Abstract
C-di-GMP, a bacterial second messenger plays a key role in survival and adaptation of bacteria under different environmental conditions. The level of c-di-GMP is regulated by two opposing activities, namely diguanylate cyclase (DGC) and phosphodiesterase (PDE-A) exhibited by GGDEF and EAL domain, respectively in the same protein. Previously, we reported a bifunctional GGDEF–EAL domain protein, MSDGC-1 from Mycobacterium smegmatis showing both these activities (Kumar and Chatterji, 2008). In this current report, we have identified and characterized the homologous protein from Mycobacterium tuberculosis (Rv 1354c) named as MtbDGC. MtbDGC is also a bifunctional protein, which can synthesize and degrade c-di-GMP in vitro. Further we expressed Mtbdgc in M. smegmatis and it was able to complement the MSDGC-1 knock out strain by restoring the long term survival of M. smegmatis. Another protein Rv 1357c, named as MtbPDE, is an EAL domain protein and degrades c-di-GMP to pGpG in vitro. Rv1354c and 1357c have seven cysteine amino acids in their sequence, distributed along the full length of the protein. Disulfide bonds play an important role in stabilizing protein structure and regulating protein function. By proteolytic digestion and mass spectrometric analysis of MtbDGC, connectivity between cysteine pairs Cys94-Cys584, Cys2-Cys479 and Cys429-Cys614 was determined, whereas the third cysteine (Cys406) from N terminal was found to be free in MtbDGC protein, which was further confirmed by alkylation with iodoacetamide labeling. Bioinformatics modeling investigations also supported the pattern of disulfide connectivity obtained by Mass spectrometric analysis. Cys406 was mutated to serine by site directed mutagenesis and the mutant MtbC406S was not found to be active and was not able to synthesize or degrade c-di-GMP. The disulfide connectivity established here would help further in understanding the structure – function relationship in MtbDGC.
Introduction
The cell-cell communication or quorum sensing plays a major role in survival and maintenance of bacteria during the stationary phase. One of the interesting aspects of quorum sensing is the coordinated response of bacteria like biofilm formation, antibiotic production, sporulation, expression of virulence factor etc. [1]. A cell produces small autoinducer molecule and simultaneously senses the concentration of the autoinducer in the cell surface [2]. Second messengers act as autoinducers and relay signals received at the cell surface to target molecules within the cells. Nucleotide derivatives, which act as second messengers have been extensively studied for their regulatory function [3]. Cyclic adenosine monophosphate (cAMP), Cyclic guanosine monophosphate (cGMP), Guanosine 3′,5′ (bis) pyrophosphate (ppGpp) are all important second messengers both in prokaryotes and eukaryotes. cGMP is commonly used in eukaryotes but has very little role in bacteria [4]. cAMP is known to activate catabolite regulatory protein (CRP), a transcription regulator of gene involved in carbon metabolism [1], [5]. ppGpp on the other hand, regulates bacterial survival during nutrient starvation [6]. Another nucleotide, Bis-(3′-5′)-cyclic dimeric guanosine monophosphate (C-di-GMP) has been found to be involved in modulating cell surface and biofilm formation in several bacteria. This molecule was first reported more than 20 years back as a positive allosteric regulator of cellulose synthesis [7], [8].
C-di-GMP is synthesized from cyclization of two GTP molecules by diguanylate cyclase (DGC) and degraded to linear diguanylic acid (pGpG) by phosphodiestrases (PDE) [9], [10]. These two opposing enzymatic activities regulate the cellular pool of c-di-GMP. The DGC and PDE activity are encoded by conserved amino acid motifs GGDEF and EAL or HD-GYP, respectively [3], [11], [12]. With the advent of whole genome sequencing it is observed that GGDEF and EAL domain are ubiquitously present in all bacteria but absent in eukaryote [13]. Gram negative bacterial genomes harbor large number of proteins belonging to GGDEF-EAL domain super family, whereas Gram positive bacteria have small number of them. For example Vibrio cholerae and Escherichia coli has 53 & 36 proteins but Bacillus subtilis and Mycobacterium smegmatis has 7 & 1 GGDEF-EAL domain proteins, respectively [14]. In many cases GGDEF and EAL domains are present in tandem and most of the proteins so far characterized have either DGC or PDE-A activity. Interestingly, the possibility of opposing enzymatic activities co-existing in a single protein has also been reported [15], [16]. Our last report on MSDGC-1 from M. smegmatis was one such example of a bifunctional protein [17].
C-di-GMP has been implicated in regulation of many cellular responses relevant to pathogenesis, such as motility, secretion, cytotoxicity and biofilm formation. Most of the studies have indicated that the higher cellular level of c-di-GMP increases the biofilm formation and promotes the sessile form of life whereas a low level promotes motility [4], [18], [19], [20]. The appealing aspect of c-di-GMP signaling is the regulation of virulence gene and the well studied example is Vibrio cholerae. The vieA, an EAL domain protein not only controls the host colonization but also regulates the Cholera toxin production [15]. Similar reports of c-di-GMP mediated expression of virulence genes and survival in host have been published for Salmonella typhimurium and Pseudomonas aeruginosa [21], [22]. Though most of the studies have been done in Gram negative bacteria, few studies in Gram positive bacteria indicate that c-di-GMP may not play an important role in their physiology [23], [24]. Contrary to this we have earlier established the presence and physiological relevance of c-di-GMP in M. smegmatis, a GC-rich Gram positive bacterium [17]. The pathogenic species of Mycobacterium genera M. tuberculosis causes tuberculosis and is one of the oldest diseases known to man. This work was the initial attempt to characterize the stress elements in M. tuberculosis and we were interested in following the role of cell-cell communication in this organism.
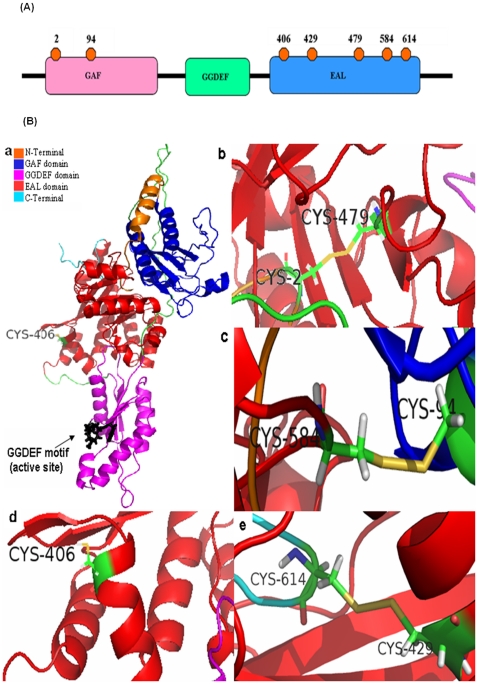
Upon sequence alignment (Figure 1) we observed that MSDGC-1 showed 60% identity at amino acid level to MtbDGC, suggesting that the protein may have a similar role to play in Mycobacterium tuberculosis. Fig. 1 shows that three cysteine residues are conserved in MtbDGC and MSDGC-1, whereas only one cysteine residue is conserved in all the three proteins. Expression of MtbDGC in E. coli leads to production of the expressed proteins in insoluble inclusion bodies. IBs must be resolubilized and refolded into an active confirmation which requires extensive trial and error method. In this report we discuss the solubilization of the protein in its native form maintaining the c-di-GMP synthetic activity.
Figure 1. Sequence alignment of MSDGC-1, MtbDGC and MtbEAL was generated by Clustal W and was adjusted manually.
Cysteines at the identical position in both the sequences are indicated by a black background.
Disulfide bonds play an important role in stabilizing the native structure and regulating its basic function [25], [26]. Free cysteines residues are also important to establish protein function, ligand binding and catalysis [27]. Here we have used chemical and proteolytic cleavage of MtbDGC and identified the disulfide linkages by alkylation and Mass spectrometry [28], [29]. We observed that in MtbDGC the disulfide linkages exist between (Cys94-Cys584), (Cys2-Cys479) and (Cys429-Cys614). The Cys406 was found to be free and interestingly when mutated to serine the protein was found to be inactive. To further understand the structure and disulfide connectivity of MtbDGC protein, we report here bioinformatics modeling of MtbDGC protein.
Materials and Methods
Bacterial strains, plasmids and oligonucleotides
Bacterial strains, plasmids and oligonucleotides used in the study are listed in Table 1. DH5α and BL21 (DE3) E. coli strains were grown in LB broth at 37°C with agitation or on a plate containing 1.5% w/v agar. Antibiotics were used at following concentration as and when required: Ampicillin (100 µg ml−1) or kanamycin (35 µg ml−1) for E. coli and kanamycin (20 µg ml−1) or Hygromycin (20 µg ml−1) for M. tuberculosis or M. smegmatis. The PCR reactions were carried out using Dynazyme EXT polymerase. All of the clones generated were confirmed by sequencing (MWG, India). Restriction enzymes used for the cloning were procured from New England Biolabs or Fermentas.
Table 1. Bacterial strains plasmids and primers used in this study.
| Strain, plasmid and primer | Characteristics | Source or Reference |
| Bacterial strains | ||
| ΔMSDGC-1 | M. smegmatis where MSDGC_2196 has been replaced with Kan cassette Kanr | (Kumar et al., 2008) |
| MtbRv1354 complimented | ΔMSDGC complemented with MtbDGC Kanr and Hygr | This work |
| pET MtbDGC | MtbRv1354c cloned in pET 21b Ampr | This work |
| pET MtbPDE | MtbRv1357c cloned in pET 21b Ampr | This work |
| Plasmids | ||
| pET21b | Cloning vector (c-terminal His tagged) Ampr | Novagen |
| pMV361 | Cloning vector with hsp60 promoter, kanr | (Stover et al., 1991) |
| Primers | Sequence (5′-3′) | |
| MtbRv1354f | AGCATCCCATATGTGCAACGACACCGCGAC | |
| MtbRv1354r | GCACCTCTCGAGAGATAACGCCGGGTCAG | |
| MtbRv1357f | ATGCAGCCCTCATATGGATCGTTGTTG | |
| MtbRv1357r | CACGCAAGCTTCTCTCTGAAAAAGTCG | |
| MtbC406F | CTGCTGGCACCGGGCTCCTTCATCCCTGTG | |
| MtbC406R | CACAGGGATGAAGGAGCCCGGTGCCAGCAG | |
Cloning of MtbDGC and MtbPDE
Genes Rv 1354c (1872bp, 623aa) and Rv 1357c (924bp, 307aa) were PCR amplified from genomic DNA of M. tuberculosis H37Rv using a set of primers, MtbRv1354f, MtbRv1354r, MtbRv1357cf and MtbRv1357cr respectively (Table 1). The amplicons Rv 1354 and Rv 1357c were cloned in pET21b vector using NdeI - XhoI and NdeI - HindIII restriction sites, respectively. The resultant plasmids were named as pETMtbDGC for Rv1354c and pETMtbPDE for Rv1357c.
Expression, isolation and purification of MtbDGC and MtbPDE from inclusion bodies
BL-21 cells carrying MtbDGC and MtbPDE plasmids were grown at 37°C in 2 L flask containing 500 ml of LB medium supplemented with 100 µg/ml of ampicillin. Protein expression was induced when cell density was reached to 0.6 at OD600 with 1 mM of isopropyl-D-thiogalactopyranoside (IPTG). After 4 h induction, cells were harvested by centrifugation at 8,000 rpm for 15 min at 4°C. Cells were lysed in lysis buffer containing 50 mM Tris-HCl at pH 7.9, 500 mM NaCl, 100 mM Dithiothreitol (DTT), 1 mM Ethylenediaminetetraacetic acid (EDTA), 6 M Urea and 20% Glycerol. Cell suspensions were sonicated at 150 kHz, using a sonicator with a 13 mm probe. This cycle was repeated three times for a total sonication time of 5 min each. Cell debris was removed by centrifugation at 12,000 rpm for 20 min at 4°C. After centrifugation the supernatant was loaded on Nickel-Nitrilotriacetic acid (Ni-NTA) column and washed with 100 column volumes of wash buffer containing 10 mM imidazole. The protein was eluted with the elution buffer containing 500 mM imidazole. Eluted protein was further dialyzed as reported below.
Refolding of MtbDGC and MtbPDE proteins with step-wise dialysis
The protein solubilized in the elution buffer containing 500 mM imidazole was subsequently dialyzed at various steps with decreasing concentration of urea in the buffer containing 50 mM Tris-HCl (pH 7.9 at 4°C), 250 mM NaCl, 10 mM DTT, 5% Glycerol. Urea concentration was varied from 6 M to 0 M and the dialysate was allowed to reach equilibrium [30]. As reported earlier disulfide containing proteins can refold even in the presence of concentrated denaturant [31]. At the final stage with no urea, precipitation of the protein was not observed. Finally the protein was dialysed against the same buffer having no DTT and assayed for its activity.
Enzymatic assays
Di-guanylate cyclase and phosphodiesterase assays were adapted from procedures described previously [17], [32]. Both the activities were followed in a buffer containing 5 µM of protein, MtbDGC, MtbPDE or MSDGC-1 and 25 mM Tris-HCl (pH 7.9), 250 mM NaCl, 10 mM MgCl2 in 50 µl volume. The reaction was triggered by the addition of a mixture of 0.1 mM Guanosine 5′-triphosphate (GTP) and α-labeled [32P]GTP (Board of Radiation and Isotope Technology (BRIT), India; 0.01 µCi µl−1 or 3500Ci mmol−1) in the case of MtbDGC. However, MtbPDE activity was checked by replacing GTP with c-di-GMP. Aliquots were withdrawn at regular time intervals and the reaction was stopped with an equal volume of 50 mM EDTA. Reaction products (2.5 µL) were separated on polyethyleneimine-cellulose plates (Merk) in 1∶1.5 (v/v) saturated NH4SO4 and 1.5 M KH2PO4 (pH 3.6) and plates were exposed to a phosphor-imager screen. The [32P] c-di-GMP, prepared as described below was used as substrate for PDE-A activity. Preparation of [32P] labelled and nonlablled c-di-GMP was done by the protocol as described earlier [10].
Western blot analysis
Western blot analysis for the detection of MtbDGC was carried out with primary antibodies raised in rabbit against the purified protein. Total cellular proteins extracted from cells grown at different time were normalized and 100 µg of the lysate was separated by Poly acrylamide gel electrophoresis (SDS-PAGE) and blotted on to a Sodium Dodecyl Sulphate -Polyvinylidende Fluoride (PVDF) membrane. The polyclonal serum generated was used as primary antibodies and a secondary antibody was purchased from Sigma Aldrich. Both were used after ten thousand fold dilution. The blots were developed with 26 mg aminoethylcarbazole ml−1 and 0.01% v/v H2O2.
Long term starvation cultures
The ΔMSDGC-1, wild type strain of M. smegmatis mc2 155 and strain developed by complementing ΔMSDGC-1 with Mtbdgc were grown in MB7H9 with 0.02% w/v glucose and 0.05% v/v Tween−80 till saturation. Cultures were declumped before plating on agar described earlier [29]. The colony forming units of these cultures were determined at regular interval of time up to 20 days. Antibiotics were omitted from the culture to rule out any possible effect of antibiotics on long term survival.
Proteolytic digestion of protein (MtbDGC)
10 µg of purified full length protein was diluted to final acetonitrile concentration of 5–10% in presence of digestion buffer (50 mM of ammonium bicarbonate) and subsequently digested at 37°C overnight using the following proteolytic enzymes in individual experiments. MtbDGC was digested with 200 ng trypsin (sequencing grade, modified trypsin (promega)), 200 ng chymoptrypsin (sequencing grade, modified trypsin (promega)) and double digested with 100 ng of trypsin and 100 ng of chymotrypsin respectively. The proteolytic digestion mixtures were desalted using C-18 column (Supelco) and subsequently analyzed by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI MS) and (Liquid Chromatography-Electrospray Ionisation) LC-ESI-MS/MS.
Reduction and alkylation of digested protein sample
10 µL of purified protein was taken in 15 µL of 0.1 M ammonium bicarbonate buffer pH 8.0 for alkylation. For the reduction, DTT was added to a final concentration of 8 mM and was incubated at 37°C for 3 h. For alkylation iodoacetamide (IAM) was added to the final concentration of 40 mM. The mixture was incubated at room temperature in the dark for 90 mins. Reaction mixture was incubated at 37°C overnight for further digestion. Two sets of reactions were carried out, one to check for alkylation and the others for reduction and followed by alkylation.
MALDI-TOF Mass spectrometry
Mass-spectra of digested gel spots were obtained by Matrix-Assisted Laser Desorption Ionization time of flight (MALDI-TOF) mass spectrometry on an Ultraflex TOF/TOF spectrometer (Bruker Daltonics). All the mass spectra were acquired in positive-ionization mode with reflectron optics. The instrument was equipped with 50 Hz pulsed nitrogen laser (λ = 337 nm) and operated under delayed extraction conditions; delayed time 90-ns, and 25 kV accelerating voltage. All peptides samples were prepared by mixing an equal amount of matrices dihydroxybenozic acid/α-cyano-4-hydroxycinnamic acid saturated in 0.1% Trifluoroacetic acid and acetonitrile (1∶1). Typically, 50–100 laser shots were used to record each spectrum.
Tandem Mass Spectrometry (LC-ESI-MS, MS/MS)
For all the disulfide connectivity studies, the proteolytic peptide mixture was analyzed by reverse-phase HPLC-ESI-MS, MS/MS (Electrospray Ionisation, ESI, Bruker Daltonics) spectrometry. Peptides were separated on HPLC (1100 series HPLC (Agilent)) system equipped with C18 column (Supelco). The column eluant was directly coupled to a HCT Ultra PTM Discovery System (ETD II- Bruker Daltonics). Mass spectra (ESI-MS) and tandem mass spectra (ESI-MS/MS) mode were recorded in positive-ion mode with a resolution of 12,000-15,000 Full Width of Half Maximum (FWHM). For tandem mass spectrometry dissociation, the mass analyzer was set to ±1 m/z. The precursor ions were fragmented in a collision cell using nitrogen as the collision gas. Spectra were calibrated in static mode using MS/MS fragment ions of standard supplied by the manufacturer. The enzymatically digested peptides were separated using mobile phase A consisted of 0.05% formic acid in 98% H2O/2% CAN (Acetonitrile) and B consisted of 0.05% formic acid in 98% ACN/2% H2O with a gradient of 5% B in first 5 mins, followed by 95% B in next 50 mins and 5% B in last 10 mins. The gradient times were varied depending upon the sample specifications.
Homology modeling, minimization and model validation
The sequence and domain information of 623 amino acid length Rv1354c was taken from swissprot (ID:P64826; http://us.expasy.org/sprot/) [33]. Protein consists of three domains named as GAF (residue 28-171), GGDEF (residue 212–345) and EAL (residue 354–609). Since no homology was found for the full length protein, Modeling of individual domain was done and joined to give the complete structure. BLAST [34] and PSI-BLAST [35] was used against National centre for biotechnology information (NCBI), Protein Data Bandk (PDB) database to find top hit as a template and target vs template sequence alignment for individual domains was done using CLUSTALW2 [36]. On the basis of sequential similarity 10 models each of EAL and GGDEF domain were modeled with the protein PDB [37] entry 2R6O (chain:A;identity:40%) and PDB entry 1W25 (chain:A;identity:40%) respectively using MODELLER release 9v7 [38] and model using PHYRE [39] server. GAF and EAL domain consists of N terminal (residue 1–27) and C terminal (residue 610–623) of full protein respectively. Considering three modeled domains as template full with the minimum DOPE score was selected. A threaded model of GAF domain was produced convergence to machine precision. Minimized structure was validated with the programs PROCHECK score (91.8%) [40], VERIFY3D (78.04%) [41] and PROSA score (−6.68) [42]. PyMOL [43] was used for 3D-structure visualization, protein was modeled using MODELLER 9v7. Top scored model was solvated in SPC216 water box (total number of water molecule: 93396) and charge neutralization was accomplished with the addition of 7 Na+. Energy minimization of the model was carried out by GROMACS 3.3.1(The Gronian Machine for Chemical Simulations) [44] using OPLS [45] force field and steepest descent method. The minimization was set to run for 10000 steps with 0.002 ps time step or until.
Alignment of proteins containing GAF, GGDEF, EAL architecture
Since the protein of interest was having three domains namely GAF, GGDEF, & EAL domain, protein sequences with the above architecture were searched in PFAM database [46]. A total of 183 sequences were found. Many of the protein sequences consisted of transmembrane or low complexity regions and there was a large variation in length among the sequences. This variation was more prominent in the region before GAF domain and the relative positioning of each domain with respect to other. Linker region between the GAF and GGDEF domain also varied in length. Each domain was separated from the corresponding sequences by taking the domain information given in the PFAM database as SWISSPROT did not provide the domain start & end information for all 183 sequences. Alignment of each of the domain sequences was done separately using ClustalW2. Although there was ambiguity in the assignment of start and end positions of each domain of proteins in PFAM and SWISSPROT databases, the SWISSPROT data was considered for modelling as it is supported by experimental methods.
Cysteine mutation using site directed mutagenesis
Third cysteine of MtbDGC at position 406 was mutated to serine amino acid using PCR based approaches. In this method two complimentary oligonucleotides containing the desired mutation were used to prime the PCR on a pET MtbDGC plasmid DNA template. Conditions used for PCR are as follows - in total volume of 50 µL template DNA 100 ng, thermostable polymerase buffer (10×) 5 uL, mutagenic primer 20 pmol, 0.6 µL of 25 mM dNTPs solution and thermostable polymerase 2–5 U. The tube was initially heated at 94°C for 2 min, followed by 19 cycles of 94°C for1 min, for annealing 54°C for 1 min and an extension step at 72°C for 8 mins, the tube is held at 72°C for 20 mins and if required at 4°C indefinitely. Subsequently digestion of the reaction mixture with DpnI was done to removes the template DNA leaving intact newly synthesized double stranded mutant PCR product designated as MtbC406S [47], [48]. The primers used in mutagenesis are listed in Table 1. The clone was confirmed by sequencing at MWG (data not shown), Bangalore. The plasmid MtbC406S was further transformed in BL-21 Escherichia coli cells. Protein was expressed and purified as described in section 2.3. Activity of the mutated protein was checked as described in section 2.5.
Circular dichorism study of MtbDGC and MtbC406S
The purified MtbDGC and MtbC406S protein after folding was further dialyzed against the CD buffer containing 10 mM phosphate buffer pH 8.0, 150 mM KCl and was filtered through a 0.2 µM PVDF filter (Milipore). 5 uM of MtbDGC and MtbC406S protein were used for CD studies. Circular dichorism spectra were recorded on a Jasco J-715 spectopolarimeter at 22°C using a cell of path - length 0.2 cm, bandwith 2.0 nm and a response time of 2s. Spectra were averaged over three scans with each sample and were recorded at a speed of 50 nm/min from 250 nm to 200 nm in a quartz cuvette.
Results and Discussion
MtbDGC possesses both DGC and PDE-A and MtbPDE possesses PDE-A activity
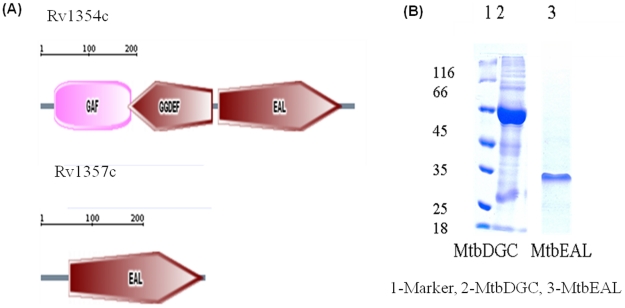
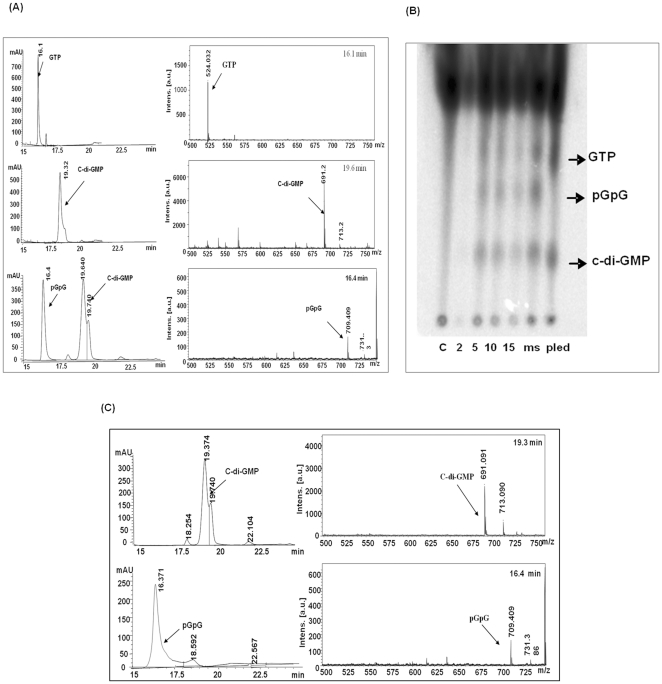
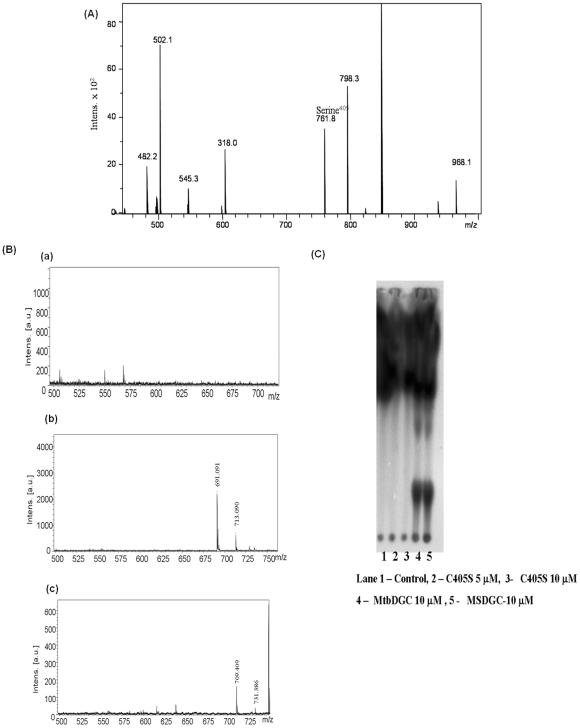
Domain organization of Rv 1354c and Rv 1357c was obtained from SMART database as shown (Figure 2A). Rv 1354c and Rv 1357c are named and referred throughout the manuscript as MtbDGC and MtbPDE, respectively. MtbDGC and MtbPDE were purified in soluble form as described in materials and methods to homogeneity as judged by SDS-PAGE profile (Figure 2B). Both the proteins were more than 80% pure (Protein id. NP-215870.1). Further digestion of the band and subsequent MALDI and MASCOT analysis confirmed the presence of MtbDGC and MtbEAL proteins (data not shown). These protein preparations were used to check the DGC and PDE-A activity. The reaction products were separated on reverse phase High performance liquid chromatography (HPLC) along with GTP and purified c-di-GMP as controls. Further, these peaks were collected and subjected to MALDI-TOF analysis. The major ions detected were m/z 542 (M+H)+ for GTP, 691 (M+H)+ and 713 (M+Na)+ for c-di-GMP, 709 (M+H)+ and 731 (M + Na)+ for pGpG, thus confirming that MtbDGC is capable of both synthesizing and degrading c-di-GMP (Figure 3A). Reaction mixture with MSDGC-1 protein (Kumar and Chatterji, 2008), a known DGC protein of Mycobacterium smegmatis was used as control. On the Thin Layer Chromatography (TLC) plate the spot at the same Rf of c-di-GMP apparently followed by another spot with Rf of pGpG. Reaction mixture with MSDGC-1 was used as a positive control and reaction mixture with cell lysate was used as the negative control (Figure 3B).
Figure 2. Representatives of C-di-GMP specific domain architectures in Mycobacteirum tuberculosis and their subsequent protein purification.
(A) Domain architecture of the protein with GGDEF and EAL domain in Mycobacterium tuberculosis. The length of each sequence is approximately to scale. (B) Purification of C- terminal His6 tag proteins Lane – 1 Marker, Lane - 2 MtbDGC and Lane – 3 MtbPDE. C-terminal His6 –tagged Rv1354 is 67.6 KDa and Rv1357c is 33.9 KDa. Purity of the preparations was determined by (10%) SDS- PAGE.
Figure 3. MtbDGC is a bifunctional protein showing both diguanylate cyclase (DGC) and phosphodiesterae acitivity (PDE-A) and MtbPDE is a functional protein showing phosphodiesterae acitivity (PDE-A).
(A) Purified His6 protein was assayed for the ability to synthesize c-di-GMP. HPLC analysis for the detection of c-di-GMP and pGpG: (a): GTP, (b): Purified c-di-GMP, (c): With protein MtbDGC, separated on reversed phase HPLC. MALDI TOF analysis of the relevant HPLC fractions, in the positive ion detection mode, ions were detected at m/z of 542 (M+H)+ for GTP, 691 (M+H)+ and 713 (M+Na)+ for c-di-GMP, 709 (M+H)+ and 731 (M + Na)+ for pGpG. (B) Purified His6 protein was assayed for the ability to degrade c-di-GMP. HPLC analysis for the detection of pGpG. (c): Purified c-di-GMP (C): With Protein (MtbPDE),. MALDI TOF analysis of the relevant HPLC fractions, in the positive ion detection mode, ions were detected at m/z = 691 (M+H)+ and 713 (M+Na)+ for c-di-GMP, 709 (M+H)+ and 731 (M + Na)+ for pGpG.
Similarly, the reaction mixture where c-di-GMP was incubated with MtbPDE showed reduction in peak area of c-di-GMP and appearance of an additional peak at 16.4 min at the same retention time of pGpG. The mass spectrum of 16.4 min peak showed major ion at m/z 709, an addition of a water molecule to c-di-GMP (Figure 3C). The purified pGpG showed the identical mass spectrum at m/z 709 confirming that the MtbPDE possesses phosphodiesterase activity.
MtbDGC is functional in vivo
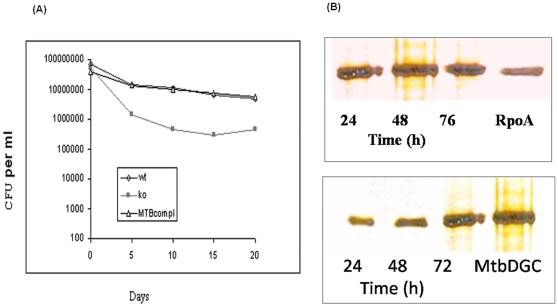
Previously we reported that the MSDGC-1 which possesses both DGC and PDE-A activities is required for long term survival in M. smegmatis, under nutritional starvation. MtbDGC is also bifunctional and homologous to MSDGC-1. It is intriguing to test the in vivo functionality of MtbDGC in the MSDGC-1 disrupted mutant ΔMSDGC-1. We complimented this mutant through chromosomal integration of MtbDGC in ΔMSDGC-1 strain using pMV361 integration vector containing Rv1354c gene. These strains were grown in carbon starved conditions and their survival was followed in terms of CFU until 20 days of incubation. Figure 4A shows that MtbDGC was able to restore the long term survival phenotype of the mutant strain as the CFU of wild type and complimented strain was comparable.
Figure 4. Complementation of ΔMSDGC-1 with MtbDGC and its long term survival.
(A) ΔMSDGC-1 a strain of M. smegmatis was complemented with MtbDGC and its growth was compared with wildtype as well as ΔMSDGC-1. Cultures were grown in MB7H9 medium with 0.02% v/v glucose and 0.05% Tween-80 as carbon source. Aliquots were withdrawn at different time interval after culture reached stationary phase (48 hours of growth) and plated on MB7H9 agar supplemented with Kanamycin containing 2% w/v glucose as carbon source. The colony forming unit were determined and plotted. (B) Level of MtbDGC increases in stationary phase. ΔMSDGC-1 was transformed with MtbDGC and expression was studied in three phases of growth i.e. exponential, early stationary and late stationary phase. The α subunit (RpoA) of RNA polymerase was used as an control because its level remains unchanged in different phases of growth. Whole cell lysates was separated on 10% SDS-PAGE and analyzed by Western blot technique probed with antibody raised against MtbDGC.
Level of MtbDGC increases in stationary phase of growth
As described earlier, the level of MtbDGC in bacteria varies according to the environmental change and the growth phase. We determined the level of MtbDGC expression in three phases of the growth, i.e. exponential, early stationary and late stationary phase. Whole cell lysate of M. tuberculosis compliment was separated on SDS-PAGE and analyzed by Western blot technique with polyclonal antibodies raised in rabbit against MtbDGC protein. An immunoreactive protein band, approximately 68 kDa in size (Figure 4B) was detected, with increased band intensity in the stationary phase. Figure 4B shows that the amount of MtbDGC was three times higher in the stationary phase culture than in the exponential phase, as detected by quantization of the blot. In each lane, the same amount of protein (100 µg) was loaded. It should be mentioned here that the growth of the bacteria was found to be slow when the dgc gene was over expressed (data not shown).
Proteolytic digestion of MtbDGC protein
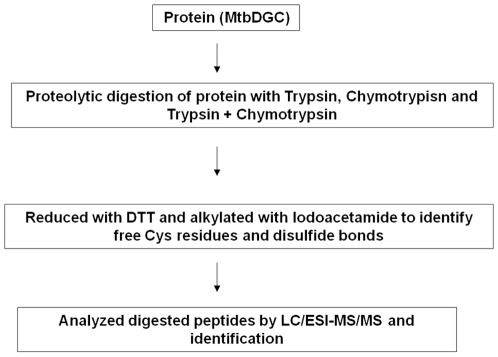
The flow chart for the protein digestion is depicted in Figure 5. Protein was digested with trypsin, chymotrypsin and in combination of both in three different sets of experiments. The ratio of concentration of proteolytic enzyme (Trypsin and Chymotrypsin) to protein affects the LC-ESI-MS/MS response. The optimal ESI signal was obtained when the ratio of enzyme to protein was 1∶ 20. Injection of 10–15 nmol of digested protein into the capillary of LC-ESI-MS/MS system produced a signal sufficient to characterize peptide containing labeled Cys and disulfide bonded Cys residues (data not shown).
Figure 5. Schematic diagram of experimental setup to study disulfide connectivity pattern.
Mass spectral characterization of free cysteine residue and disulfide bonds
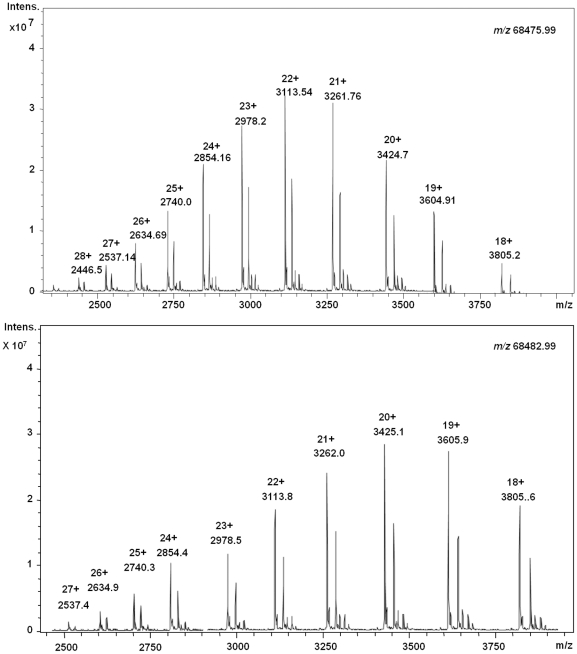
In order to identify number of disulfide bonds and free cysteine in the intact MtbDGC protein, LC-ESI-MS of intact protein was studied, which confirmed m/z of 68475.99 (His6 protein). The protein was further reduced with 8 mM DTT and mass difference of seven Daltons m/z 68482.99 (His6 protein) was obtained confirming the presence of at least three disulfide bonds in the protein (Figure 6A and 6B).
Figure 6. LC-ESI-MS of intact MtbDGC protein.
(A) LC-ESI-MS of Histidine tagged intact MtbDGC protein and corresponding inset zoom of deconvoluted mass m/z 68475.99 of the protein. (B) LC-ESI-MS of DTT reduced MtbDGC intact protein and corresponding inset zoom of deconvoluted mass (m/z 68482.99). The shift in seven Dalton was observed in comparison of unreduced protein.
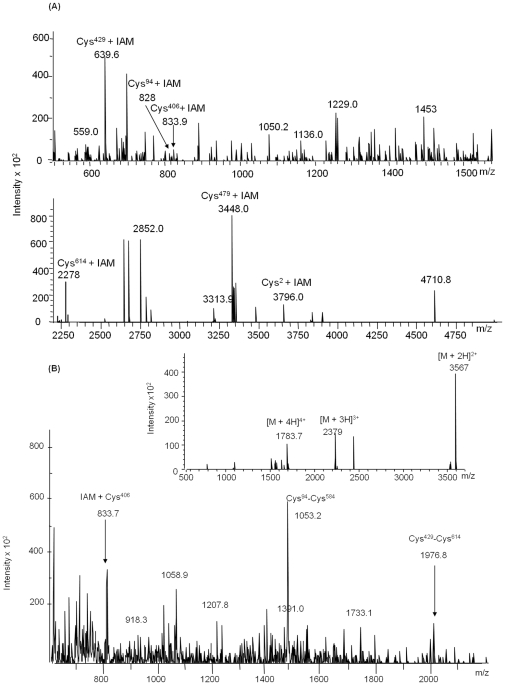
A common problem in detecting peptides which contain free cysteine residues is that the sulfhydryl group reacts with other chemical in the solution. Thus, the cysteine residues in the protein are alkylated by alkylting reagent like iodoacetamide, iodoacetic acid or 4-venyl pyridiene. Here, we used iodoacetamide to alkylate the samples, using five fold molar excess of iodoacetamide, and we were able to detect all seven cysteine containing peptides in proteolytic digested protein in MALDI as well as by LC-ESI-MS analysis. A sample that displayed an isotopic distribution of the required peptides was further used for the studies. Figure 7(A) shows a representative mass spectrum from DTT (200-fold molar excess) reduced and iodoacetamide labeled protein after digestion with a combination of trypsin and chymotrypsin. Ions at m/z 3796.2 (aa. 1–35), 828 (aa. 92–98), 833.9 (aa. 400–407), 693.6 (aa. 428–432), 3448.8 (aa. 461–494), 341.3 (aa. 584–585) and 1452.6 (aa 610–623) represents Cys2, Cys94, Cys406, Cys429, Cys479 containing peptides, respectively, matching with the calculated value as depicted in table 2. Figure 7(A) was compared with mass spectrum depicted in Figure 7(B) generated by iodoacetamide labeled, oxidized protein after double digestion with trypsin and chymotrypsin. Singly charged [M + H]+ 833.7, 1053.2 and doubly charged [M +2H]2+ 3566.5 and 1399.5 ions corresponds to the calculated mass of free cysteine peptide Cys406 and disulfide connected dipeptide Cys94-Cys584, Cys2-Cys479 and Cys429-Cys614, respectively, as depicted in table 3. Each fragment that contained iodoacetamide labeled Cys406 residue showed a mass shift of 57 Da (Mass of iodoacetamide) compared with the mass of unlabelled peptide.
Figure 7. LC-ESI-MS of MtbDGC in reduced and oxidized condition, after digestion with chymotrypsin and trypsin.
(A). LC-ESI-MS of DTT reduced and iodoacetamide alkylated MtbDGC digested with trypsin and chymotrypsin. The singly charged ions at m/z 3796.2, 828, 833.7, 639.6, 3448.0 and 2775.0 show the mass of cysteine peptides at Cys2, Cys94, Cys406, Cys429, Cys479 and Cys614 respectively (B) LC-ESI-MS of unreduced and iodoacetamide alkylated MtbDGC digested with trypsin and chymotrypsin. The singly charged ions at m/z 833.7, 1053.2 and 1399.5 [M +2H]2+ show the free Cys405 residue at third position and disulfide bond at Cys94-Cys584 and Cys429-Cys614 position respectively. Corresponding insect zoom shows the disulfide bonded Cys2-Cys478, the dominant ion is the doubly charged ion of [M +2H]2+ at m/z 3566.5.
Table 2. Theoretically and experimental mass of peptides generated from reduced, alkylated and digested samples of MtbDGC.
| Position of Cysteine in Protein | Expected Mass(m/z) | Observed Mass(m/z) | ||
| Unlabelled | Labeled | Unlabelled | Labeled | |
| Digested by Trypsin + Chymotrypsin | ||||
| Cys2 (aa- 1-35) | 3739.2 | 3796.2 | 3739.2 | 3796.2 |
| Cys94 (aa-92-98) | 770.9 | 827.9 | 770.9 | 828.9 |
| Cys406 (aa-400-407) | 776.9 | 833.9 | 776.9 | 833.7 |
| Cys429 (aa-428-432) | 582.6 | 639.6 | 582.6 | 639.6 |
| Cys479 (aa-461-494) | 3391.8 | 3449.8 | 3391.8 | 3448.0 |
| Cys584 (aa-584-586) | 284.3 | 341.3 | 284.3 | 341.3 |
| Cys614 (aa-610-629) | 2218.51 | 2275.51 | 2218.0 | 2275.0 |
| Digested by Chymotrypsin | ||||
| Cys2 (aa-1-39) | 4185.7 | 4242.7 | 4185.7 | 4242.0 |
| Cys94 (aa-92-112) | 1257.4 | 1313.4 | 1256.0 | 1313.4 |
| Cys406 (aa-395-407) | 1396.6 | 1453.4 | 1396.0 | 1453.0 |
| Cys429 (aa-424-432) | 1107.3 | 1164.3 | 1107.3 | 1164.0 |
| Cys479 (aa-461-513) | 5342.0 | 5399.0 | 5342.0 | 5399.0 |
| Cys584 (aa-562-590) | 2591.9 | 2648.9 | 2591.9 | 2648.6 |
| Cys614 (aa-594-629) | 4151.8 | 4208.8 | 4151.8 | 4208.0 |
| Digested by Trypsin | ||||
| Cys2 (aa-1-35) | 3739.2 | 3796.1 | 3739.6 | 3796.6 |
| Cys94 (aa-81-98) | 2023.3 | 2080.3 | 2025.1 | 2097.8 |
| Cys406 (aa-400-422) | 2355.7 | 2412.7 | 2353.0 | 2410.0 |
| Cys429 (aa-423-427) | 2204.4 | 2261.4 | 2201.0 | 2258.0 |
| Cys479 (aa-448-494) | 4997.5 | 5054.5 | 4998.0 | 5559.0 |
| Cys584 (aa-584-586) | 440.5 | 497.52 | 441.2 | 498.2 |
| Cys614 (aa-610-629) | 2218.5 | 2275.5 | 2218.5 | 2275.5 |
Table 3. Theoretically and experimental mass of peptides generated from intact proteins samples of MtbDGC upon oxidation.
| Position of Cysteine in Protein | Expected Mass(m/z) | Observed Mass(m/z) | ||
| Unlabelled | Labeled | Unlabelled | Labeled | |
| Digested by Trypsin + Chymotrypsin | ||||
| (Cys2 - Cys479) | 7129.0[(3739.2+3391.8) -2 = 7129.0] | 7129.0 | [M+2H]2+: 3566.5 | [M+2H]2+: 3566.5 |
| (Cys94 - Cys584) | 1053.2[(770.9+284.3) -2 = 1053.2] | 1053.2 | [M+H]+: 1053.2 | [M+H]+: 1053.2 |
| Cys406 | 776.9 | 833.9 | [M+H]+776.9 | [M+H]+: 833.7 |
| (Cys429- Cys614) | 2799.1[(582.6+2218.5) -2 = 2799.1] | 2856.1 | [M+H]+: 2799.1 | [M+H]+: 2856.0 |
| Digested by Chymotrypsin | ||||
| (Cys2 - Cys479) | 9525.7[(4185.7+5342) – 2 = 9525.7] | 9525.7 | [M+3H]3+3176.9 | [M+3H]3+3177.9 |
| (Cys94 - Cys584) | 3847.9[(1257.4+2591.9) – 2 = 3847.9] | 3847.9 | [M+H]+: 3846.2 | [M+H]+: 3846.2 |
| Cys406 | 1396.6 | 1453.6 | [M+H]+1396.0 | [M+H]+: 1453.4 |
| (Cys429- Cys614) | 5259.1[(1107.3+4151.8) – 2 = 4434.2] | 5316.1 | [M+H]+2629.5 | [M+H]+: 2629.5 |
| Digested by Trypsin | ||||
| (Cys2 - Cys479) | 9145.0[(3739.2+4997.7) – 2 = 8734.7] | 8791.7 | [M+3H]3+: 2911.5 | [M+3H]3+: 2911.5 |
| (Cys94 - Cys584) | 244384.0[(2202+2184) – 2 = 4384.0] | 4384.0 | [M+2H]2+: 2461.4 | [M+2H]2+2461.4 |
| Cys406 | 2355.7 | 2412.7 | 2353.0 | 2410.0 |
| (Cys429- Cys614) | 4420.9[(2204.4+2218.5) – 2 = 4420.9] | 4477.9 | [M+H]+:4420.9 | [M+H]+4477.9 |
To evaluate and confirm the pattern obtained from trypsin and chymotrypisn treated double digested protein, MtbDGC protein was also digested with chymotrypsin or trypsin. The m/z 4242.7 (aa. 1–39), 1313 (aa. 92–112), 1453.0 (aa. 395–407), 1164.0 (aa. 424–432), 5399.0 (aa. 461–513), 2648.9 (aa. 562–590) and (His6) 3385.2 (aa. 594–629) ions are in good agreement with the calculated mass of free cysteine containing peptide labeled with iodoacetamide (Table 2) when digested with Chymotrypsin (data not shown). The predicted disulfide linked peptides Cys429-Cys614, Cys94-Cys584 and Cys2-Cys479 were also detected in chymotrypsin digested and reduced protein. The m/z for 3176.9 [M +3H]3+, 3846.2, 4491.6 [M + H]H+ (table 3) ions represents dipeptide linked Cys2-Cys479, Cys94-Cys584 and Cys429-Cys614 respectively. The m/z 1453.0 for the [M + H]+ ion shows free cysteine at Cys406. Similar results were also obtained with trypsin digested protein (data not shown).
Disulfide connectivity
Disulfide connected dipeptide were analyzed by LC-ESI-MS/MS and individual peptides were identified based on the information obtained by tandem mass spectra. The technique involves the breakage of peptide backbone generating predominantly b and y-type ions [49]. The LC-ESI-MS/MS has been shown to give ambiguous result due to thiol exchange reaction. To overcome this problem all the analysis was done in the presence of alkylting agent iodoacetamide. To prove that the disulfide bonding is not altered, alkylation was done before or after protein was proteolyticaly digested.
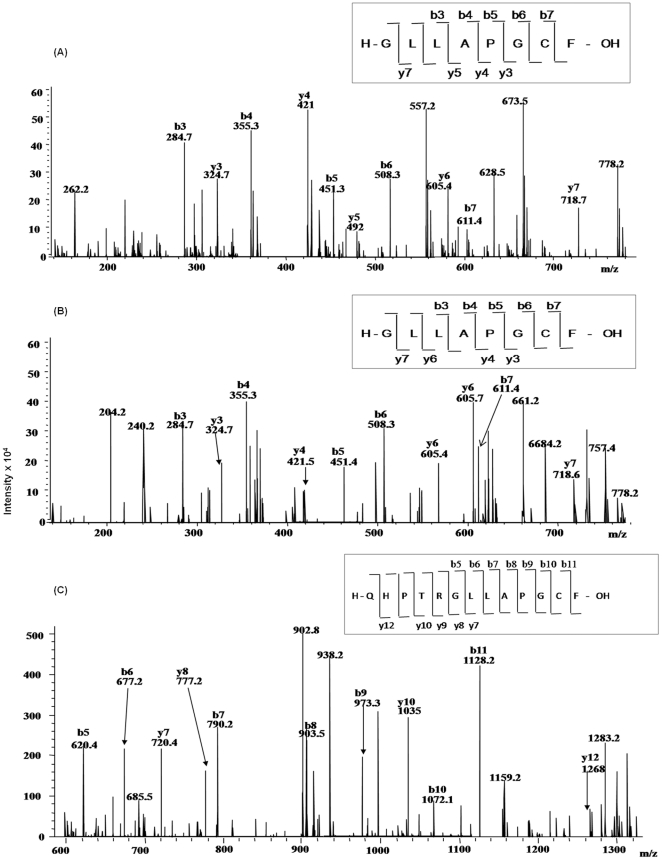
MS/MS analysis of singly charged [M + H]+ ions at m/z 776.6 (aa. 400–407) obtained from trypsin and chymotrypsin digested protein after DTT treatment confirmed Cys406 as a free cysteine containing peptide, Figure 8A. The Collision induced dissociation (CID) spectrum of the linearized peptide showed successive b and y-ions of m/z 284.7 (b3), 355 (b4), 451.3 (b5), 508.3 (b6), 611.4 (b7), 324.7(y3), 421 (y4), 492 (y5) and 718.7 (y7). The mass difference between the successive b or y ions represented the residual mass of amino acid as obtained by theoretical digestion. More than 50% of the predicted amino acid sequence obtained by MS/MS of the peptide matched with Mascot search data base. Figure 8B represents the free Cys406 peptide ion m/z 833.7 obtained from chymotrypsin and trypsin digested and iodoacetamide labeled protein in unreduced condition. The mass differences between the successive b or y ions are in agreement with the calculated mass. To further confirm that Cys406 is free in MtbDGC protein was digested with chymotrypsin in oxidized condition and was analyzed by LC/ESI/MS-MS. Singly charged [M +H]+ ion at m/z 1396.0 confirmed peptide containing Cys406. MS/MS of unreduced, trypsin digested and iodoacetamide labeled sample also confirmed Cys406 to be a peptide containing free cysteine (Figure 8C). Spectra were also acquired to locate free cysteine in reduced condition for trypsin and Chymotrypsin digested protein (data not shown).
Figure 8. LC-ESI-MS/MS of peptide containing free Cysteine406.
(A). LC-ESI-MS/MS of m/z 776.6, containing free Cys406 from DTT treated, trypsin and chymotrypsin digested MtbDGC. Inset shows the sequence derived from this MS/MS spectrum. (B) LC-ESI-MS/MS of m/z 833.7, containing free Cys406 from unreduced trypsin and chymotrypsin digested MtbDGC. Inset shows the sequence derived from this MS/MS spectrum. (C) LC-ESI-MS/MS of m/z 1396, containing free Cys406 from DTT treated, chymotrypsin digested MtbDGC. Inset shows the sequence derived from this MS/MS spectrum.
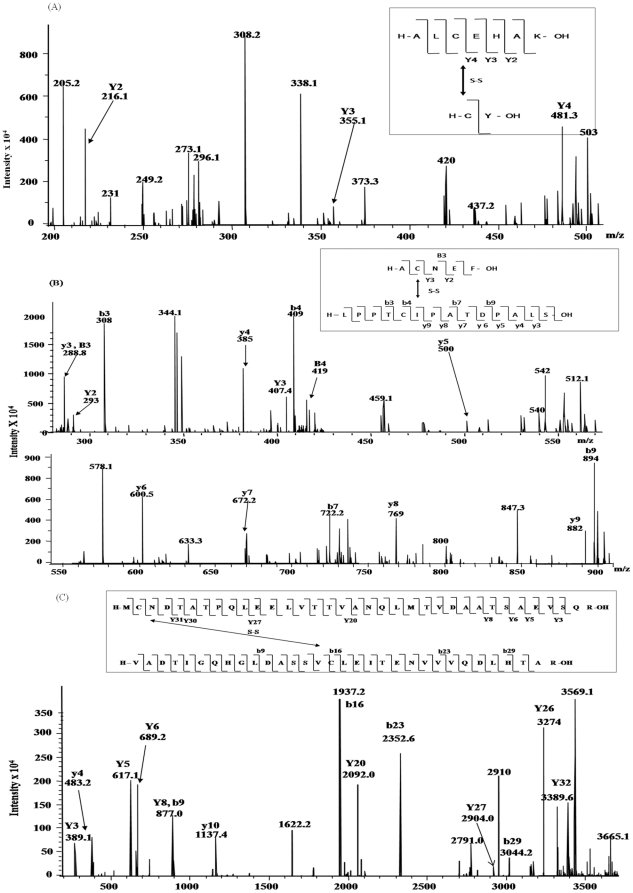
MS/MS analysis of the doubly charged [M +2H]2+ ion at m/z 3566.5, obtained from chymotryptic and tryptic digested sample, alkylated with iodoacetamide, confirmed that the peptide containing Cys2 (m/z 3739.2) was linked to the peptide containing Cys479 (m/z 3391.8) (Figure 9A). The CID spectrum of dipeptide containing Cys2 and Cys479 showed successive b and y type ions of m/z 389.1 (Y3), 483.4 (y4), 617.7 (Y5), 689.2 (y6), 877.0 (y8, b9), 1137.4 (y10), 1937.2 (b16), 2092.0 (Y20), 2352.6 (b23), 2904.0 (Y27), 3044.2 (b29), 3274 (y26) and 3389.6 (Y32). Evidence was also obtained for another expected disulfide pair (Cys94-Cys584). As shown in Figure 9B, the MS/MS of singly charged ion at m/z 1053.2 confirms that peptide containing Cys94 (m/z 770) is linked with peptide containing Cys584 (m/z 284.3). The CID spectrum showed successive b and y type ions of m/z 288.8 (B3), 293 (Y2), 407.2 (Y3), 308 (b3), 409 (b4), 722.2 (b7), 894 (b9), 288.8 (y3), 385 (y4), 500 (y5), 600.5 (y6), 672.2 (y7), 769 (y8) and 882 (y9). As shown in Figure 9C, the tandem mass spectra obtained from the ion m/z 1976.8 confirm the structure of peptide containing the third disulfide bridge between Cys429-Cys614. Figure 9C shows that the prominent fragments are derived from the singly charged ion of dipeptide containing Cys429 and Cys614. From MS/MS analysis it can be seen that only minor fragmentation occurred within the amino acid sequence between disulfide linked cysteine residues. The data showed no other disulfide linked peptides, other than the expected. It is consistent with the fact that it is difficult to cleave peptides that contain –S-S- bonds as fragmentation requires cleavage of double bonds [28]. No evidence was found for the fragmentation of the amide bonds and formation of linearized sequences by cleavage of peptide bonds inside the disulfide ring [50].
Figure 9. LC-ESI-MS/MS of disulfide connected dipeptide.
(A) LC-ESI-MS/MS m/z 3566.5 (doubly charge), containing disulfide bonded Cys2-Cys479 peptide from trypsin and chymotrypsin digested MtbDGC. Inset shows the sequence derived from this MS/MS spectrum. (B) LC-ESI-MS/MS of m/z 1053.2, containing disulfide bonded Cys94-Cys584 peptide from trypsin and chymotrypsin digested intact unreduced MtbDGC. Inset shows the sequence derived from this MS/MS spectrum. (C) LC-ESI-MS/MS of m/z 2799.1, containing disulfide bonded Cys429-Cys614 peptide from trypsin and chymotrypsin digested MtbDGC. Inset shows the sequence derived from this MS/MS spectrum.
Similar dissociation fragmentation processes have already been reported earlier for intramolecular disulfide-bonded peptides [26]. The obtained data confirmed the disulfide connectivity within intramolecular disulfide bond, but not intermolecular disulfide bond. Similar results to those shown in Figure 9(A–C) were observed for chymotrypsin and trypsin digested samples treated with or without iodoacetamide.
Sequence alignment of proteins containing GAF, GGDEF, EAL domains and homology modeling of Rv1354c
There is no structure available for full length GAF-GGDEF-EAL bifunctional protein. However, to probe the disulfide connectivity, even an approximate structure would be very useful. Structure obtained by modeling studies (Figure 10) showed a good correspondence between the predicted disulfide connectivity obtained by mass spectral analysis.
Figure 10. Modeled MtbDGC.
(A) Distribution of seven cysteines on full length MtbDGC. (B) Modeled MtbDGC with the disulfide bonds. (a) Modeled protein with exposed Cys406 residue. GGDEF motif (residue number 261–265) and is shown in black color. (b) Disulfide bond between Cys residue Cys2-Cys479 (c) Disulfide bond between Cys residue 894-584. (d) free and exposed Cys406 (e) disulfide between Cys residue 429-614.
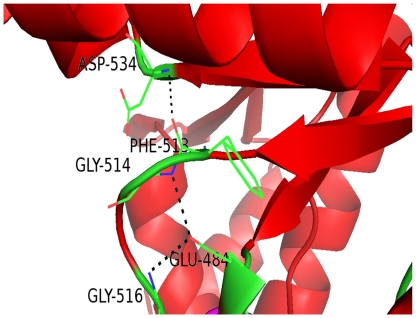
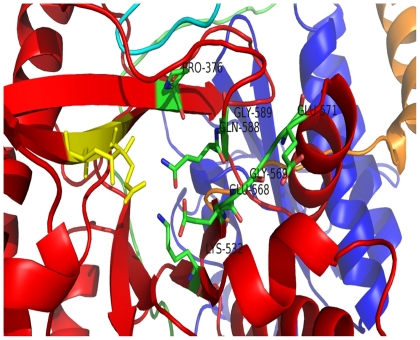
Schmidt and coworkers [51] first noticed that the catalytically active EAL domains seem to contain a conserved motif that was later confirmed to contain loop 6 [DFG(T/A)GYSS], loop between the α6-helix and β6-strand and one of the residues (Asp) for Mg2+ binding [51], [52]. Alignment of the EAL domain sequences shows a conserved motif [DFG(A/S/T)(A/G)(Y/F)(S/T)(S/T/G/A/N)] or also called loop 6 (Figure S1), apart from conserved EAL motif and many conserved acidic residues. Similar result was found when EAL domain (PF00563) family (total of 8082 sequences) in PFAM database, was analyzed (data not shown). Out of three domains named as GAF (residue 28–171), GGDEF (residue 212–345) and EAL (residue 354–609), EAL domain is the biggest with (β/α)10 barrel fold with 10 α-helix and β-strands each assigned by STRIDE [53]. Percentage consensus of G2, G4, F3, S7 and S8 in the loop 6 was found to be 88%, 79%, 80%, 65% and 61% respectively highlighting the importance of both Glycine residues along with Phenylalanine in the loop. Above result is based on the alignment of 8082 sequences belonging to EAL domain family. G514 and G516 are involved in the side chain - main chain hydrogen bonding with E484, while F513 and D534 residues are involved in the main chain-main chain hydrogen bonding (Figure 11) [54]. Hence, it can be concluded that the above three residues are important for the positioning of the loop 6. Depending upon the involvement of residues of loop 6 in forming hydrogen bond, they can be divided into two parts: i) D512F513G514T515G516 (ii) Y517S518A519. The first half gives stability and proper positioning to the loop, while the latter half is flexible. Alignment of EAL domain sequences obtained from the GAF, GGDEF, EAL architecture shows very high conservancy of P376, E484, G505, L523, K532, E568, G569, V570, E571,Q588G589. As discussed earlier E484 is involved in the hydrogen bonding with two glycines of loop 6. On mapping P376, K532, E568, G569,E571, Q588, G589 on the modeled structure, it was found that these residues are in close proximity with E389A390 L391 motif (Figure 12) may also play an important role in coordinating with Mg2+ions.
Figure 11. Part of EAL domain with the residues of loop 6.
Hydrogen bond between residues is shown in the black broken line.
Figure 12. EAL motif shown in yellow color and the conserved residues in close proximity to the motif is highlighted.
GGDEF domain sequences were also aligned in the same fashion as the EAL domain sequences and were found to have high conservancy of G232, D237, G307 & R259 apart from GGDEF261–265 motif (Figure S2). When the above three residues were mapped on the structure, D237 is found to be on the surface, possibly making it important in oligomerization. High conservancy of G232 can be related with the need of neglecting satiric clashes and the proper orientation of the two consecutive helices. R259 may help in the stabilizing the GTP-GGDEF domain complex as it is close to the GGDEF motif.
P131 & G140 of GAF domain shows high conservancy (Figure S3). Both of them are part of beta strands and at the same time they are involved in van der Waals interaction with R381 and N54 respectively.
Although the part of linker region between GAF and GGDEF domain (linker 1; residue 184–211) is considered as a part of GGDEF domain, yet while analyzing it was considered as a part of linker region (according to SWISSPROT linker 1 ranges from residue 172–211) and analysis shows the high conservancy of D187, T190 & N194, R195(Figure S4). All are polar residues and may play important role in stabilizing the oligomer of the proteins.
Several N-terminal residues (residue 1–28) are involved in either main chain-main chain (M1-V565, C2-V565,R583-D4,A6-D581) or side chain-main chain (T7-T165,Q9-A159) interaction with the residues of GAF and EAL domain. At the same time C2 forms a disulfide bond with C479, showing the importance of the N-terminal residues in stabilizing the GAF-EAL interface. Although C-terminal residues are not involved in the formation of hydrogen bond, C614 forms a disulfide bond with C429. Hydrogen bonds were found between the GAF and EAL domain residues also (data not shown). Interestingly enough GAF domain does not interact with the GGDEF domain, but interactions were found between the linker 1(residue 172–211).
EAL-GGDEF interface has lesser number of interactions compared to that of the GAF-EAL interface. Few main chain-main chain & side chain-main chain hydrogen bonds were found (data not shown).
Van der Waals interaction between two atoms was evaluated using CONTACT program of CCP4 program suit by setting the limit of distance below 4.5 Å [55]. The result shows that although EAL is the last domain, yet it is in contact with both GAF and GGDEF domains.
The proposed structure of Rv 1354c protein shows well exposed GGDEF motif. Disulfide bond length was found to be 2.3 A° (Cys2-Cys479, Cys429-Cys614) and 2.2 A° (Cys94-Cys584). Cys406 residue of EAL domain is more solvent assessible than other six cysteine residues.
Construction of Cysteine mutant by site directed mutagenesis
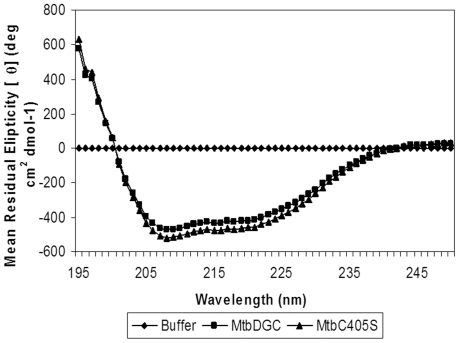
From the Mass spectral data it was confirmed that cysteine at position three (Cys406) is free. To further elucidate whether Cys406 has any role in the enzymatic activity of the protein, the Cys406 was mutated to serine. Cysteine mutation was confirmed by LC-MS and the mutated protein showed the mass of 68459.93 Da (histidine tagged) (data not shown). MtbC406S was further reduced with 8 mM of DTT and the reduced protein showed the mass difference of seven Da confirming the presence of only three cysteine bond and Cys3 (Cys406) is free (data not shown). The chymotrypin and trypsin digested MtbC406S shows the change of 16 Da in the peptide bearing cysteine at position three confirming the mutation of Cys406 to Serine406 (aa. 400–407) (Figure 13A). MS/MS analysis of mutated peptide (aa. 400–407) obtained from trypsin and chymotrypsin digested protein showed the mutation of cysteine to serine (data not shown). In order to check the activity of c-di-GMP in MtbC406S protein, the mass spectrometric as well as thin layer chromatographic analysis were done as shown in Figure 13B and Figure 13C. It is clear from the data that Cys406 which is a free cysteine plays a critical role in the regulation of enzymatic activity and by mutating cysteine to serine, protein activity is lost. However, when CD spectrum of the mutated protein was compared with MtbDGC protein (Figure 14) and no difference was found in the folding pattern of the protein. This indicates that Cys406 plays an important role in c-di-GMP regulation but does not play any significant role in the folding of the protein.
Figure 13. Functional activity of MtbC406S mutated protein.
(A) LC-ESI-MS of DTT reduced MtbC406S digested with trypsin and chymotrypsin. The m/z 760.9 shows the substitution of Cys406 to Serine406. (B) (a) MtbC406S protein: MALDI TOF analysis, in the positive ion detection mode. No peaks are obtained at 691 (M+H)+ and 713 (M+Na)+ for c-di-GMP, 709 (M+H)+ and 731 (M + Na)+ for pGpG (b) MtbDGC protein for control: MALDI TOF analysis, in the positive ion detection mode, ions were detected at 691 (M+H)+ and 713 (M+Na)+ for c-di-GMP (c) 709 (M+H)+ and 731 (M + Na)+ for pGpG.
Figure 14. Circular Dichorism spectra of MtbDGC and MtbC406S.
Concluding remarks
The cyclic dinucleotide 3′, 5′ cyclic guanylic acid (c-di-GMP) has been characterized as an important second messenger that affects a range of physiological traits in bacterial species. Many reports have come in recent past, from diverse branches of bacteria utilizing c-di-GMP signaling pathways [56], [57]. Prior to our study in Mycobacterium smegmatis (Kumar and Chatterji, 2008), nothing was known about the role of c-di-GMP in Mycobacteria. We reported that MSDGC-1 protein is a bifunctional protein and can synthesize and degrade c-di-GMP in vitro. This study was undertaken to follow the role of C-di-GMP in M. tuberculosis. In this work we investigate the homologue of MSDGC-1, MtbDGC in M. tuberculosis H37Rv. We reveal the following findings: (1). M. tuberculosis has one protein with GAF-GGDEF-EAL (MtbDGC) and another with only EAL domain (MtbPDE). (2). MtbDGC is a bifunctional multidomain protein exhibiting both the activity of DGC and PDE-A. (3). MtbPDE is a single EAL protein exhibiting only PDE-A activity. (4) Presence of three disulfide bond and one free cysteine in MtbDGC protein. (5) Upon mutation of free cysteine at position 406 to serine, synthesis of C-di-GMP was completely abolished.
All the known examples of GGDEF-EAL domain either have DGC or PDE-A activity. However, it is difficult to predict the dominant activity from the primary sequence [56], [58]. In Rhodobacter sphaeroides and Vibrio parahaemolyticuss GGDEF/EAL protein shows both DGC and PDE-A activities as a function of environmental cue [59], [60], but no clear cut distinction of the activities were possible. In this regard mycobacterial protein (MSDGC-1 and MtbDGC) is the first such protein which shows two activities at a time in vitro. It appears that both M. tuberculosis and M. smegmatis proteins have similar function; the role of additional EAL domain protein in M. tuberculosis is yet to be addressed.
M. tuberculosis DGC protein reported here was found to be predominantly expressed at the stationary phase suggesting its role in survival at stationary phase. This hypothesis was substantiated by complementation of MSDGC-1 knockout strain of M. smegmatis by mtbdgc. From sequence analysis and presence of dual activities it appears that both M. smegmatis and M. tuberculosis DGC will have similar function in the respective host. However, the only difference is presence of seven cysteines in MtbDGC. Comparison of the sequences of DGC protein revealed that three cysteines are conserved in MSDGC-1 out of seven in MtbDGC. It is interesting to study the presence of high numbers of cysteines in MtbDGC with regards to folding and regulation of c-di-GMP signaling at enzymatic level.
In order to address above hypothesis we studied disulfide bonded cysteine and free cysteine in MtbDGC. Disulfide bonded cysteine might play and crucial role in folding of MtbDGC, whereas free cysteine may be involved in regulation activity of enzyme. Extensive mass spectrometric analysis confirmed the presence of three disulfide bonded Cys94-Cys584, Cys2-Cys479 and Cys428-Cys614 and one free cysteine at position Cys406. Data obtained from mass spectrometric analysis was further confirmed by homology modeling.
Structure obtained by homology modeling revealed presence of three disulfide bonds and one free cysteine with exposed GGDEF motif. The homology model presented here, is the first example which shows that the last (EAL) domain comes in the middle interacting with the other two (GAF & GGDEF) domains. Probably it may hint in the possible mechanism regulating the activities of such proteins having both GGDEF & EAL domains to avoid a possible futile cycle. Apart from conserved motifs and polar residues, significant number of conserved Glycine residues was also found. Almost all conserved residues are either involved in the van der Waals interaction or in the hydrogen-bonding and are important in maintaining the orientation of the domains and thereby the shape of the protein.
Cys406 was found to be distinct far from the remaining six cysteines and does not participate in any disulfide connectivity. As we hypothesized that free cysteine might have regulatory role. We mutated Cys406 to Serine and enzymatic activity was assayed. C-di-GMP synthesis activity was completely abolished in mutated protein. The exact role of Cys406 in mechanism of c-di-GMP synthesis still remains to be investigated. To our best knowledge this is the first example of regulation of enzymatic activity by free cysteine in GAF-GGDEF-EAL protein.
Supporting Information
Alignment of EAL domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and [DFG(A/S/T)(A/G)(Y/F)(S/T)(S/T/G/A/N)] motif is shown in the box. Protein of interest contains DFGTGYSA motif.
(TIF)
Alignment of GGDEF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and G232, D237, G307, R259 & GGDEF261-265 motif is shown in box. Residue number of residues corresponds to the number in the protein chosen.
(TIF)
Alignment of GAF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and P131 & G140 are boxed.
(TIF)
Alignment of GGDEF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and D187, T190 & N194R195 are shown in box.
(TIF)
Acknowledgments
We thank Satisha, Sunita Prakash and Dr. Suman Thakur for technical help in performing the Mass spectrometry experiments, Dr. Manish Kumar for helping in MtbDGC complementation and MtbDGC cloning experiments and Indra Mani Sharma for reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is financially supported by the Department of Biotechnology, Government of India. KG would like to thank the Department of Biotechnology and PK would like to thank DBT-BINC, Government of India, for the fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Miller MB, Bassler BL. Quorum sensing in bacteria. Ann Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Redfield JR. Is quorum sensing a side effect of diffusion sensing? Trends in Microbiol. 2002;10:365–70. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 3.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanlylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–48. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr Opin Micorbiol. 2004:185–91. doi: 10.1016/j.mib.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Guo YL, Seebacher T, Kurz U, Linder JU, Schultz JE. Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J. 2001;20:3667–75. doi: 10.1093/emboj/20.14.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avarbock A, Avarbock D, The JS, Buckstein M, Wang Z, et al. Functional regulation of the opposing (p)ppGpp synthethase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochem. 2005;44:9913–23. 20. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 7.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–81. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 8.Weinhouse H, Sapir S, Amikam D, Shilo V, Volman G, et al. C-di-GMP-binding protein, anew factor regulatory cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–11. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 9.Jenal U, Malone J. Mechanisms of Cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–07. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 10.Sudarshan N, Lee ER, Weinberg Z, Moy RH, Kim JN, et al. Riboswitches in Eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–13. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan RP, Yvonne F, Jean FL, Lisa CC, Stephen S, et al. Cell–cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006;103:6712–17. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Ryjenkow DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic Diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1972–78. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Micorobiol. 2005 doi: 10.1186/1471-2180-5-35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regine H. Principles of c-di-GMP signalling in bacteria. Nat Rev Micorbiol, 2009;7:263–73. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 15.Tischler AD, Camilli A. Cyclic Diguanylate regulates Vibrio cholerae Virulence gene expression. Infect Immun. 2005;73:5873–82. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defense and killing of macrophages: role of cyclic di GMP. Mol Microbiol. 2005;56:1234–45. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 17.Kumar M, Chatterji D. C-di-GMP a second messenger required for long term survival, but not for biofilm formation in Mycobacterium smegmatis. Micorbiol. 2008;154:2942–55. doi: 10.1099/mic.0.2008/017806-0. [DOI] [PubMed] [Google Scholar]
- 18.Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–39. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 19.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Micorbiol. 2009;12:1–7. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Simm R, Morr M, Kadar A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic-di-GMP levels and transition from sensibility to motility. Mol Micorbiol. 2004;53:1123–34. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 21.Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defense and killing of macrophages: role of cyclic di GMP. Mol Microbiol. 2005;56:1234–45. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 22.Kulesekara H, Lee V, Brencic A, Liberati N, Urbach J. Analysis of Pseudomonas aeruginosa Diguanylate Cyclases and Phosphodiesterases Reveals a Role for Bis-(3′-5′)- Cyclic-GMP in Virulence. Proc Natl Acad Sci USA. 2006;103:2839–44. 22. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland LM, Sinéad TO'D, Ryjenkov DA, Gomelsky L, Slater SR, et al. A Staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol. 2008;190:5178–89. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang F, Xue T, Sun H, Xing L, Zhang S. The Staphylococcus aureus GGDEF domain-containing protein, GdpS, influences protein A gene expression in a cyclic diguanylic acid-independent manner. Infect Immun. 2009;77:2849–56. doi: 10.1128/IAI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutton RJ, Boyd D, Berkem M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–38. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman JJ, Wallis TP, Pitt JJ. Protein disulfide bond determination by Mass Spectrometry. Mass Spectrom Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Watson JT. A novel methodology for assignment of disulfide bond pairings in protein. Protein Sci. 1997;6:391–95. doi: 10.1002/pro.5560060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yen T-Y, Joshi RK, Hui Y, Seto NOL, Palcic MM, et al. Characterization of cysteine residues and disulfide bonds in proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2000;35:990–02. doi: 10.1002/1096-9888(200008)35:8<990::AID-JMS27>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Lu CH, Chen YC, Yu CS, Hwang JK. Predicting disulfide connectivity patterns. Proteins: Structure Function and Bioinformatics. 2007;67:262–270. doi: 10.1002/prot.21309. [DOI] [PubMed] [Google Scholar]
- 30.Bernardez Clark ED, Schwartz E, Rudolph R. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 1999;309:217–36. doi: 10.1016/s0076-6879(99)09017-5. [DOI] [PubMed] [Google Scholar]
- 31.Takagi T, Kubo K. Sodium dodecyl sulfate-protein polypeptide complexes in 8M urea with special reference to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochem Biophys Acta. 1997;578:68–75. doi: 10.1016/0005-2795(79)90114-4. [DOI] [PubMed] [Google Scholar]
- 32.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Gen Dev. 2004;18:715–27. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL. Nucl Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The protein data bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restrains. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 39.Kelley LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 40.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Tirado-Rives J. The OPLS Force Field for Proteins. Energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK – a program to check the stereochemical quality of protein structures. J Ap Cryst. 1993;26:283–291. [Google Scholar]
- 43.Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 44.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucl Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC [Google Scholar]
- 46.Finn RD, Mistry J, Tate J, Coggill C, Heger A, et al. The Pfam protein families database. Nucl Acids Res Database Issue. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng WP, Nickoloff JA. Site directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 48.Barik S. Site directed mutagenesis in vitro by megaprimer PCR. Methods Mol Biol. 1996;57:203–15. doi: 10.1385/0-89603-332-5:203. [DOI] [PubMed] [Google Scholar]
- 49.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Cell Biol. 2004;5:699–11. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 50.Mormaan M, Eble J, Schwoppe C, Mesters R, Berdel W. Fragmentation of intra peptide and inter peptide disulfide bonds of proteolytic peptides by nano ESI collision induced dissociation. Anal Bioannal Chem. 2008;392:831–838. doi: 10.1007/s00216-008-2258-7. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP- specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinig M, Frishman D. STRIDE: a Web server for secondary structure assignment known atomic coordinates of proteins. Nucl Acids Res. 2004;32:W500–2. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald IK, Thornton JM. Satisfying Hydrogen Bonding Potential in Proteins. JMB. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 55.Collaborative Computational Project, Number 4. “The CCP4 Suite: Programs for Protein Crystallography”. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL etal. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinus; genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–25. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salona C, Garcia B, Latasa C, Arana-Toldea A, Zorraquiano V, et al. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci USA. 2009;106:7997–02. doi: 10.1073/pnas.0812573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulasekara H, Lee V, Brencic A, Liberation N, Urbach J, et al. Analysis of Pseudomonas aeruginosa, diguanylate cyclases and phosphodiesterase reveals a role forbis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci USA. 2006;103:2839–44. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarutina M, Ryienkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem. 2006;281:34751–58. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 60.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates c-di-GMP regualtion of gene expression relevant to growth on surfaces. J Bacteriol. 2008;2008; 190:851–60. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of EAL domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and [DFG(A/S/T)(A/G)(Y/F)(S/T)(S/T/G/A/N)] motif is shown in the box. Protein of interest contains DFGTGYSA motif.
(TIF)
Alignment of GGDEF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and G232, D237, G307, R259 & GGDEF261-265 motif is shown in box. Residue number of residues corresponds to the number in the protein chosen.
(TIF)
Alignment of GAF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and P131 & G140 are boxed.
(TIF)
Alignment of GGDEF domain from the protein having GAF, GGDEF, EAL architecture. Protein of our interest is highlighted and D187, T190 & N194R195 are shown in box.
(TIF)