Abstract
Pyridoxal phosphate (PLP) dependent enzymes catalyze many different types of reactions at the α-, β-, and γ-carbons of amine and amino acid substrates. Dialkylglycine decarboxylase (DGD) is an unusual PLP dependent enzyme that catalyzes two reaction types, decarboxylation and transamination, in the same active site. A structurally-based, functional model has been proposed for the DGD active site, which maintains that R406 is important in determining substrate specificity through interactions with the substrate carboxylate while W138 provides specificity for short-chain alkyl groups. The mechanistic roles of R406 and W138 were investigated using site directed mutagenesis, alternate substrates, and analysis of steady-state and half-reaction kinetics. Experiments on the R406M and R406K mutants confirm the importance of R406 in substrate binding. Surprisingly, this work also shows that the positive charge of R406 facilitates catalysis of decarboxylation. The W138F mutant demonstrates that W138 indeed acts to limit the size of the subsite C binding pocket, determining specificity for 2,2-dialkylglycines with small side chains as predicted by the model. Finally, work with the double mutant W138F/M141R shows that these mutations expand substrate specificity to include L-glutamate and lead to an increase in specificity for L-glutamate over 2-aminoisobutyrate of approximately eight orders of magnitude compared to WT DGD.
Pyridoxal phosphate (PLP1) dependent enzymes catalyze a wide variety of reactions types at the α-, β-, and γ-carbons of amine and amino acid substrates. It was hypothesized by Dunathan (1) that PLP dependent enzymes maintain their reaction specificity through stereoelectronic effects. According to this hypothesis, a given enzyme maintains a specific orientation of the common external aldimine intermediate, aligning the scissile bond parallel to the p orbitals of the extended π system thereby maximizing orbital overlap and resonance interactions with the activated bond. The binding constraints that control orientation of the substrate in the active site contribute to both reaction specificity and substrate specificity. Thus, reaction specificity and substrate specificity are especially interrelated in PLP dependent enzymes.
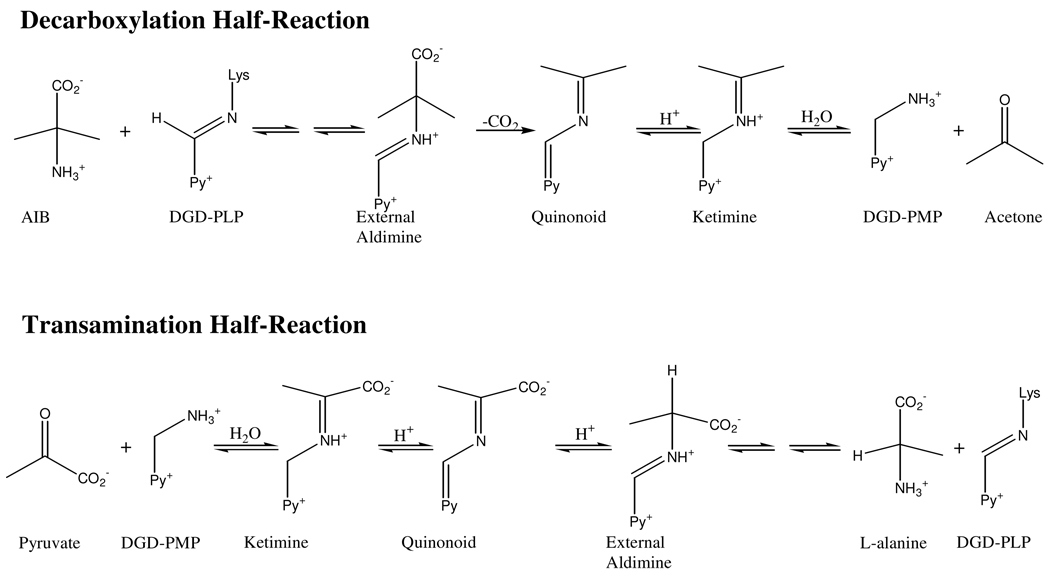
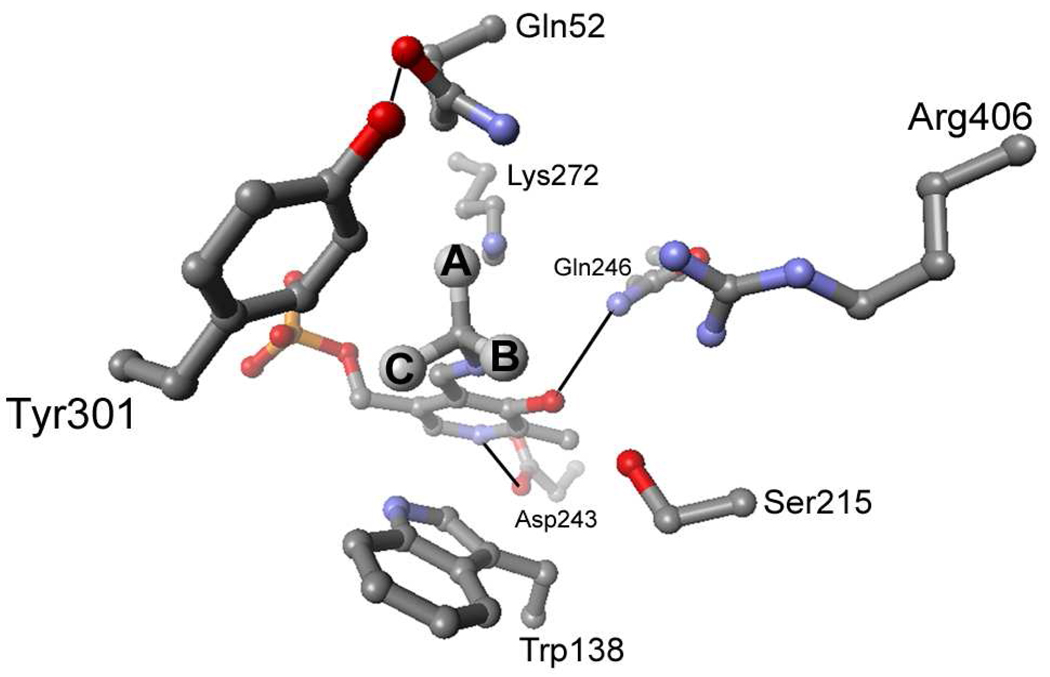
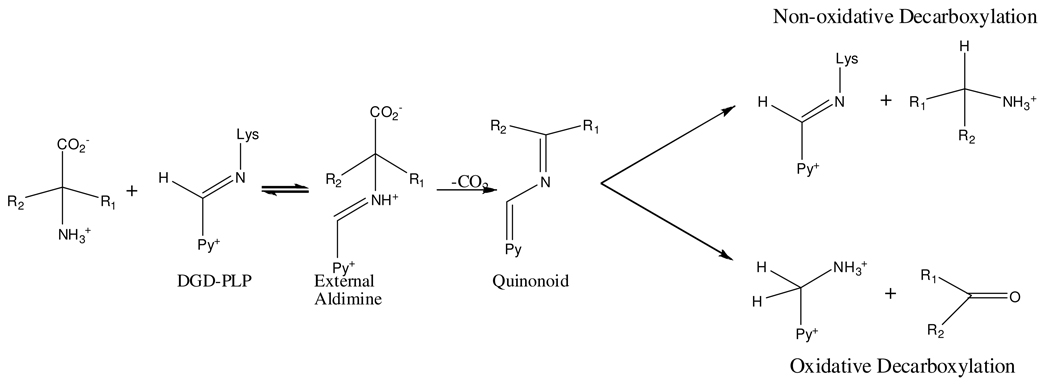
This is particularly true for dialkylglycine decarboxylase (DGD), which is an unusual PLP dependent enzyme that catalyzes decarboxylation and transamination in the same active site in distinct half-reactions (Scheme 1). A functional active site model of DGD has been proposed (2) and validated (3). In this model there are three subsites: the A subsite, near Q52 and K272, is the stereoelectronically activated position in which bond breaking and making occur; the B subsite, near R406 and S215, which can bind a carboxylate or an alkyl group; and the C subsite, near W138 and M141, which can bind an alkyl group only and is less sterically tolerant than the B subsite (Figure 1). The model suggests R406 is important in determining substrate specificity through interactions with the substrate carboxylate while W138 maintains specificity for short-chain alkyl groups by limiting the size of the substrate side chain binding pocket.
Scheme 1.
Figure 1.
Active site structure of DGD showing the positions of the A, B, and C subsites of the functional model.
Herein, steady-state and half-reaction kinetics are presented for the R406M and R406K mutants, which confirm the importance of R406 in substrate binding. This work also demonstrates that R406 plays an unexpected role in decarboxylation catalysis. Results with the W138F mutant support the binding subsite model in which W138 limits the size of the C subsite binding pocket, providing specificity for 2,2-dialkylglycines with small side chains. Finally, work with the double mutant W138F/M141R shows that these mutations lead to an expansion of the substrate specificity to include L-glutamate as a decarboxylation substrate.
Experimental Procedures
Materials and Preparation of Mutants
All chemicals were purchased from Sigma unless otherwise noted. The Quikchange site directed mutagenesis protocol (Strategene) was used to introduce the desired mutations. The pBTac (Boehringer Mannheim) plasmid containing the WT DGD gene was used as a template for all reactions except for the W138F/M141R double mutant which used the same plasmid containing the W138F DGD gene. The following primer pairs used to introduce the desired mutations, with the converted codon underlined: R406M: 5′-GGG CGG CGT GTT CAT GAT CGC GCC GCC GCT GAC G-3′ and 5′-CGT CAG CGG CGG CGC GAT CAT GAA CAC GCC GCC C-3′; R406K: 5′-GGG CGG CGT GTT CAA AAT CGC GCC GCC GCT GAC G-3′ and 5′-CGT CAG CGG CGG CGC GAT TTT GAA CAC GCC GCC C-3′; W138F: 5′-CGG CTT CGC GCA GTC GTT TCA CGG GAT GAC GGG-3′ and 5′-CCC GTC ATC CCG TGA AAC GAC TGC GCG AAG CCG-3′; and W138F/M141R: 5′-CGT TTC ACG GGC GCA CGG GCG CGG CC-3′ and 5′-GGC CGC GCC CGT GCG CCC GTG CCC CG-3′. Due to the high GC content of the DGD gene it was necessary to use 5% (v/v) DMSO in PCR reactions. After transformation into E. coli JM109 using standard procedures (4), the mutant plasmid was isolated using a commercial kit (Qiagen) and the full coding region sequenced to confirm that the desired mutation had been incorporated.
Expression and Purification of Mutants
For overexpression, 750 mL TB was inoculated with 5 mL overnight culture and grown at 37 °C until OD600 = 0.4. The cells were placed on ice for 20 minutes, expression induced with 500 µM IPTG, and cell growth continued at 33 °C for 8–10 hours. The cells were pelleted by centrifugation, resuspended in lysis buffer (20 mM TEA-HCl pH 7.8, 50 mM KCl, 20 µM PLP, 1 M ammonium sulfate) and stored at −80 °C. The thawed cell suspension was sonicated and the cell debris removed by centrifugation. The cell free extract was brought to 2.2 M ammonium sulfate, stirred at 4 °C for 1 hour and the precipitated protein removed by centrifugation. The pellet was dissolved and dialyzed into 20 mM TEA-HCl pH 7.8, 20 µM PLP and the protein solution was loaded onto a 50 mL Q Sepharose Fast Flow column (Pharmacia). A gradient of 100–400 mM KCl in 20 mM TEA-HCl pH 7.8 eluted protein. The fractions were analyzed by SDS-PAGE and those containing DGD were pooled, concentrated, brought to 1M in KCl and loaded onto a 50 mL phenyl Sepharose column (Pharmacia). The protein was eluted with a gradient of 1-0 M KCl in 20 mM TEA-HCl pH 7.8. The purest fractions, as judged by SDS-PAGE, were pooled, concentrated and dialyzed into 50 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 20 µM PLP. The protein was aliquoted, flash frozen and stored at −80°C. Protein concentration was determined using both the extinction coefficient determined for DGD and the DC protein assay kit from Bio-Rad with IgG as a standard.
Steady-state Assays
Oxidative decarboxylation of 2-aminoisobutyric acid (AIB), D,L-α-methyl serine (AMS) and D,L-α-methyl glutamate (AMG) was followed by coupling the ketone produced to the 2° ADH reaction as previously described (3). Loss of NADPH absorbance at 340 nm was followed on a Kontron UVIKON 9420. Reaction mixtures contained 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 1 unit/mL 2° ADH, 20 µM PLP and 300 µM NADPH. Stock solutions of all substrates were prepared in 20 mM TEA-succinate and the pH of the solution adjusted to 7.8. Pyruvic acid was titrated to pH 7.8 with TEA and used as the α-keto acid substrate unless otherwise noted. For assays using AMS or AMG, the ketone product of oxidative decarboxylation was tested and confirmed to be a substrate for 2° ADH. Additionally, it was confirmed that the rate of decarboxylation was linearly dependent on DGD concentration for these assays. Initial rates were plotted versus substrate concentration and fitted to the Michaelis-Menten equation.
Oxidative decarboxylation of D- or L-glutamate to produce succinic semialdehyde was followed using the NADP-dependent succinic semialdehyde dehydrogenase (SSDH) reaction, monitoring the increase of NADPH absorbance at 340 nm as previously described (5). The reaction mixtures contained 30 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 5 mM DTT, 1 mM EDTA, 20 µM PLP, 500 µM NADP+, 5 mM α-ketoglutarate and 3 units/mL SSDH. Initial rates were plotted versus substrate concentration and fitted to the Michaelis-Menten equation.
R406M and R406K Steady-state Kinetics
The steady-state kinetic parameters, kcat, KAIB and Kpyr were determined using the 2° ADH coupled assay. Briefly, one substrate was held near saturation while the other was varied. It was not possible to fully saturate R406M with AIB or AMS due to the solubility limits of these compounds and the large Km values.
Non-oxidative Decarboxylation End-point Assays for R406M
Assays to identify the amine products of non-oxidative decarboxylation were performed using a previously reported HPLC-based method (6). Reactions with 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, 53 mM pyruvate, 10 µM R406M and either 500 mM AIB or 133 mM L-alanine were allowed to react at 25 °C for 12–16 hours. The protein was denatured by adding 2 µL glacial acetic acid to the 200 µL reaction and the precipitated protein was removed by centrifugation. The reaction mixture was then derivatized with o-phthaldialdehyde (OPA) prior to HPLC analysis. Briefly, a 100 µL sample was mixed with 100 µL freshly prepared OPA reagent (4 mg solid OPA, mixed with 4.5 mL 0.1 M borate pH 10.4, 15 µL 30% (v/v) Brij-35 detergent and 10 µL β-mercaptoethanol), allowed to react for 1 minute at room temperature then acidified with 2 M acetic acid to pH 4.5. The derivatized sample was immediately run on a Hibar LiChrosorb C18 column using an Agilent 1100 HPLC connected to a Perkin-Elmer 650-15 fluorescence detector. The HPLC method (6) used 50 mM sodium acetate pH 5.7, 5% (v/v) THF (Solvent A); methanol (Solvent B); acetonitrile (Solvent C). The elution gradient was as follows: 90% A, 10% B to 35% A, 65% B from 0–15 minutes; 35% A, 65% B from 15–20 minutes; 35% A, 65% B to 50% B, 50% C from 20–25 minutes; 50% B, 50% C from 25–30 minutes. The flow rate was 1 mL/min and the products were monitored using fluorescence (excitation = 340 nm, emission = 455 nm).
Stopped-flow Multiwavelength Analysis of the AIB Decarboxylation Half-reaction in R406M
The AIB decarboxylation half-reaction was followed by rapidly mixing PLP-enzyme (in 20 µM enzyme in 50 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 20 µM PLP) against an AIB solution prepared in the same buffer in an Applied Photophysics SX.18MV-R stopped-flow spectrometer. Coenzyme spectral changes were followed from 200–800 nm using a diode array detector. The data from 300–600 nm was globally fitted using Specfit. A four-exponential model was used for the R406M data. The apparent rate constants were fitted to eq 1 for the first process seen in R406M.
| (1) |
Spectral Analysis of R406M Decarboxylation Half-reactions with AC5C and AC6C
Coenzyme spectral changes from 300–600 nm were followed using a Hewlett Packard 8453 UV-vis spectrophotometer. Reaction conditions were 50 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 20 µM PLP, 20 µM enzyme and varying concentrations of 1-aminocyclopentane-1-carboxylate (AC5C) or 1-aminocyclohexane-1-carboxylate (AC6C). The multiwavelength data for both substrates was globally fitted to a two-exponential model using Specfit. The apparent rate constants obtained were plotted versus substrate concentration and fitted to eq 1.
Analysis of Products of R406M Reaction with L-alanine
Lactate dehydrogenase (LDH) was used to test for pyruvate resulting from transamination of L-alanine. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, 20 µM R406M and 150 mM L-alanine. The reaction was incubated at 25 °C for 12 hours, at which time 2 unit/mL LDH and 300 µM NAD+ were added and the reaction monitored for an increase in absorbance indicating the formation of pyruvate. A negative control without enzyme showed that no appreciable amount of pyruvate formed due to non-enzymatic transamination under these conditions.
An end-point assay using yeast alcohol dehydrogenase (YADH) was used to test for acetaldehyde, the product of oxidative decarboxylation of L-alanine. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, 20 µM R406M, 53 mM pyruvate and 100 mM L-alanine. The reaction was incubated at 25 °C for 12 hours at which time 2 units/mL YADH and 200 µM NADH were added and the reaction monitored for a decrease in absorbance due to formation of acetaldehyde. A negative control without enzyme was included. Any change in absorbance significantly greater than observed in the negative control was used to estimate the upper limit for activity with the substrate.
An HPLC end-point assay was used to identify ethylamine, the product of non-oxidative decarboxylation of L-alanine. The reaction was prepared as described above for the oxidative decarboxylation end-point assay. After incubation the enzyme was denatured with acid and removed by centrifugation. The sample was subsequently derivatized with OPA and run on the HPLC as described above. A negative control without enzyme was included to determine any non-enzymatic decarboxylation.
W138F Steady-state Kinetics
The values of kcat, and the KM values for AIB (KAIB) and pyruvate (Kpyr), were determined using the 2° ADH coupled assay as described above. KAIB and Kpyr were determined by holding pyruvate at 50 µM (~10 × Kpyr) and AIB at 200 mM (~5 × KAIB). Studies to determine the catalytic efficiency of W138F with α-keto acids of varying side chain lengths used pyruvate, α-ketobutyrate, α-ketovalerate, α-ketocaproate. In these assays the AIB concentration was held at 200 mM (5 × KAIB) and the concentration of α-keto acid was varied. The data were fitted to the Michaelis-Menten equation.
Inhibition studies with D-phenylglycine were performed using saturating pyruvate concentration and holding the concentration of AIB at KAIB (40 mM). The data were fitted assuming competitive inhibition (eq 2), as previously observed with WT DGD (3).
| (eq 2) |
End-point Assays with W138F
W138F was tested for decarboxylation activity with D- and L-phenylglycine using end-point assays coupling the L-alanine formed from the reaction to L-alanine dehydrogenase. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 1 mM pyruvate, 20 µM PLP, 28 µM W138F and 10 mM D- or L-phenylglycine. The reactions were incubated at 25 °C for 12 hours at which time 300 µM NAD+ and 2 units/mL L-alanine dehydrogenase were added and the reaction monitored for an increase in absorbance indicating the formation of L-alanine. In all cases, a sample without enzyme was included as a negative control.
Spectral Analysis of W138F Decarboxylation of L-phenylglycine
Coenzyme spectral changes from 300–600 nm were followed as described above for the spectral analysis of AC5C and AC6C decarboxylation. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, 20 µM W138F and varying concentrations of L-phenylglycine. A two-exponential model using Specfit best fitted the data.
Steady-state Kinetics with W138F/M141R
Reaction with AIB using the 2° ADH coupled assay gave the steady-state kinetic parameters kcat and KAIB. Kpyr was not determined. Instead, it was assumed that Kpyr in W138F/M141R is essentially the same as in W138F, as observed for KAIB. When determining KAIB, pyruvate was held at a saturating concentration (1 mM) based on the Kpyr for W138F. The SSDH coupled assay was used to study activity with L-glutamate. Reaction conditions were 30 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 5 mM DTT, 1 mM EDTA, 20 µM PLP, 500 µM NADP+, 5 mM α-ketoglutarate, 3 units/mL SSDH and varying concentrations of L-glutamate.
Inhibition studies with D-glutamate and AMG were performed using the 2° ADH coupled assay with AIB decarboxylation. Pyruvate was used at a saturating concentration and AIB was held at KAIB (40 mM) while the concentration of inhibitor was varied. The data was fitted assuming competitive inhibition using eq 2.
End-point Assays with D- and L-glutamate
Decarboxylation of D- and L-glutamate in WT, W138F and W138F/M141R was studied using the SSDH coupled assay. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, and 1 mM α-ketoglutarate. D- and L-glutamate were held at 250 mM while 20 µM W138F, 45 µM W138F/M141R or 5 µM WT was used. Samples were incubated at 25 °C for 12–16 hours at which time 5 mM DTT, 1 mM EDTA, 500 µM NADP+, and 3 units/mL SSDH were added. The reactions were monitored for an increase in absorbance indicating the formation of succinic semialdehyde resulting from oxidative decarboxylation of glutamate. In all cases, a negative control without enzyme was also included. Any change in absorbance significantly greater than observed in the negative control was used to estimate the upper limit for activity with the substrate.
Non-oxidative Decarboxylation End-point Assays for W138F/M141R
Non-oxidative decarboxylation of D- or L-glutamate to form γ-aminobutyric acid (GABA) was monitored using HPLC, using the method described above. Reaction conditions were 100 mM TEA-succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, and 1 mM α-ketoglutarate and 200 mM either D- or L-glutamate. The 200 µL reaction was incubated at 25 °C for 12–16 hours, after which enzyme was denatured with 2 µL glacial acetic acid and removed by centrifugation. The sample was derivatized with OPA and analyzed using the HPLC method described above. Negative controls without enzyme were included to check for background decarboxylation.
Results
R406M and R406K Steady-State Kinetics
The steady-state kinetic parameters are given in Table 1. Both mutants display nonlinear kinetic traces indicative of a fast enzyme form that converts to a slow form, as observed previously with WT (7). The data presented are for the fast form of the enzyme, with the exception of Kpyr for R406M which was determined for the slow enzyme form due to the experimental difficulty of determining this parameter for the fast enzyme form. The R406M mutant shows an 80-fold decrease in kcat relative to WT, while kcat in R406K only decreases 10-fold. The Km values for AIB and pyruvate have increased. In R406M, KAIB is 100-fold larger than for WT while Kpyr shows a smaller increase of ~35-fold. As expected, substrate binding in R406K in not weakened to the degree seen in R406M; KAIB increases ~10-fold while Kpyr increases ~80-fold in R406K. HPLC analysis of the reaction products find that R406M does not non-oxidatively decarboxylate AIB to form isopropylamine (data not shown).
Table 1.
Steady-state Kinetic Parameters for R406 Mutantsa
| AIB |
AMS |
|||||||
|---|---|---|---|---|---|---|---|---|
| kcat (s−1) |
KAIB (mM) |
Kpyr (mM) | kcat/KAIB (M−1s−1) |
kcat (s−1) |
Kams (mM) |
Kpyr (mM) |
kcat/Kams (M−1s−1) |
|
| R406M | 0.22 (0.02) | 360 (61) | 3.5b (0.5) | 0.6 (0.1) | 0.074 (0.004) | 452 (73) | 3.7 (0.6) | 0.16 (0.03) |
| R406K | 1.6 (0.1) | 33 (6) | 8.8 (0.8) | 49 (9) | 0.76 (0.05) | 62 (11) | 6.7 (1.1) | 12 (2) |
| WT | 17.7 (0.5)c | 2.2 (0.2) | 0.098(0.008) | 8000 (800) | 4.00 (0.09) | 4.5 (0.3) | nd | 890 (60) |
Errors are given in parentheses. nd, not determined.
Kpyr was determined for the slow form of R406M DGD.
WT values for reaction with AIB taken from (16).
The mutants using AMS as a substrate show trends in kcat and Km similar to those observed with AIB. With AMS as a substrate, the fast and slow forms of the enzyme are not observed. The rate decreases approximately 55-fold in R406M and 5-fold in R406K compared to WT, while Kams increases 100-fold for R406M and 14-fold for R406K. These results suggest that R406 interacts strongly with both amino and keto acid substrates by forming a hydrogen bond/salt bridge to their carboxylate groups, as expected based on X-ray crystallographic structures (8). The decrease in the decarboxylation rate constant, however, is unexpected and suggests that R406 plays a direct role in the decarboxylation half-reaction in addition to substrate binding.
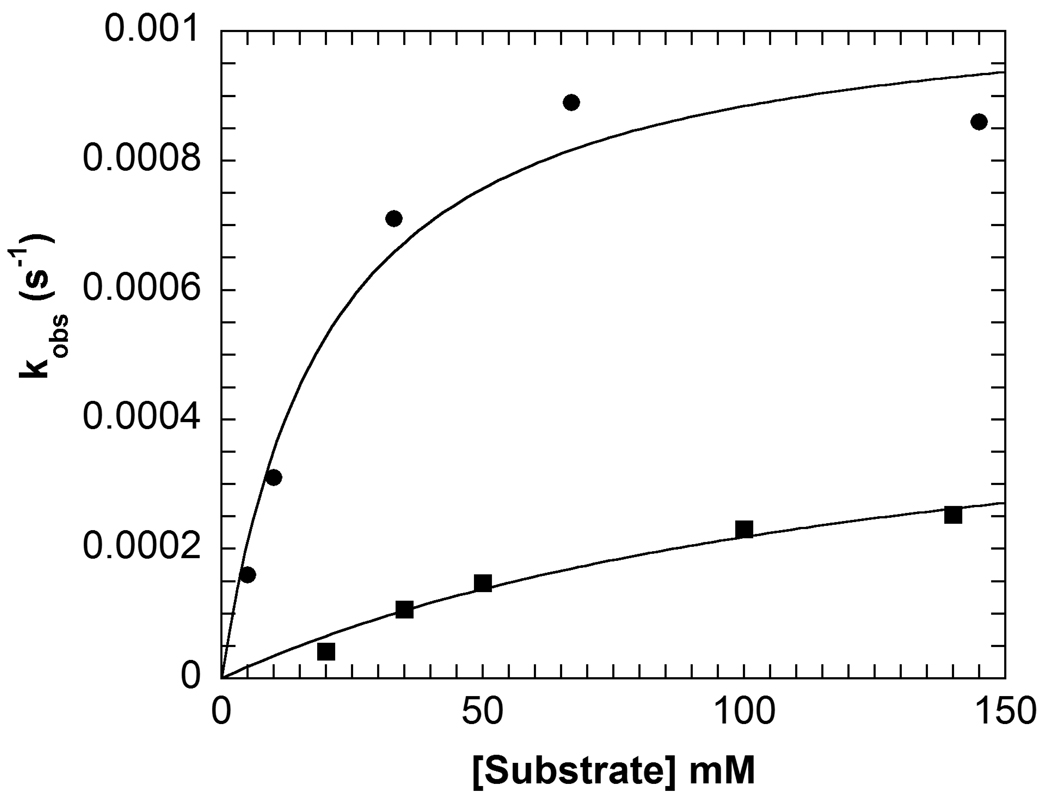
The decarboxylation half-reaction with the cyclic substrates, AC5C and AC6C, was investigated in R406M using the spectral changes of the PLP cofactor. The spectral changes were fitted to a one-exponential equation for both substrates. This fitting (Figure 2) gave kmax and the apparent dissociation constant, Kapp (Table 2). The rate of decarboxylation for these substrates decreases significantly, 105-fold for AC5C and 4,600-fold for AC6C compared to WT. The apparent dissociation constants are slightly increased for the substrates; ~4-fold for AC5C and ~3-fold for AC6C.
Figure 2.
Concentration dependence of the observed rate constant for the decarboxylation half-reaction with AC5C (circles) and AC6C (squares) in R406M obtained from analysis of multiwavelength data, the lines represent fits to the Michaelis-Menten equation. Experimental conditions: 20 µM enzyme, 100 mM TEA succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, varying concentrations of AC5C or AC6C.
Table 2.
Kinetic Parameters for Alternate Substratesa
| AC5C | AC6C | |||
|---|---|---|---|---|
| kmax (s−1) |
Kapp (mM) |
kmax (s−1) |
Kapp (mM) |
|
| R406M | 9.4 (0.9) × 10−4 | 15 (6) | 5 (1) × 10−4 | 140 (50) |
| WTb | 0.98 (0.03)c | 4.0 (0.8) | 2.3 (0.2)c | 53 (150) |
Errors are given in parentheses.
WT values taken from (Sun 37, 3865).
The values given for WT are steady-state kcat and Km values; it is assumed that the decarboxylation half-reaction is rate limiting in WT for AC5C and AC6C, so that kcat = kmax.
Spectral Analysis of the AIB Decarboxylation Half-Reaction in R406M
The spectral changes seen in the AIB decarboxylation half-reaction in R406M are complex, and accounted for by a serial four-step model (data not shown). Although the spectral changes are complex and the exact chemical nature of each process is not clear, fitting the concentration dependence of the apparent rate constants shows clearly that the decarboxylation half-reaction in R406M is not rapid. For example, the kinetic parameters for the fastest process, for which the spectral changes are consistent with decarboxylation to form the PMP form of the enzyme, are kmax1 = 0.46 ± 0.04 s−1, Kapp1 = 800 ± 160 mM. These are in reasonably good agreement with the steady-state values found for the fast enzyme form of R406M: kcat = 0.22 ± 0.02 s−1 and KAIB = 360 ± 61 mM.
Analysis of the Products of R406M Reaction with L-alanine
The product(s) of reaction with L-alanine were investigated using several assays. Assays using lactate dehydrogenase showed clearly that pyruvate from transamination of L-alanine was formed when R406M was incubated with L-alanine. Assays using YADH were used to evaluate if R406M catalyzed the oxidative decarboxylation of L-alanine to acetaldehyde. Even after 12 hours there was no formation of acetaldehyde under the conditions tested. To exclude the possibility that R406M catalyzes non-oxidative decarboxylation of L-alanine, HPLC analysis was performed. Under no conditions was ethylamine, the product of non-oxidative decarboxylation, observed. Together these results show that R406M is capable of catalyzing only the transamination of L-alanine and that no decarboxylation occurs with this substrate. Again, this implies an important role for R406 specifically in decarboxylation since WT can decarboxylate alanine. (3)
W138F Steady-state Kinetics
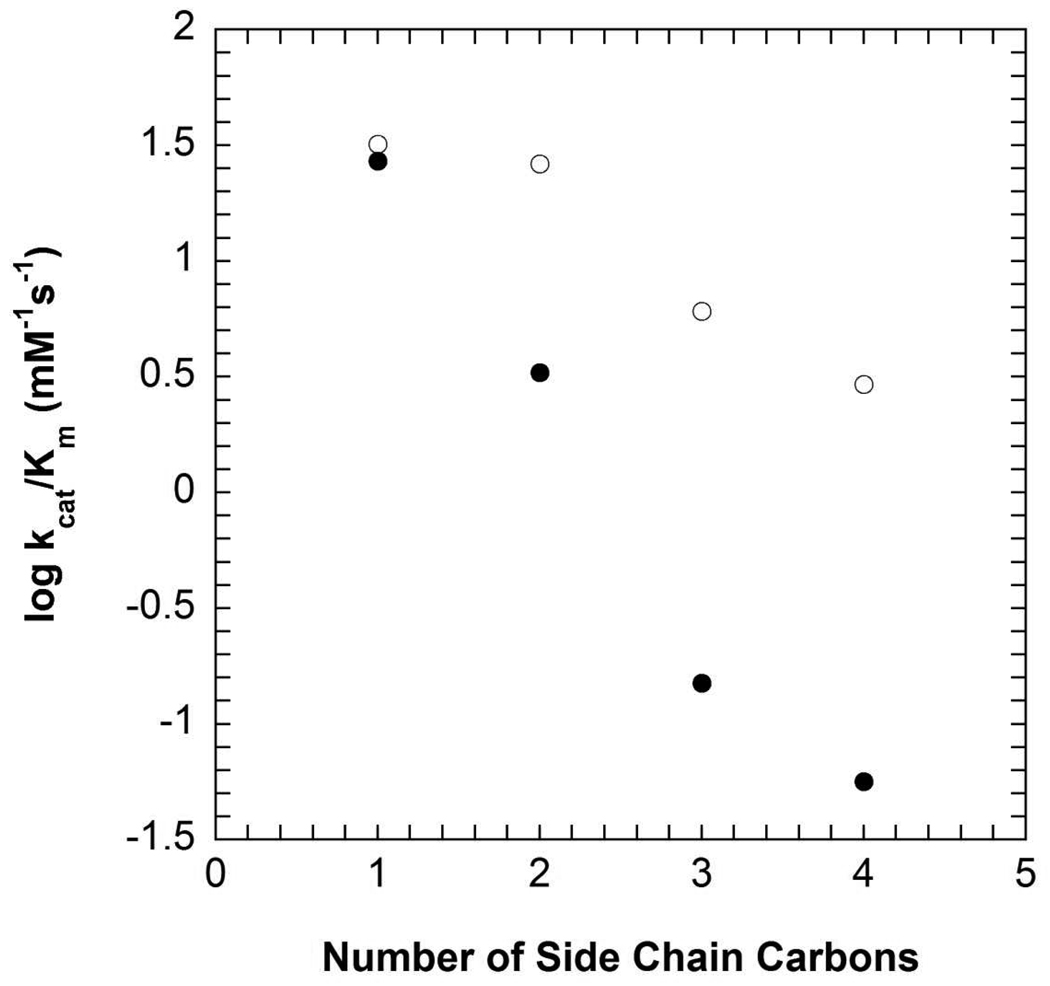
The steady-state kinetic parameters for W138F are given in Table 3. W138F shows ~100-fold decrease in kcat compared to WT, while KAIB increases approximately 20-fold. In W138F the catalytic efficiency (kcat/Kα-keto acid) decreases approximately one order of magnitude as the side chain length of the α-keto acid increases from one carbon (pyruvate) to four carbons (α-ketocaproate). In WT kcat/Kα-keto acid decreases three orders of magnitude (Figure 3) over the same range (3). Figure 3 plots the logarithm of kcat/Kα-keto acid vs. the number of side chain carbons; the former corresponds to the free energy of activation for reaction of free PMP enzyme with α-keto acid, while the latter is expected to be a measure of the energy of interaction of the α-keto acid side chain with the C subsite. The negative correlation implies that repulsive interactions between the α-keto acid side chain with the C subsite raise the free energy of activation as the size of the α-keto acid is increased.
Table 3.
Steady-State Kinetic Parameters for Binding and Oxidative Decarboxylation of Alternate Substrates in W138F Mutantsa
| D- glutamate |
L-glutamate | AIB | ||||||
|---|---|---|---|---|---|---|---|---|
| Kib (mM) |
kcat (s−1) |
KL-glu (mM) |
kcat/KL-glu (M−1s−1) |
kcat (s−1) |
KAIB (mM) |
kcat/KAIB (M−1s−1) |
Ratio kcat/KL-glu: kcat/KAIB |
|
| W138F/M141R | 89 (10) | 0.0022 (0.0003) |
270 (74) |
0.008 (0.002) |
0.0040 (0.0002) |
41 (5) |
0.10 (0.01) |
0.08 |
| W138F | 4 × 10−6c | 0.179 (0.007) |
37 (4) |
4.8 (0.6) |
||||
| WT | > 500 | 6 × 10−6c | 480d (40) |
8 × 10−6 | 17.7e (0.5) |
2.2e (0.2) |
8000e (800) |
1 × 10−9 |
Errors are given in parentheses.
Ki was determined assuming competitive inhibition against AIB in the AIB decarboxylation reaction.
Based on the total absorbance change observed in the SSDH end-point assay.
The Km value reported is the Ki obtained assuming competitive inhibition against AIB.
Data for WT AIB decarboxylation taken from (17).
Figure 3.
Rate constants for oxidative decarboxyation of AIB using α-ketoacids with linear side chains of one to four carbons in length for WT (closed circles) and W138F (open circles). Experimental conditions: 100 mM TEA succinate pH 7.8, 100 mM dipotassium succinate, 200 mM AIB, 20 µM PLP, 1 unit/mL 2° ADH, 300 µM NADPH.
W138F can decarboxylate L-phenylglycine, as observed for WT (3). End-point assays using L-alanine dehydrogenase (to detect alanine formed from pyruvate in the reaction mixture) show L-phenylglycine undergoes oxidative decarboxylation (data not shown). The reactivity with D-phenylglycine is quite different. In contrast to WT, which shows no inhibition by D-phenylglycine (3), W138F is inhibited by D-phenylglycine, Ki = 50 mM. The inhibition observed was not due to pH or salt effects.
Steady-State Kinetics of W138F/M141R
The steady-state kinetic parameters for W138F/M141R with AIB, L-glutamate and D-glutamate are given in Table 3. For reaction with AIB, the double mutant shows a ~4000-fold decrease in kcat relative to WT but only a ~45-fold decrease relative to W138F. The apparent dissociation constant for AIB remains essentially the same for the double mutant as for W138F, increasing ~20-fold relative to WT.
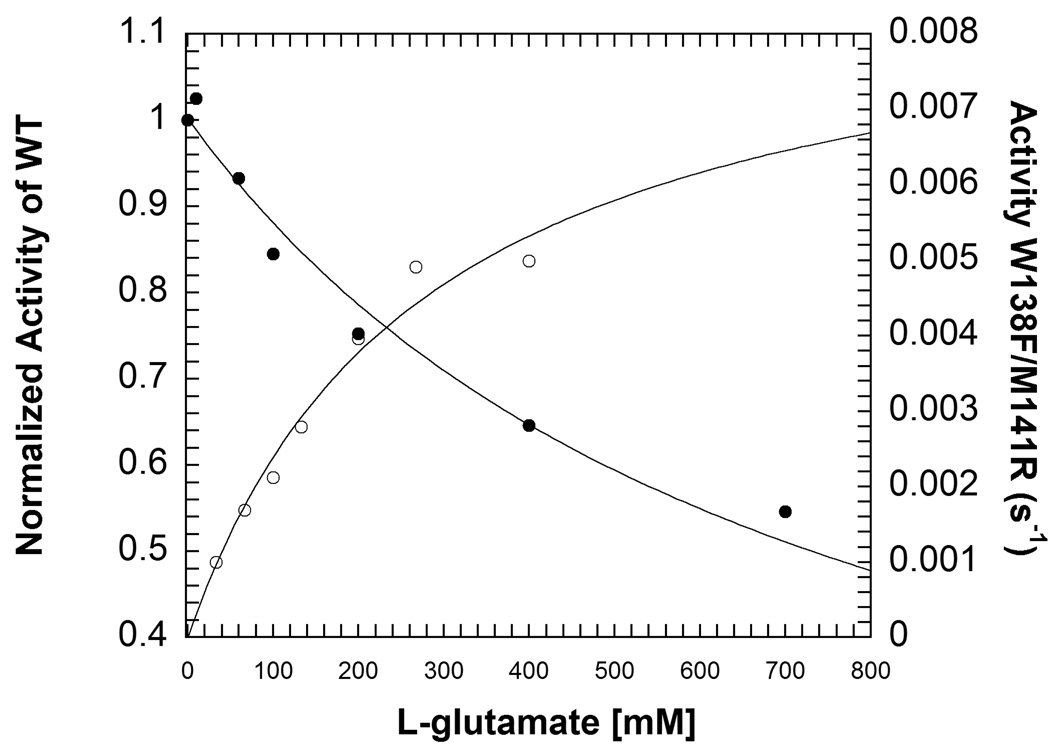
W138F/M141R catalyzes oxidative decarboxylation of L-glutamate (Figure 4). The rate of L-glutamate decarboxylation increases approximately 550-fold compared to WT and W138F. The parameters for W138F/M141R are kcat = 0.0022 ± 0.0003 s−1, KL-glu = 270 ± 74 mM. Although WT does not catalyze decarboxylation of L-glutamate at a rate fast enough to fully characterize the reaction, inhibition by L-glutamate of AIB decarboxylation can be measured. The inhibition constant (Ki) for L-glutamate with WT is 480 ± 40 mM (Figure 4). HPLC assays show that a small amount of non-oxidative decarboxylation, ~9%, occurs with L-glutamate in W138F/M141R (data not shown).
Figure 4.
Comparison of inhibition of WT by L-glutamate (closed circles) and activity in the W138F/M141R mutant (open circles). The lines represent fits to eq. 2 for WT and the Michaelis-Menten equation for W138F/M141R. Experimental conditions for WT: 100 mM TEA succinate pH 7.8, 100 mM dipotassium succinate, 20 mM AIB, 2 mM pyruvate, 20 µM PLP, 1 unit/mL 2° ADH, 300 µM NADPH and varying concentrations of L-glutamate. Experimental conditions for W138F/M141R: 30 mM TEA-succinate pH 7.8, 50 mM dipotassium succinate, 5 mM DTT, 1 mM EDTA, 20 µM PLP, 500 µM NADP+, 5 mM α-ketoglutarate, 3 units/mL SSDH and varying concentrations of L-glutamate.
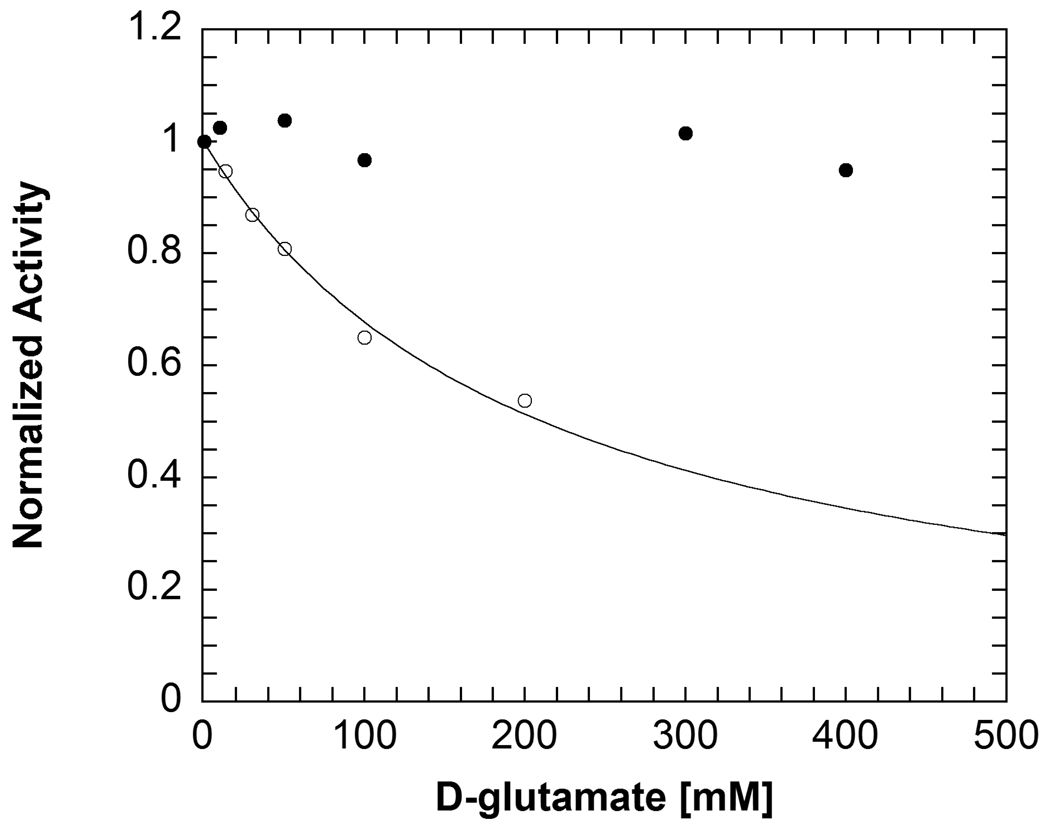
End-point assays show that W138F/M141R, W138F and WT do not use D-glutamate as a substrate. Assays with SSDH find that oxidative decarboxylation does not occur. Likewise, HPLC-based assays to identify GABA, the product of non-oxidative decarboxylation, also find that decarboxylation does not occur with D-glutamate in W138F/M141R. Inhibition studies find that W138F/M141R binds D-glutamate, with Ki = 89 ± 10 mM (Figure 5). In contrast, WT is not inhibited by D-glutamate even at concentrations up to 500 mM (Figure 5).
Figure 5.
Comparison of inhibition of WT (closed circles) and W138F/M141R (open circles) by D-glutamate. The line represents a fit to eq. 2. Inhibition studies were performed using the 2° ADH coupled assay with AIB decarboxylation. Pyruvate was used at a saturating concentration and AIB was held at KAIB while the concentration of D-glutamate was varied. Experimental conditions: 100 mM TEA succinate pH 7.8, 100 mM dipotassium succinate, 20 µM PLP, 1 unit/mL 2° ADH, 300 µM NADPH.
W138F/M141R does not decarboxylate AMG based on end-point assays using 2° ADH. Controls with levulinic acid, the product of oxidative decarboxylation of AMG, show that 2° ADH can use it as a substrate. Inhibition studies find that although W138F/M141R does not decarboxylate AMG, it can bind to the enzyme active site, Ki = 136 ± 20 mM.
Discussion
R406 is Important to Catalysis of Decarboxylation
Decarboxylases often have a hydrophobic active site that destabilizes the charged carboxylate ground-state thereby promoting CO2 loss. Examples of PLP dependent decarboxylases that employ this strategy include ornithine decarboxylase (9, 10), dopa decarboxylase (11) and arginine decarboxylase (12). The presence of R406 in the relatively hydrophilic DGD active site is therefore unusual, but required of the transamination half-reaction in the normal catalytic cycle. Based on the reaction specificity of DGD, it was originally proposed that R406 is necessary for proper substrate binding in the transamination half-reaction but hindered the decarboxylation half-reaction (2). The R406M mutant was expected to relieve the ground-state stabilization R406 affords in the decarboxylation half-reaction and, by this simple reasoning, to increase the decarboxylation rate constant. Although the loss of the interaction between the substrate carboxylate and R406 was expected to weaken substrate binding, the mutation was also expected to shift the binding of the substrate carboxylate to favor the activated A subsite (Figure 1) over the nonactivated B subsite when substrate is bound, thereby increasing kdecarb.
As expected, the steady-state results show that substrate binding has been significantly weakened in R406M (Table 1). However, the expected increase in the rate of decarboxylation is not observed. Although R406M still maintains the strict specificity for oxidative decarboxylation with AIB observed in WT (3), the steady-state results indicate that decarboxylation is impaired in R406M.
The steady-state results do not rule out the possibility that decarboxylation is rapid but the overall catalytic cycle is limited by slow transamination. To address this possibility, the AIB decarboxylation half-reaction was studied in isolation using the spectral properties of the PLP and PMP cofactors. Although the spectral traces were kinetically complex, these studies clearly show that the maximal rates of the observed processes are not significantly faster than the observed steady-state rate. This rules out the possibility of rapid decarboxylation in R406M and supports a catalytic role for R406 in decarboxylation in addition to substrate binding.
The decarboxylation of alternate substrates in R406M also suggests a catalytic role for R406. The alternate substrates 1-aminocyclopentane-1-carboxylic acid (AC5C) and 1-aminocyclohexane-1-carboxylic acid (AC6C) bind with the cyclic component of their structures in subsites B and C and with the carboxylate in subsite A, based on the X-ray structures of their phosphonate analogues in complex with DGD (13). From the functional model of the DGD active site, one expects these substrates to undergo rapid decarboxylation because the α-carboxylate must occupy the stereoelectronically activated A subsite. However, previous pre-steady state studies with these substrates found no increase in the decarboxylation rate constant compared to AIB. (14) This was attributed in part to ground state stabilization of the α-carboxylate by R406. Removing the positive charge in the R406M mutant was expected to relieve the ground state stabilization and increase the rate of decarboxylation. Contrary to initial predictions, experiments with these alternate substrates suggest that the R406M mutation does not enhance decarboxylation by removing electrostatic ground-state stabilization from interaction of R406 with the α-carboxylate. In fact, a large decrease in the rate of decarboxylation with AC5C and AC6C compared to WT is observed with R406M (Table 2). These results suggest that R406 facilitates decarboxylation.
The reaction with L-alanine also suggests that R406 is important to decarboxylation. In R406M, it was expected that the mutation would increase the portion of substrate bound with its carboxylate in the activated A subsite and therefore increase the amount of decarboxylation of L-alanine in R406M compared to WT. In contrast to this prediction, no significant amount of decarboxylation, either oxidative or non-oxidative (Scheme 2), occurs in R406M with L-alanine. Instead L-alanine exclusively undergoes transamination in R406M, supporting the proposal that R406 plays an important role in the catalysis of decarboxylation.
Scheme 2.
The much larger decreases seen in the rate of decarboxylation with AC5C and AC6C (>4000-fold) than with AIB (~100-fold) in R406M may be the result of the superposition of two opposing trends in the decarboxylation of AIB and AMS. It is possible that as predicted, AIB and AMS enjoy some degree of rate acceleration due to preferred binding of the substrate carboxylate in the activated A subsite but also suffer from the absence of R406. With the cyclic substrates, there should be no major differences in their orientation and binding between R406M and WT because the cycloalkane occupies subsites B and C. The difference seen in the rate of decarboxylation with these substrates between R406M and WT therefore provides a simpler measure of the effect of the R406M mutation than reactions with either AIB or AMS, which can bind in at least two different conformations in the active site.
A Positive Charge in the 406 Position is Sufficient for Efficient Catalysis
To further elucidate the role of R406, the R406K mutant was made and characterized (Table 1). The Km values for AIB and AMS increase slightly, further reinforcing the importance of R406 in substrate carboxylate binding. The R406K mutant suggests that a positive charge in the 406 position is required for efficient catalysis in DGD, as shown by a decrease in kcat of only ~10-fold compared to WT.
The R406 mutants demonstrate that the simple model in which R406 hinders decarboxylation due to ground-state stabilization is incomplete. Studies with R406K show that the positive charge of a lysine largely rescues the activity lost in the R406M mutant, arguing against a specific role for the guanidino group of R406. In contrast to the original model in which the substrate carboxylate must be completely in the A subsite, interacting only with Q52 and nearly perpendicular to the PLP ring for decarboxylation to occur, the work with the R406 mutants suggest a more subtle catalytic strategy is employed.
It is likely that decarboxylation starts from a position between the A and B subsites, where the substrate carboxylate can enjoy interactions with both R406 and Q52. As the reaction proceeds and negative charge develops on Cα, the carboxylate moves into a position more nearly perpendicular to the PLP ring, taking advantage of the stabilization that stereoelectronic effects provide as the driving force for such interactions increases. This refined model is supported by X-ray crystallographic studies of DGD in which the phosphonate group of an aminophosphonate inhibitor is found interacting with both R406 and Q52 (13). In this model, the methionine residue in the R406M mutant is unable to hold the substrate carboxylate in an orientation from which decarboxylation can occur, leading to the dramatic decrease in the observed rate of decarboxylation. In contrast, the positive charge of the lysine is sufficient to position the carboxylate between the A and B subsites and efficient catalysis occurs.
W138 Limits the Size of the C Subsite
Based on X-ray crystallographic structures (2, 13) it has been proposed that W138 limits the size of the C subsite making DGD specific for short-chain alkyl groups. Comparison with γ-aminobutyrate aminotransferase (GABA-AT), a related aminotransferase, that binds substrates with longer side chains shows that GABA-ATs have a phenylalanine or tyrosine in the corresponding position. On this basis, the W138F mutation is proposed to open the C subsite, allowing this mutant to bind substrates that place larger side chains in the C subsite.
This prediction is borne out in studies with α-keto acids of increasing side chain length (Figure 3). Compared to WT, a smaller decrease in the catalytic efficiency is observed in W138F with increasing α-keto acid side chain length. Since an α-keto acid must bind with its side chain in the C subsite, this demonstates a less restrictive C subsite in W138F. Enlargement of the C subsite in W138F is further supported by inhibition studies with D-phenylglycine. The WT enzyme does not bind D-phenylglycine, but the more spacious C subsite in W138F allows D-phenylglycine to bind to the mutant enzyme.
The Combined W138F/M141R Mutations Introduce New Substrate Specificity
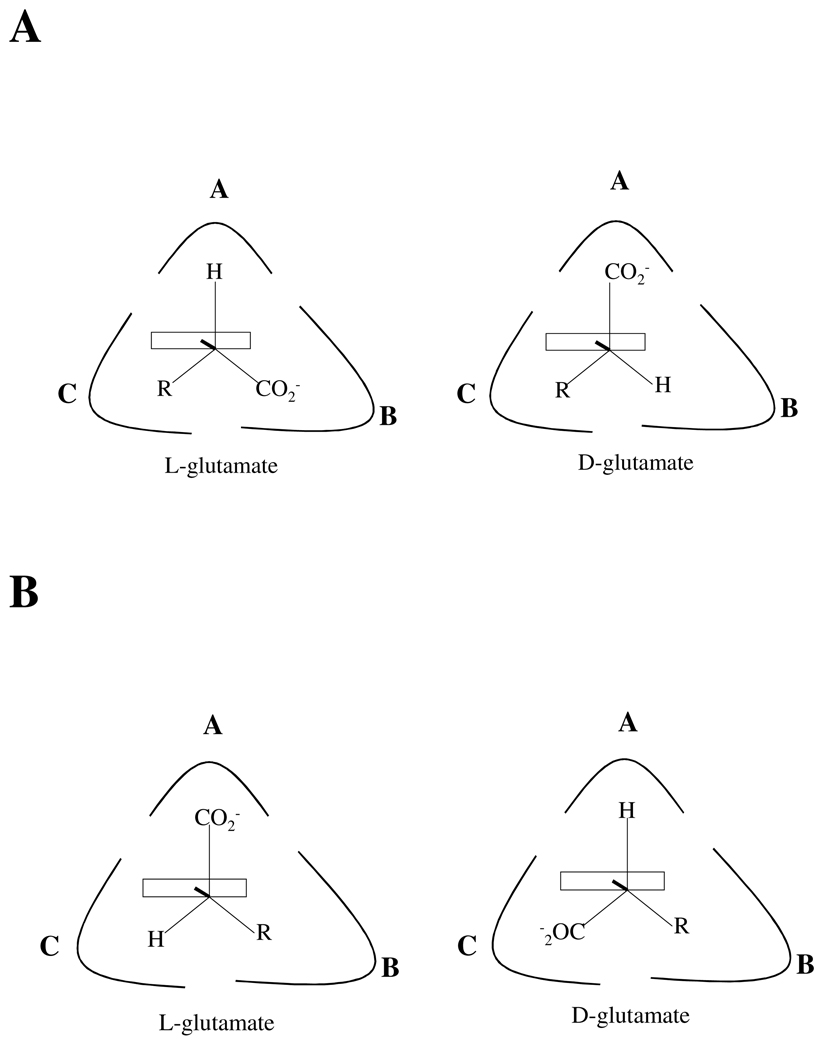
The W138F/M141R mutant was made to redesign the C subsite, using GABA-AT as a model to introduce new substrate specificity for D-glutamate. It was thought that the mutations would anchor the D-glutamate side chain in the redesigned C subsite (Figure 6A), placing the carboxylate in the activated A subsite from which decarboxylation would occur. L-glutamate would also bind to the enzyme with the substrate side chain in the redesigned C subsite, but the stereochemistry would require the carboxylate to occupy the B subsite; therefore, decarboxylation was not expected to occur with L-glutamate.
Figure 6.
Diagram of different binding conformations of L- and D-glutamate in the DGD active site. (A) Expected binding of L- and D-glutamate in the W138F/M141R active site. (B) Revised model of L- and D-glutamate binding in the W138F/M141R active site based on the results presented herein.
Steady-state assays show unexpected reactivities for D- and L-glutamate. The double mutant does not decarboxylate D-glutamate, although inhibition studies with D-glutamate show that it can bind to the mutant enzyme. However, W138F/M141R can decarboxylate L-glutamate. It has been shown that decarboxylation occurs from the stereoelectronically activated A subsite in DGD (3) and that stereoelectronic effects accelerate the rate of decarboxylation by at least six orders of magnitude compared to decarboxylation that occurs without the benefit of stereoelectronic stabilization (15). Combined with the reactivity of L- and D-glutamate this strongly suggests that these substrates are not binding with the glutamate side chain anchored in the redesigned C subsite in W138F/M141R DGD, as initially expected.
A model consistent with the structural and experimental data is shown in Figure 6B. In this model the glutamate side chain binds in the B subsite, while the mutations made to the C subsite allow binding of the α-carboxylate. The model is consistent with the observed activity with L-glutamate and the binding but lack of activity with D-glutamate. Both L- and D glutamate bind with the side chain in the B subsite, with L-glutamate this places the α-carboxylate in the A subsite. However, with the side chain anchored in the B subsite, D-glutamate binds with the α-carboxylate in the redesigned C subsite, a position that is not activated and therefore decarboxylation does not occur. Modeling of W138F/M141R suggest that the α-carboxylate can interact with the positively charged side chain R141 in the C subsite.
This model makes several predictions that are supported by experiment. The first is that WT does not bind D-glutamate, since the W138F/M141R mutations confer the ability to bind the α-carboxylate in the C subsite. Indeed, there is no significant inhibition seen with D-glutamate up to concentrations of 500 mM. The model also predicts that WT can bind L-glutamate, placing the side chain carboxylate in the B subsite. Experimentally, it is found that L-glutamate binds to the enzyme with Ki = 480 mM. The tighter binding of L-glutamate to W138F/M141R compared to WT may be related to the increase in the rate of L-glutamate decarboxylation in the double mutant compared to WT. Although W138F/M141R is able to use L-glutamate as a substrate, it is not efficient in its use of this substrate (kcat = 0.0022 s−1, kcat/Km L-glu = 0.08 M−1s−1). The changes have however, lead to significant changes in substrate specificity. A ~1000-fold increase in L-glutamate decarboxylation efficiency is observed for W138F/M141R compared to WT, and the specificity of the redesigned enzyme for L-glutamate has increased by approximately eight orders of magnitude in the mutant.
It was hypothesized by Dunathan (1) that a given PLP dependent enzyme maintains its reaction specificity by maintaining a specific orientation of the common external aldimine intermediate, aligning the scissile bond parallel to the p orbitals of the extended π system thereby maximizing orbital overlap and resonance interactions with the activated bond (see position labeled “A” in Figure 1). The binding constraints that control orientation of the substrate in the active site contribute to both reaction specificity and substrate specificity. The interrelation between reaction and substrate specificity is underlined by the work presented here with the R406 mutants. These mutants confirm the importance of R406 in substrate binding and also demonstrate the importance of the positive charge R406 provides in decarboxylation catalysis. Although W138 is not directly involved in catalysis, studies with α-ketoacids and the W138F mutant shows that this residue limits the size of the C subsite binding pocket, providing specificity for 2,2-dialkylglycines. Finally, by changing two residues, W138F/M141R, a mutant enzyme with expanded substrate specificity was created. In W138F/M141R the specificity for L-glutamate compared to the natural substrate AIB has increased by approximately eight orders of magnitude compared to the WT enzyme.
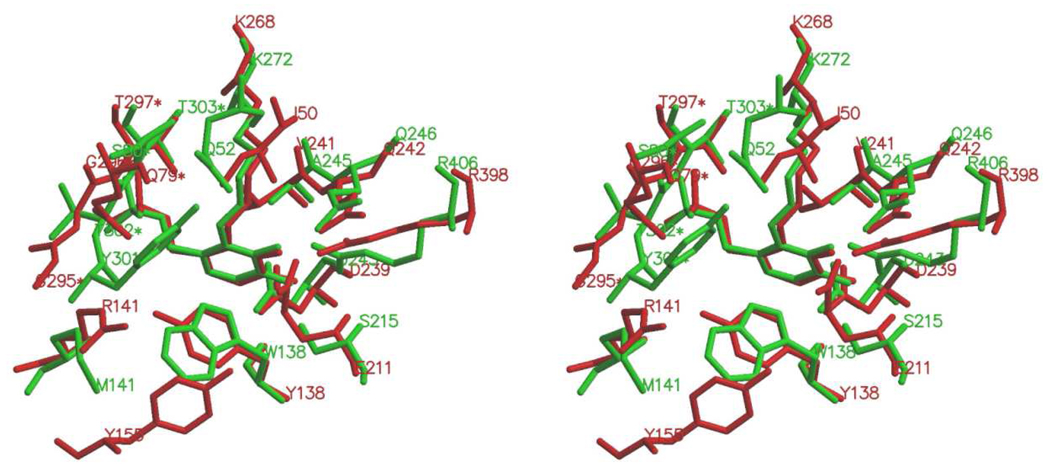
Figure 7.
Overlay of the active sites of unbound E. coli GABA-AT (red) and DGD (green). This figure was prepared with Swiss-PdbViewer and MOLSCRIPT.
Acknowledgments
Supported by grant GM54779 from the National Institutes of Health
Footnotes
Abbreviations used: DGD, 2,2-dialkylglycine decarboxylase; PLP, pyridoxal 5’-phosphate; AIB, 2-aminoisobutric acid; TB, terrific broth; IPTG, isopropyl-β-D-thioglactoside; TEA, triethanolamine; AMS, D,L-α-methyl serine; AMG, D,L-α-methylglutamate; 2°ADH, NADPH dependent secondary alcohol dehydrogenase; SSDH, succinic semialdehyde dehydrgenase; DTT, dithiothreitol; OPA, o-phthaldialdehyde; AC5C, 1-aminocyclopentane-1-carboxylic acid; AC6C, 1-aminocyclohexane-1-carboxylic acid; LDH, lactate dehydrogenase; YADH, yeast alcohol dehydrogenase; GABA, γ-aminobutyric acid; WT, wild type DGD; GABA-AT, γ-aminobutyric acid aminotransferase
References
- 1.Dunathan HC. Conformation and Reaction Specificity in Pyridoxal Phosphate Enzymes. Proc Natl Acad Sci U S A. 1966;55:712–716. doi: 10.1073/pnas.55.4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toney MD, Hohenester E, Keller JW, Jansonius JN. Structural and Mechanistic Analysis of Two Refined Crystal Structures of the Pyridoxal Phosphate-Dependent Enzyme Dialkylglycine Decarboxylase. J Mol Biol. 1995;245:151–179. doi: 10.1006/jmbi.1994.0014. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Zabinski RF, Toney MD. Reactions of Alternate Substrates Demonstrate Stereoelectronic Control of Reactivity in Dialkylglycine Decarboxylase. Biochemistry. 1998;37:3865–3875. doi: 10.1021/bi972055s. [DOI] [PubMed] [Google Scholar]
- 4.Sambrook J, Russell DW. Molecular Cloning : A Laboratory Manual. 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 5.Liu W, Peterson PE, Langston JA, Jin X, Zhou X, Fisher AJ, Toney MD. Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia Coligamma-Aminobutyrate Aminotransferase. Biochemistry. 2005;44:2982–2992. doi: 10.1021/bi048657a. [DOI] [PubMed] [Google Scholar]
- 6.Christenson SD, Wu W, Spies MA, Shen B, Toney MD. Kinetic Analysis of the 4-Methylideneimidazole-5-One-Containing Tyrosine Aminomutase in Enediyne Antitumor Antibiotic C-1027 Biosynthesis. Biochemistry. 2003;42:12708–12718. doi: 10.1021/bi035223r. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X, Kay S, Toney MD. Coexisting Kinetically Distinguishable Forms of Dialkylglycine Decarboxylase Engendered by Alkali Metal Ions. Biochemistry. 1998;37:5761–5769. doi: 10.1021/bi973010u. [DOI] [PubMed] [Google Scholar]
- 8.Malashkevich VN, Strop P, Keller JW, Jansonius JN, Toney MD. Crystal Structures of Dialkylglycine Decarboxylase Inhibitor Complexes. J Mol Biol. 1999;294:193–200. doi: 10.1006/jmbi.1999.3254. [DOI] [PubMed] [Google Scholar]
- 9.Jackson LK, Brooks HB, Myers DP, Phillips MA. Ornithine Decarboxylase Promotes Catalysis by Binding the Carboxylate in a Buried Pocket Containing Phenylalanine 397. Biochemistry. 2003;42:2933–2940. doi: 10.1021/bi026795z. [DOI] [PubMed] [Google Scholar]
- 10.Grishin NV, Osterman AL, Brooks HB, Phillips MA, Goldsmith EJ. X-Ray Structure of Ornithine Decarboxylase from Trypanosoma Brucei: The Native Structure and the Structure in Complex with Alpha-Difluoromethylornithine. Biochemistry. 1999;38:15174–15184. doi: 10.1021/bi9915115. [DOI] [PubMed] [Google Scholar]
- 11.Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural Insight into Parkinson's Disease Treatment from Drug-Inhibited Dopa Decarboxylase. Nat Struct Biol. 2001;8:963–967. doi: 10.1038/nsb1101-963. [DOI] [PubMed] [Google Scholar]
- 12.O'Leary MH, Piazza GJ. Medium Effects in Enzyme-Catalyzed Decarboxylations. Biochemistry. 1981;20:2743–2748. doi: 10.1021/bi00513a007. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Rogers CJ, Fisher AJ, Toney MD. Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition. Biochemistry. 2002;41:12320–12328. doi: 10.1021/bi026318g. [DOI] [PubMed] [Google Scholar]
- 14.Sun S, Bagdassarian CK, Toney MD. Pre-Steady-State Kinetic Analysis of the Reactions of Alternate Substrates with Dialkylglycine Decarboxylase. Biochemistry. 1998;37:3876–3885. doi: 10.1021/bi972056k. [DOI] [PubMed] [Google Scholar]
- 15.Fogle EJ, Liu W, Woon ST, Keller JW, Toney MD. Role of Q52 in Catalysis of Decarboxylation and Transamination in Dialkylglycine Decarboxylase. Biochemistry. 2005;44:16392–16404. doi: 10.1021/bi051475b. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Toney MD. pH Studies on the Mechanism of the Pyridoxal Phosphate-Dependent Dialkylglycine Decarboxylase. Biochemistry. 1999;38:311–320. doi: 10.1021/bi981455s. [DOI] [PubMed] [Google Scholar]