus, more in-depth multivariate and mechanistic clinical analyses were limited. Moreover, no recommendations for monitoring viral persistence were available; therefore, only a subset of severely ill patients admitted to the ICU was evaluated. Despite that, an important connection between clinical and laboratory information was studied, revealing the continuous evolution of H1N1pdm HA sequences and the stability of the NA gene in severely ill patients [14].
In conclusion, this study provides evidence that severe H1N1pdm infection is associated with significant morbidity and mortality in cancer patients. In these patients, viral persistence without the emergence of antiviral resistance may occur during the clinical course of the disease. This result has major implications for the clinical management of H1N1pdm infections and infection control strategies. Our study may provide insights into H1N1pdm shedding and might contribute to the development of new guidelines to manage cancer patients with H1N1pdm infection.
Methods
Ethics statement
The Ethics Committee (Comitê de Ética em Pesquisa; CEP; http://www.inca.gov.br/conteudo_view.asp?id=2380) at the Instituto Nacional de Câncer (INCa), Rio de Janeiro, Brazil, headed by Dr. Adriana Scheliga approved the study under protocol #18/2010 and waived the need for informed consent.
Design and setting
This was a prospective cohort study conducted in the Hospital do Câncer-I, Instituto Nacional de Câncer (HC-I-INCa), Rio de Janeiro, Brazil, from July 8th to October 1st, 2009. The HCI-INCa is a 160-bed comprehensive cancer center primarily for the population of Rio de Janeiro and neighboring states. The present study was strictly observational, and every clinical decision was at the discretion of the attending physician.
Patients, data collection and definitions
All patients with a definite diagnosis of cancer requiring hospital admission for any reason and who displayed influenza-like illness were evaluated. Patients in complete remission from cancer for more than five years were not considered.
Data were collected using a standardized case report form that included demographic data, clinical presentation, comorbidities, cancer status, use of immunosuppressive therapies, time course of acute illness, need for intensive care, use of antivirals, adjunctive therapies, advanced life support and in-hospital mortality (supporting information; SI). Patients were included if they had a fever (>37.8°C) and/or respiratory influenza-like illness and confirmed influenza A H1N1pdm diagnosis (by at least one of three assays, IFI, rRT-PCR or cell culture, and according to case definitions from the WHO [46]). Patients were treated according to the Brazilian Public Health guidelines [47].
Sample collection and analysis
Nasopharyngeal Dacron-swab specimens were collected from all patients and placed onto transport medium (Hanks solution with 100 U/mL penicillin and 100 µg/mL streptomycin) at the initial evaluation. Tracheal aspirates were also obtained if the patient required tracheal intubation. Patients' clinical samples were directly tested for a panel of respiratory viruses using an IFI assay for influenza A (respiratory virus panel; Biotrin, Mount Merrion, Co. Dublin, Ireland.). Specimens were also sent to the Brazilian National Influenza Center (IOC/Fiocruz) for H1N1pdm confirmation using rRT-PCR, which was performed in accordance with the current guidelines from the WHO/CDC [46]. Viral shedding was evaluated in the subset of patients that remained under mechanical ventilation for longer than seven days and in those patients with persistent hypoxemia and pulmonary infiltrate. These patients received a specific number to which their sample number was appended. That is, the first sample was “s1”, and subsequent specimens were numbered consecutively (Table S12). Viral secretion was evaluated using both cell culture and rRT-PCR assays until it was negative. Virus isolation was performed in Madin-Darby canine kidney (MDCK) cells and/or embryonated eggs (see Text S1 and Table S12). The functional antiviral assay was performed using the NA-Star kit (Applied Biosystems, CA), according to the manufacturer's instructions. The RT-PCR protocol for sequencing using the Sanger method and pyrosequencing [48] are presented in the SI, as well as the phylogenetic analysis.
Blood samples were routinely sent for bacterial culturing, as were tracheal aspirates if the patient was intubated.
Statistical analysis
Standard descriptive statistics were used to describe the study population. Continuous variables were reported as the mean ± standard deviation or median (range) as appropriate. Univariate analysis was used to identify factors associated with hospital mortality. Two-sample t-tests and a chi-square or Fisher's exact test were also used. Two-tailed P values <0.05 were considered statistically significant.
Supporting Information
Clinical investigation and Influenza virus assays.
(0.06 MB DOC)
Diagnostic tests for Influenza A virus.
(0.04 MB DOC)
Patient's characteristics and outcomes according to age range.
(0.04 MB DOC)
Prevalence of underlying malignancies.
(0.03 MB DOC)
Use of Chemotherapy, corticosteroids and granulocyte colony stimulating factor previous to H1N1pdm infection.
(0.04 MB DOC)
Patient's Neutropenia.
(0.04 MB DOC)
Oseltamivir treatments.
(0.03 MB DOC)
Focus of bacterial infection in cancer patients with Influenza A H1N1pdm.
(0.03 MB DOC)
Bacteria isolates from cancer patients with Influenza A H1N1pdm.
(0.03 MB DOC)
Organ Dysfunctions 72 h after H1N1pdm diagnosis.
(0.03 MB DOC)
Frequency of signs and symptoms at clinical suspicion.
(0.03 MB DOC)
Pulmonary infiltrates at Influenza diagnosis and hospital discharge, by chest radiography.
(0.03 MB DOC)
Clinical and Viral Characteristics of the followed-up cohort.
(0.08 MB DOC)
Phylogenetic tree of HA gene from the classical swine, Eurasian swine, American Avian and human seasonal lineages. The bootstrap probability is indicated for each interior branch, all values below 80% are hidden. The scale bar indicates the number of amino acid changes per site. Colored circles indicate the samples from our study. This tree is unrooted. Each Influenza HA lineage is displayed beside their respective clade.
(0.12 MB TIF)
Phylogenetic tree of NA gene from the followed-up cohort. The bootstrap probability is not indicated for each interior branch since it is below 85%. The scale bar indicates the number of amino acid changes per site. The tree is rooted by California/07/2009 NA sequence.
(0.18 MB TIF)
Acknowledgments
We are indebted to the Instituto Nacional de Câncer, Fiocruz, FAPERJ and CNPq for their support. The authors thank Marc-Alain Widdowson (CDC/CCID/NCIRD) for critical review of the manuscript. Alicia Fry (CDC Atlanta epidemiology intelligence officer) was also extremely helpful in discussions on virus shedding.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported by Instituto Nacional de Câncer (www.inca.gov.br), Instituto Oswaldo Cruz/Fiocruz (www.ioc.fiocruz.br) and Brazilian Ministry of Health/Decit (http://dtr2001.saude.gov.br/sctie/decit/index.htm). Dr. Soares, Dr. Bozza and Dr. Siqueira were supported in part by individual research grants from CNPq (www.cnpq.br). Dr. Souza used, in part, individual research grants from FAPERJ (www.faperj.br). FAPERJ/SESDEC/CNPq provided the grant #E-26/110.778/2010 (PPSUS - DECIT/SCTIE/MS). Thanks are also due to IOC/Fiocruz for providing MM a PhD fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dominguez-Cherit G, Lapinsky SE, Macias AE, Pinto R, Espinosa-Perez L, et al. Critically Ill patients with 2009 influenza A(H1N1) in Mexico. Jama. 2009;302:1880–1887. doi: 10.1001/jama.2009.1536. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Zarychanski R, Pinto R, Cook DJ, Marshall J, et al. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. Jama. 2009;302:1872–1879. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 3.Ministério da Saúde. 2009;22 Informe epidemiológico: Influenza pandêmica (H1N1) 2009 - Update 10 Available: http://portal.saude.gov.br/portal/arquivos/pdf/boletim_influenza_se_47.pdf. Accessed 2009 december. [Google Scholar]
- 4.Lapinsky SE. H1N1 novel influenza A in pregnant and immunocompromised patients. Crit Care Med. 2009;38:e52–e57. doi: 10.1097/CCM.0b013e3181c85d5f. [DOI] [PubMed] [Google Scholar]
- 5.Kunisaki KM, Janoff EN. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect Dis. 2009;9:493–504. doi: 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casper C, Englund J, Boeckh M. How I treat influenza in patients with hematologic malignancies. Blood. 2010;115:1331–1342. doi: 10.1182/blood-2009-11-255455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha BA, Thekkel V, Krilov L. Nosocomial swine influenza (H1N1) pneumonia: lessons learned from an illustrative case. J Hosp Infect. 2010;74:278–281. doi: 10.1016/j.jhin.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Hajjar LA, Mauad T, Galas FR, Kumar A, da Silva LF, et al. Ann Oncol 28: In press -; 2010. Severe novel influenza A (H1N1) infection in cancer patients. doi;10.1093/annonc/mdq254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redelman-Sidi G, Sepkowitz KA, Huang CK, Park S, Stiles J, et al. 2009 H1N1 influenza infection in cancer patients and hematopoietic stem cell transplant recipients. J Infect. 2010;60:257–263. doi: 10.1016/j.jinf.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Seiter K, Nadelman RB, Liu D, Ahmed T, Montecalvo MA. Novel influenza A (H1N1) in patients with hematologic malignancies. J Clin Oncol. 2010;28:e27–e29. doi: 10.1200/JCO.2009.25.8889. [DOI] [PubMed] [Google Scholar]
- 11.Takiyama A, Wang L, Tanino M, Kimura T, Kawagishi N, et al. Sudden death of a patient with pandemic influenza (A/H1N1pdm) virus infection by acute respiratory distress syndrome. Jpn J Infect Dis. 2010;63:72–74. [PubMed] [Google Scholar]
- 12.Casper C, Englund J, Boeckh M. How we treat influenza in patients with hematologic malignancies. Blood. 2010;115:1331–1342. doi: 10.1182/blood-2009-11-255455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouphael NG, Talati NJ, Vaughan C, Cunningham K, Moreira R, et al. Infections associated with haemophagocytic syndrome. Lancet Infect Dis. 2007;7:814–822. doi: 10.1016/S1473-3099(07)70290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Oseltamivir-resistant pandemic (H1N1) 2009 influenza virus, Weekly Epidem. Record. 2009;30 Available: http://www.who.int/wer/2009/wer8444.pdf. Accessed 2009 October. [Google Scholar]
- 15.McMinn P, Carrello A, Cole C, Baker D, Hampson A. Antigenic drift of influenza A (H3N2) virus in a persistently infected immunocompromised host is similar to that occurring in the community. Clin Infect Dis. 1999;29:456–458. doi: 10.1086/520243. [DOI] [PubMed] [Google Scholar]
- 16.To KK, Chan KH, Li IW, Tsang TY, Tse H, et al. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. Human infection with pandemic (H1N1) 2009 virus: Updated interim WHO guidance on global surveillance. 2009;28 Available: http://www.who.int/csr/resources/publications/swineflu/surveillance/en/index.html. Accessed 2009 April. [Google Scholar]
- 18.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melidou A, Gioula G, Exindari M, Chatzidimitriou D, Diza E, et al. Molecular and phylogenetic analysis of the haemagglutinin gene of pandemic influenza H1N1 2009 viruses associated with severe and fatal infections. Virus Res. 2010;151:192–199. doi: 10.1016/j.virusres.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization. Protocol of virus sequencing. 2009;12 Available: http://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf. Accessed 2009 May. [Google Scholar]
- 21.Centers of Disease Control and Prevention (CDC) Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients - Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:893–896. [PubMed] [Google Scholar]
- 22.Witkop CT, Duffy MR, Macias EA, Gibbons TF, Escobar JD, et al. Novel Influenza A (H1N1) outbreak at the U.S. Air Force Academy: epidemiology and viral shedding duration. Am J Prev Med. 2010;38:121–126. doi: 10.1016/j.amepre.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Cao B, Li XW, Mao Y, Wang J, Lu HZ, et al. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N Engl J Med. 2009;361:2507–2517. doi: 10.1056/NEJMoa0906612. [DOI] [PubMed] [Google Scholar]
- 24.Li IW, Hung IF, To KK, Chan KH, Wong SS, et al. The natural viral load profile of patients with pandemic swine-origin influenza A H1N1 2009 (pH1N1) and the effect of oseltamivir treatment. Chest. 2010;137:759–768. doi: 10.1378/chest.09-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleury H, Burrel S, Balick Weber C, Hadrien R, Blanco P, et al. Prolonged shedding of influenza A(H1N1)v virus: two case reports from France 2009. Euro Surveill 14: pii = 19434. 2009;28 Available: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19434. Accessed 2009 December. [PubMed] [Google Scholar]
- 26.Redelman-Sidi G, Sepkowitz KA, Huang CK, Park S, Stiles J, et al. H1N1 Influenza Infection In Cancer Patients And Hematopoietic Stem Cell Transplant Recipients. J Infect. 2010;60:257–263. doi: 10.1016/j.jinf.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Webb SA, Pettila V, Seppelt I, Bellomo R, Bailey M, et al. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361:1925–1934. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 28.Azoulay E, Thiery G, Chevret S, Moreau D, Darmon M, et al. The prognosis of acute respiratory failure in critically ill cancer patients. Medicine (Baltimore) 2004;83:360–370. doi: 10.1097/01.md.0000145370.63676.fb. [DOI] [PubMed] [Google Scholar]
- 29.Soares M, Caruso P, Silva E, Teles JM, Lobo SM, et al. Characteristics and outcomes of patients with cancer requiring admission to intensive care units: a prospective multicenter study. Crit Care Med. 2010;38:9–15. doi: 10.1097/CCM.0b013e3181c0349e. [DOI] [PubMed] [Google Scholar]
- 30.Hayden FG, Belshe R, Villanueva C, Lanno R, Hughes C, et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis. 2004;189:440–449. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- 31.Cowling BJ, Chan KH, Fang VJ, Lau LL, So HC, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010;362:2175–2184. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seville MT, Blair JE, Vikram HR, Kusne S. H1N1 Influenza in Hospitalized Transplant Recipients. Transplantation. 2010;90:571–574. doi: 10.1097/TP.0b013e3181e86af0. [DOI] [PubMed] [Google Scholar]
- 33.Hien TT, Boni MF, Bryant JE, Ngan TT, Wolbers M, et al. Early pandemic influenza (2009 H1N1) in Ho Chi Minh City, Vietnam: a clinical virological and epidemiological analysis. PLoS Med. 2010;7:e1000277. doi: 10.1371/journal.pmed.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Serres G, Rouleau I, Hamelin ME, Quach C, Skowronski D, et al. Contagious period for pandemic (H1N1) 2009. Emerg Infect Dis. 2010;16:783–788. doi: 10.3201/eid1605.091894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling LM, Chow AL, Lye DC, Tan AS, Krishnan P, et al. Effects of early oseltamivir therapy on viral shedding in 2009 pandemic influenza A (H1N1) virus infection. Clin Infect Dis. 2010;50:963–969. doi: 10.1086/651083. [DOI] [PubMed] [Google Scholar]
- 36.Mora M, Rodriguez-Castellano E, Pano-Pardo JR, Gonzalez-Garcia J, Navarro C, et al. Influenza A pandemic (H1N1) 2009 virus and HIV. Emerg Infect Dis. 2010;16:1175–1176. doi: 10.3201/eid1607.091339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J, Ma J, Wang Q. Evolutionary trends of A(H1N1) influenza virus hemagglutinin since 1918. PLoS One. 2009;4:e7789. doi: 10.1371/journal.pone.0007789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J Infect Dis. 2006;193:760–764. doi: 10.1086/500465. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi S, Kimura F, Kobayashi A, Sato K, Motoyoshi K. Influenza virus reactivation after remission with oseltamivir treatment in a patient undergoing nonmyeloablative bone marrow transplantation. J Infect Chemother. 2008;14:308–310. doi: 10.1007/s10156-008-0615-2. [DOI] [PubMed] [Google Scholar]
- 42.Baz M, Abed Y, McDonald J, Boivin G. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis. 2006;43:1555–1561. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 43.Nichols WG, Guthrie KA, Corey L, Boeckh M. Influenza infections after hematopoietic stem cell transplantation: risk factors, mortality, and the effect of antiviral therapy. Clin Infect Dis. 2004;39:1300–1306. doi: 10.1086/425004. [DOI] [PubMed] [Google Scholar]
- 44.Kidd IM, Down J, Nastouli E, Shulman R, Grant PR, et al. H1N1 pneumonitis treated with intravenous zanamivir. Lancet. 2009;374:1036. doi: 10.1016/S0140-6736(09)61528-2. [DOI] [PubMed] [Google Scholar]
- 45.Ministério da Saúde. Diretrizes para o enfrentamento à pandemia de influenza A (H1N1): ações da atenção primária à saúde. 2009;25 Available: http://portal.saude.gov.br/portal/arquivos/pdf/protocolo_influenzaa_aps_atualizado.pdf. Accessed 2010 January. [Google Scholar]
- 46.World Health Organization. CDC protocol of realtime RTPCR for influenza A(H1N1). 2009;30 Available: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf. Accessed 2009 April. [Google Scholar]
- 47.Ministério da Saúde. Protocolo de manejo clínico e vigilância epidemiológica da influenza. 2009;05 Available: http://portal.saude.gov.br/portal/arquivos/pdf/protocolo_de_manejo_clinico_05_08_2009.pdf. Accessed 2009 August. [Google Scholar]
- 48.Deyde VM, Sheu TG, Trujillo AA, Okomo-Adhiambo M, Garten R, et al. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob Agents Chemother. 2010;54:1102–1110. doi: 10.1128/AAC.01417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical investigation and Influenza virus assays.
(0.06 MB DOC)
Diagnostic tests for Influenza A virus.
(0.04 MB DOC)
Patient's characteristics and outcomes according to age range.
(0.04 MB DOC)
Prevalence of underlying malignancies.
(0.03 MB DOC)
Use of Chemotherapy, corticosteroids and granulocyte colony stimulating factor previous to H1N1pdm infection.
(0.04 MB DOC)
Patient's Neutropenia.
(0.04 MB DOC)
Oseltamivir treatments.
(0.03 MB DOC)
Focus of bacterial infection in cancer patients with Influenza A H1N1pdm.
(0.03 MB DOC)
Bacteria isolates from cancer patients with Influenza A H1N1pdm.
(0.03 MB DOC)
Organ Dysfunctions 72 h after H1N1pdm diagnosis.
(0.03 MB DOC)
Frequency of signs and symptoms at clinical suspicion.
(0.03 MB DOC)
Pulmonary infiltrates at Influenza diagnosis and hospital discharge, by chest radiography.
(0.03 MB DOC)
Clinical and Viral Characteristics of the followed-up cohort.
(0.08 MB DOC)
Phylogenetic tree of HA gene from the classical swine, Eurasian swine, American Avian and human seasonal lineages. The bootstrap probability is indicated for each interior branch, all values below 80% are hidden. The scale bar indicates the number of amino acid changes per site. Colored circles indicate the samples from our study. This tree is unrooted. Each Influenza HA lineage is displayed beside their respective clade.
(0.12 MB TIF)
Phylogenetic tree of NA gene from the followed-up cohort. The bootstrap probability is not indicated for each interior branch since it is below 85%. The scale bar indicates the number of amino acid changes per site. The tree is rooted by California/07/2009 NA sequence.
(0.18 MB TIF)