Abstract
Background
Glyceraldehyde 3-phosphate dehydrogenases (GAPDHs) are cytoplasmic glycolytic enzymes, which although lacking identifiable secretion signals, have also been found localized to the surface of several bacteria (and some eukaryotic organisms); where in some cases they have been shown to contribute to the colonization and invasion of host tissues. Neisseria meningitidis is an obligate human nasopharyngeal commensal which can cause life-threatening infections including septicaemia and meningitis. N. meningitidis has two genes, gapA-1 and gapA-2, encoding GAPDH enzymes. GapA-1 has previously been shown to be up-regulated on bacterial contact with host epithelial cells and is accessible to antibodies on the surface of capsule-permeabilized meningococcal cells. The aims of this study were: 1) to determine whether GapA-1 was expressed across different strains of N. meningitidis; 2) to determine whether GapA-1 surface accessibility to antibodies was dependant on the presence of capsule; 3) to determine whether GapA-1 can influence the interaction of meningococci and host cells, particularly in the key stages of adhesion and invasion.
Results
In this study, expression of GapA-1 was shown to be well conserved across diverse isolates of Neisseria species. Flow cytometry confirmed that GapA-1 could be detected on the cell surface, but only in a siaD-knockout (capsule-deficient) background, suggesting that GapA-1 is inaccessible to antibody in in vitro-grown encapsulated meningococci. The role of GapA-1 in meningococcal pathogenesis was addressed by mutational analysis and functional complementation. Loss of GapA-1 did not affect the growth of the bacterium in vitro. However, a GapA-1 deficient mutant showed a significant reduction in adhesion to human epithelial and endothelial cells compared to the wild-type and complemented mutant. A similar reduction in adhesion levels was also apparent between a siaD-deficient meningococcal strain and an isogenic siaD gapA-1 double mutant.
Conclusions
Our data demonstrates that meningococcal GapA-1 is a constitutively-expressed, highly-conserved surface-exposed protein which is antibody-accessible only in the absence of capsule. Mutation of GapA-1 does not affect the in vitro growth rate of N. meningitidis, but significantly affects the ability of the organism to adhere to human epithelial and endothelial cells in a capsule-independent process suggesting a role in the pathogenesis of meningococcal infection.
Background
Neisseria meningitidis is an obligate human commensal that is spread from person to person by droplet infection. The organism colonizes the nasopharyngeal mucosa in an asymptomatic manner, a condition known as carriage [1]. Under certain circumstances the bacteria can invade the epithelial layers to gain access to the bloodstream, which can result in a wide spectrum of clinical syndromes ranging from transient bacteraemia to rapidly fatal sepsis. Bacteria may also interact with cerebrovascular endothelial cells and cross the blood-cerebrospinal fluid barrier to cause meningitis [2]. To reach the meninges, N. meningitidis must interact with two cellular barriers and adhesion to both epithelial and endothelial cells are crucial stages of infection. Adhesion to both cell types is complex and remains poorly understood, but initial attachment is mediated by type IV pili, which is followed by contact-dependent down-regulation of pili and capsule: structures that otherwise hinder intimate adhesion, in a process that may involve the CrgA protein [3]. Intimate interaction between bacterial membrane components and their respective host cell surface receptors may subsequently lead to uptake of the bacterial cells (reviewed in [4]).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a glycolytic enzyme which catalyzes the conversion of glyceraldehyde 3-phosphate to 1, 3-diphosphoglycerate. The most common form is the NAD+-dependent enzyme (EC 1.2.1.12) found in all organisms studied so far and which is usually located in the cytoplasm. In addition to its metabolic function, studies have demonstrated that GAPDH is present on the surface of several microbial pathogens and may facilitate their colonization and invasion of host tissues by interacting directly with host soluble proteins and surface ligands. Surface localization of GAPDH was first demonstrated in the Gram-positive pathogen, Streptococcus pyogenes. In this organism, surface-exposed GAPDH binds several mammalian proteins including the uPAR/CD87 membrane protein on pharyngeal cells [5-8], regulates intracellular host cell signalling events [9] and contributes to host immune evasion [10]. GAPDH was subsequently identified on the surface of other Gram-positive bacteria including staphylococci [11,12], S. agalactiae [13], S. pneumoniae [14] and Listeria monocytogenes [15]. In addition, surface localization of GAPDH has been reported in enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli; the protein of these pathogens has been observed to bind to human plasminogen and fibrinogen, suggesting a role in pathogenesis [16]. Similar to the surface-localized GAPDHs from other species, the EHEC and EPEC GAPDH proteins possess NAD-ribosylating activity [17]. In Mycoplasma genitalium, surface-associated GAPDH is important for adhesion to human mucin [18], and in Lactobacillus plantarum, a normal inhabitant of the human gastrointestinal tract, GAPDH was shown to be involved in adherence to gastric mucin and Caco-2 cells [19,20]. Interestingly, the major fimbriae of Porphyromonas gingivalis bind to GAPDH on the surface of several oral streptococci, and this interaction is important for colonization of the oral cavity [21]. Fungi also express GAPDH on their cell surface, for example, the GAPDH of Candida albicans was shown to be associated with the cell wall and involved in mediating adhesion to fibronectin, laminin and plasminogen [22-24]. GAPDH has also been found on the surface of the single-celled protozoan, Trichomonas vaginalis, and shown to bind extracellular matrix components, including fibronectin [25].
The N. meningitidis MC58 genome sequence contains two putative GAPDH-encoding genes (gapA-1 and gapA-2) which share 50% nucleotide identity [26]. Expression of GapA-1 (but not GapA-2) on the meningococcal cell surface was previously found to be up-regulated following contact with human epithelial cells, although no function was ascribed to this observation [27]. Two other cytoplasmic glycolytic enzymes, despite lacking identifiable secretion signals, anchoring motifs or hydrophobic membrane-spanning regions (hence the term 'anchorless proteins'), have been found localized to the surface of N. meningitidis. These are enolase, which acts to recruit plasminogen onto the bacterial surface [28], and fructose-1, 6-bisphosphate aldolase (FBA), which we have recently demonstrated is required for optimal adhesion to human cells [29]. The aim of this study was to determine whether GapA-1 can influence the interaction of meningococci and host cells.
Methods
Bacterial strains and growth conditions
E. coli TOP10F' and BL21(DE3)pLysS (Table 1) were used for the expression of 6 × histidine-tagged recombinant GapA-1 encoded by plasmid pDT-GapA1 (Table 1). E. coli JM109 was used as host for the construction of mutagenic and complementation plasmids, pSAT-8 and pSAT-14 respectively. E. coli strains were grown at 37°C in LB broth or on LB agar supplemented, where appropriate, with ampicillin (100 μg ml-1), kanamycin (30 μg ml-1) or erythromycin (200 μg ml-1). Strains of Neisseria (Table 1 and Additional file 1) were grown at 37°C, air plus 5% CO2, on Brain Heart Infusion (BHI) agar supplemented with 1% Vitox (Thermo Fisher Scientific, Waltham, MA) and kanamycin (50 μg ml-1) or erythromycin (5 μg ml-1) where appropriate.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | Cloning strain | Promega, Madison, WI |
| TOP10F' | Cloning strain | Invitrogen, Carlsbad, CA |
| BL21(DE3)pLysS | Expression strain | Invitrogen, Carlsbad, CA |
| N. meningitidis | ||
| MC58 | wild-type serogroup B strain | [26] |
| MC58ΔgapA-1 | gapA-1 deletion and replacement with kanamycin cassette | This study |
| MC58ΔgapA-1 gapA-1Ect | MC58ΔgapA-1 complemented with an ectopic copy of gapA-1 | This study |
| MC58ΔsiaD | siaD deletion and replacement with erythromycin cassette | C. Tang Imperial College |
| MC58ΔsiaD ΔgapA-1 | siaD and gapA-1 deficient strain generated from MC58ΔsiaD using pSAT-8 | This study |
| Plasmids | ||
| pCRT7/NT-TOPO | Cloning vector encoding resistance to ampicillin | Invitrogen, Carlsbad, CA |
| pDT-GapA1 | MC58 gapA-1 gene cloned in pCRT7-TOPO | This study |
| pGEM-T Easy | Cloning vector encoding resistance to ampicillin | Promega, Madison, WI |
| pSAT-6 | 3-kb fragment spanning the MC58 gapA-1 region cloned in pGEM-T Easy | This study |
| pJMK30 | Source of kanamycin resistance cassette | [43] |
| pSAT-8 | pSAT-6 containing the kanamycin resistance cassette in the same orientation as the deleted gapA-1 gene | This study |
| pSAT-12 | Complementation vector containing cbbA and encoding resistance to erythromycin | [29] |
| pSAT-14 | pSAT-12 containing gapA-1 in place of the deleted cbbA | This study |
DNA manipulation
Genomic DNA was extracted from N. meningitidis using the DNeasy Tissue kit (Qiagen, Crawley, UK). Plasmid DNA was prepared using the QIAprep Spin kit (Qiagen, Crawley, UK). All enzymes were purchased from Roche Diagnostics (Indianapolis, IN) and used according to the manufacturer's instructions. DNA sequencing was carried out at the School of Biomedical Sciences (University of Nottingham) on an ABI 377 automated DNA sequencer.
Preparation of recombinant GapA-1 and αGapA-1 rabbit polyclonal antiserum
The gapA-1 gene was amplified from N. meningitidis MC58 using oligonucleotide primers NMB0207(F) and NMB0207(R) (Table 2). The amplicon was ligated into pCRT7/NT-TOPO and the resulting plasmid, pDT-GapA1, used to transform E. coli BL21(DE3)pLysS. Transformants were grown to log phase, induced for 3 h with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and harvested by centrifugation. Recombinant 6 × histidine-tagged GapA-1 was then affinity-purified under denaturing conditions. Briefly, the culture pellet was dissolved in 20 ml lysis buffer (100 mM NaH2PO4, 10 mM Tris-Cl, 10 mM Imidazole and 8 M Urea, pH 8.0) and disrupted by sonication using a MSE Soniprep 150 for 10 cycles (each cycle consisting of a 10 s burst followed by a 10 s cooling period). The cell lysates was then mixed with 1 ml HisPur™ Cobalt Resin (Thermo Fisher Scientific, Waltham, MA) and incubated overnight at 4°C. The lysate-resin mixture was then applied to the column, and washed with 100 mM NaH2PO4, 10 mM Tris-Cl, 20 mM Imidazole, 8 M Urea, pH 6.3. Bound proteins were then eluted in elution buffer (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M Urea, pH 4.5). Eluted fractions were resolved by SDS-PAGE, and recombinant GapA-1 excised from the gel, transferred to Mini D-Tube dialyzers (Merck Biosciences, Darmstadt, Germany) and electro-eluted according to the recommendations of the manufacturer. Recombinant GapA-1 was then concentrated using YM-30 Centrifugal filter units (Millipore, Billerica, MA). To generate rabbit antiserum against purified recombinant GapA-1, a New Zealand White female rabbit was immunized subcutaneously four times at 2-week intervals with 30 μg of protein emulsified in Freund's complete (first immunization only) or incomplete adjuvant.
Table 2.
List of primers used in this study
| Primer | DNA sequence* | Restriction site |
|---|---|---|
| Expression | ||
| NMB0207(F) | CGCGGATCCATGGGCATCAAAGTCGCCATC | BamHI |
| NMB0207(R) | CGCGTCGACTTATTTGAGCGGGCGCACTTC | SalI |
| Mutagenesis | ||
| NMB0207(R)FL | GAGAACTGTCATGCGTATTCC | |
| NMB0207(F)FL | CCAAACCCAATGCCGCGAATG | |
| gapA1_M1(IR) | GCGAGATCTGCAACAAACCGTC | BglII |
| gapA1_M2(IF) | GCGAGATCTGGTTTGTTCCTTTGTTGAGGG | BglII |
| Complementation | ||
| pSAT-12iPCR(IF) | CGCAGATCTGATACCCCCGATGAC | BglII |
| pSAT-12iPCR(IR) | CGCAGATCTCATTTGTGTC TCCTTGG | BglII |
| gapA1_Comp(F)2 | CGCGGATCCATGGGCATCAAAGTC | BamHI |
| gapA1_Comp(R)2 | CGCGGATCCTTTGTTTGACGGTTTGTTG | BamHI |
*All primers were designed from the N. meningitidis MC58 genome sequence. Sequences in bold identify restriction enzyme sites.
SDS-PAGE and immunoblotting
Proteins were electrophoretically separated using 10% polyacrylamide gels (Mini-Protean III; Bio-Rad, Hercules, CA) and were stained using SimplyBlue Safestain™ (Invitrogen, Carlsbad, CA) or transferred to nitrocellulose membranes as previously described [30]. Membranes were probed with mouse anti-pentahistidine antibody (Qiagen, Crawley, UK) or rabbit primary antibody diluted 1:10,000 & 1:1000 respectively in blocking buffer (5% [wt/vol] non fat dry milk, 0.1% [vol/vol] Tween 20 in 1 × phosphate-buffered saline [PBS]) and incubated for 2 h. After being washed in PBS with 0.1% Tween 20 (PBST), membranes were incubated for 2 h with 1:30,000-diluted goat anti-mouse (or anti-rabbit) IgG-alkaline phosphatase conjugate (Sigma-Aldrich, St. Louis, MI). After washing with PBST, blots were developed using BCIP/NBT-Blue liquid substrate (Sigma-Aldrich, St. Louis, MI).
Construction of MC58ΔgapA-1
A ca. 3 kb fragment of DNA consisting of the gapA-1 gene and flanking DNA was amplified using NMB0207(F)FL and NMB0207(R)FL (Table 2) from N. meningitidis MC58 chromosomal DNA. The amplified DNA was cloned into pGEM-T Easy to generate pSAT-6 (Table 1). This was then subject to inverse PCR using primers gapA1_M1(IR) and gapA1_M2(IF) (Table 2) resulting in the amplification of a 5 kb amplicon in which the gapA-1 coding sequence was deleted and a unique BglII site had been introduced. The BglII site was used to introduce a kanamycin resistance cassette from pJMK30 (Table 1) in place of gapA-1. One of the resulting plasmids, pSAT-8, containing the resistance cassette in the same orientation as the deleted gene, was confirmed by restriction digestion and sequencing and subsequently used to mutate meningococcal strains by natural transformation and allelic exchange as previously described [31]. Mutation of gapA-1 was confirmed by PCR analysis and immunoblotting.
Complementation of gapA-1
Plasmid pSAT-12, which we previously used to complement the meningococcal cbbA gene [29] was subjected to inverse PCR using the primers pSAT-12iPCR(IF) and pSAT-12iPCR(IR) (Table 2). This resulted in deletion of the cbbA coding sequence but leaving the upstream cbbA-promoter sequence intact and introduced a unique BglII site to facilitate the cloning of gapA-1 downstream of the promoter. The gapA-1 coding sequence was amplified from strain MC58 using the primers gapA1_Comp(F)2 and gapA1_Comp(R)2 (Table 2) incorporating BamHI-sites into the amplified fragment. The BamHI-digested fragment was then introduced into the BglII site to yield pSAT-14. This vector therefore contained the gapA-1 coding sequence under the control of the cbbA promoter and downstream of this, an erythromycin resistance gene. These elements were flanked by the MC58 genes NMB0102 and NMB0103. pSAT-14 was then used to transform MC58ΔgapA-1 by natural transformation, thus introducing a single chromosomal copy of gapA-1 under the control of the cbbA promoter and the downstream erythromycin resistance cassette in the intergenic region between NMB0102 and NMB0103. Insertion of the gapA-1 gene and erythromycin resistance cassette at the ectopic site was confirmed by PCR analysis and sequencing.
Flow cytometry
These experiments were performed essentially as previously described [29]. Briefly, 1 × 107 CFU aliquots of N. meningitidis were incubated for 2 h with rabbit anti-GapA-1-specific polyclonal antiserum (RαGapA-1) (1:500 diluted in PBS containing 0.1% BSA, 0.1% sodium azide and 2% foetal calf serum) and untreated cells were used as a control. Cells were washed with PBS and incubated for 2 h with goat anti-rabbit IgG-Alexa Fluor 488 conjugate (Invitrogen, Carlsbad, CA; diluted 1:50 in PBS containing 0.1% BSA, 0.1% sodium azide and 2% foetal calf serum). Again, untreated cells were used as a control. Finally, the samples were washed before being fixed in 1 ml PBS containing 0.5% formaldehyde. Samples were analyzed for fluorescence using a Coulter Altra Flow Cytometer. Cells were detected using forward and log-side scatter dot plots, and a gating region was set to exclude cell debris and aggregates of bacteria. A total of 50,000 bacteria (events) were analyzed.
Association and invasion assays
Association and invasion assays were performed essentially as previously described [29]. Briefly, human brain microvascular endothelial (HBME) or HEp-2 cells were grown to confluence in 24-well tissue culture plates and infected with approximately 1 × 106 CFU of log-phase meningococci (multiplicity of infection of 10 bacteria per cell) and incubated for 2 h (association) or 4 h (invasion) in 5% CO2 at 37°C. To assess total cell association, monolayers were washed, then disrupted and homogenized in 1 ml 0.1% saponin in PBS. To assess invasion, monolayers were further incubated in DMEM containing gentamicin (100 μg ml-1) for 2 h. Prior to further steps, aliquots of the gentamicin-containing supernatants were plated out to confirm killing of extra-cellular bacteria. Furthermore, the susceptibility of all meningococcal strains to gentamicin at 100 μg ml-1 was confirmed prior to testing. Monolayers were then washed, disrupted and homogenized in 1 ml 0.1% saponin in PBS. Meningococci were enumerated by serial dilution of the homogenized suspensions and subsequent determination of colony-forming units by plating aliquots from appropriate dilutions of the lysates on agar. All association and invasion assays were repeated at least three times. Statistical significance was measured using a two-tailed Student t-test.
Protein and nucleic acid sequence analysis
Public databases containing previously published protein and DNA sequences were searched using the BLAST and PSI-BLAST programs available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The genome database of N. meningitidis MC58 was interrogated at http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=gnm. Sequence homology data were obtained using the CLUSTALX software (http://www.clustal.org/). Protein secretion signals were analyzed using the SignalP 3.0 server available at http://www.cbs.dtu.dk/services/SignalP/[32]. GenBank accession numbers for the gapA-1 sequences analyzed in this study are as follows: YP_97432562 (FAM18), YP_00160027 (ST-4821 strain 053442), YP_002341615 (Z2491), YP_208807 (gonococcal strain FA1090) and ZP_03723143 (N. lactamica ATCC 23970).
Results
Sequence analysis of gapA-1, flanking DNA and GapA-1 protein
In N. meningitidis strain MC58, gapA-1 (locus tag NMB0207) is located downstream of, and in the opposite orientation to, aat (NMB0206) encoding the leucyl/phenylalanyl-tRNA-protein transferase and upstream of, and in the same orientation as, NMB0208, which encodes an electron transport protein, ferredoxin (4Fe-4S-type). A similar genomic arrangement is present in the meningococcal strains Z2491 [33], FAM18 [34] and 053442 [35]. The sequences of gapA-1 in these strains are >97% identical to the MC58 gapA-1 gene. Additionally, gapA-1 orthologues are found in the gonococcal strain FA1090 (99% identical) and N. lactamica strain ST640 (93% identical). At the amino acid level, the highly conserved GAPDH active site was identified (153ASCTTNCL160), and GapA-1 shows significant homology to GAPDH enzymes from higher organisms, including the human GAPDH enzyme (45% identity). Despite its demonstrated presence on the bacterial surface [27], GapA-1 of N. meningitidis was not predicted to be an exported protein by the SignalP-HMM and -NN programs.
Cloning, expression and purification of recombinant GapA-1
The gapA-1 gene from MC58 was cloned into the expression vector pCRT7/NT-TOPO to facilitate the expression and subsequent purification of 6 × histidine-tagged recombinant GapA-1 (Figure 1a). This was used to generate RαGapA-1. Immunoblot analysis confirmed that RαGapA-1 and anti-pentahistidine antibodies both reacted to the purified recombinant GapA-1 (Figure 1b &1c).
Figure 1.
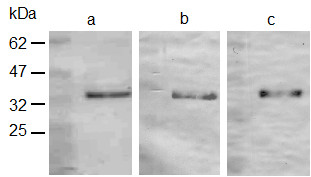
SDS-PAGE and immunoblot analysis of recombinant GapA-1. SDS-PAGE analysis confirms the purity of the recombinant GapA-1 purified under denaturing conditions (a). Immunoblot analysis shows that recombinant GapA-1 is recognized by RαGapA-1 (b) and anti-pentahistidine antibodies (c).
Construction of an N. meningitidis gapA-1 null mutant strain
To examine the roles of GapA-1 in the meningococcus, a gapA-1 knockout derivative of N. meningitidis MC58 was generated. Immunoblotting using RαGapA-1 showed that GapA-1 could be detected in whole cell lysates of wild-type but not MC58ΔgapA-1 (Figure 2, lanes 1 & 2) confirming that GapA-1 was expressed under the conditions used and that expression had been abolished in the mutant. This analysis further confirmed that the RαGapA-1 sera did not recognize GapA-2 (37-kDa) under the conditions used. To further confirm that the immuno-reactive protein was GapA-1, a wild-type copy of gapA-1 was introduced in trans into MC58ΔgapA-1 using plasmid pSAT-14 (Table 1). Introduction of gapA-1 at an ectopic site restored GapA-1 expression (Figure 2, lane 3). Further immunoblot analyses using a panel of 14 N. meningitidis strains (Additional file 1) including representatives of differing serogroups and MLST-types showed that GapA-1 expression was conserved across all strains (data not shown). Expression was also conserved in N. gonorrhoeae FA1090 (data not shown). These data complement in silico predictions that GapA-1 is universally present and suggests that GapA-1 is constitutively-expressed across pathogenic Neisseria species.
Figure 2.
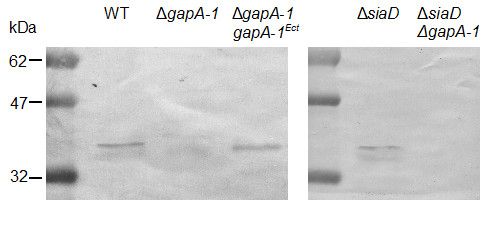
Immunoblot analysis of whole cell proteins from N. meningitidis using RαGapA-1. Analysis of MC58 wild-type, ΔgapA-1 mutant derivative and complemented mutant reveals the absence of GapA-1 in the ΔgapA-1 mutant preparation. Similar analysis shows the abolition of GapA-1 expression in the MC58ΔsiaD ΔgapA-1 mutant compared to the parental MC58ΔsiaD strain.
Meningococcal GapA-1 is only surface-accessible to antibodies in the absence of capsule
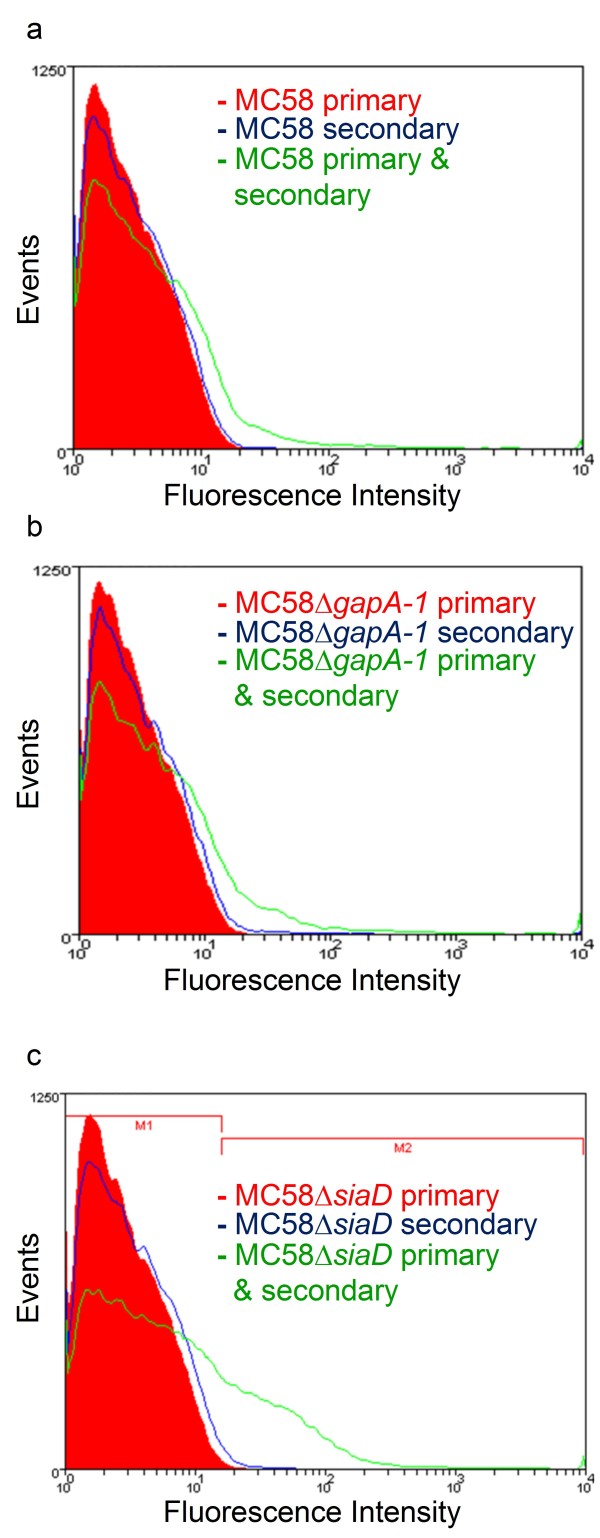
Grifantini et al showed using flow cytometry that GapA-1 was accessible to specific antibodies on the surface of meningococci [27]. However, the methodology used involved pre-treatment of the cells with 70% ethanol to permeabilize the capsule, making it unclear whether GapA-1 was accessible to antibodies in encapsulated bacteria. We used RαGapA-1 antibodies to probe wild-type, MC58ΔgapA-1 and MC58ΔsiaD (capsule-deficient) strains and analyzed them by flow cytometry (Figure 3). MC58 wild-type and MC58ΔgapA-1 treated with RαGapA-1 followed by anti-rabbit IgG-Alexa Fluor 488 conjugate showed no demonstrable shift in fluorescence signal compared to the same strains incubated with RαGapA-1 or secondary antibody alone showing that GapA-1 was not detectable on whole cells of these strains (Figure 3a &3b). However, identical experiments using MC58ΔsiaD demonstrated a clear shift in fluorescence when cells were treated with RαGapA-1 followed by anti-rabbit IgG-Alexa Fluor 488 conjugate (Figure 3c). This demonstrated that, in the absence of capsule, surface exposed GapA-1 was accessible to antibody. From the MC58ΔsiaD cells probed with both antibodies, 25% were found in the M2 region (Figure 3c), suggesting that in broth-grown cells unexposed to human epithelial cells only a minority of the population had GapA-1 was present on the cell surface. Pre-immune sera showed no reactivity against wild-type MC58 or MC58ΔsiaD, and RαGapA-1 specifically recognized only GapA-1 in immunoblot experiments confirming that the binding of RαGapA-1 to MC58ΔsiaD observed by flow cytometry was GapA-1 specific.
Figure 3.
Flow cytometry of MC58 wild-type (a), MC58ΔgapA-1 (b) or MC58ΔsiaD (c) for GapA-1 surface localization. Cells were stained with RαGapA-1 (primary alone), anti-rabbit IgG-Alexa Fluor 488 conjugate (secondary alone) or both. Fluorescence was displayed as a histogram. In panel c, the histogram area in M2 represents the population of fluorescently labelled meningococci.
GapA-1 is required for optimal adhesion to host cells
The capacity of the wild-type, GapA-1 mutant and complemented mutant strains to associate with, and invade into human brain microvascular endothelial (HBME) cells were then determined. GapA-1 deficient meningococci had a significantly reduced capacity to adhere to monolayers of HBME cells (Figure 4). No significant reduction was observed in the ability of the GapA-1 mutant to invade monolayers of HBME cells (data not shown). Similar results were also obtained using HEp-2 cells confirming that the effect was not limited to endothelial cells (data not shown). To confirm that the observed effects were not due to an impairment of in vitro growth, the growth rate of the strains was compared by measuring the optical density at 600 nm (OD600) and determining the viable counts of broth cultures sampled during exponential growth over 24 h in triplicate on three separate occasions. No significant difference between strains was observed (data not shown).
Figure 4.
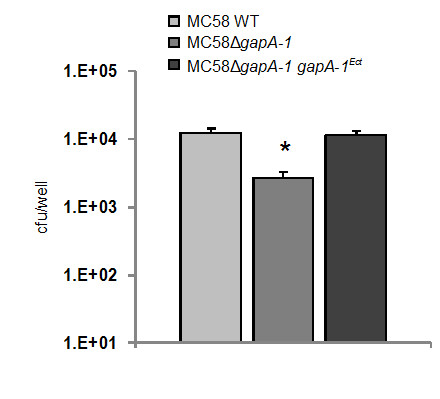
MC58ΔgapA-1 has a reduced ability to associate with HBMEs compared to the wild-type or complemented strains. The number of GapA-1-deficient meningococci associating was significantly lower than the wild-type (*P = 0.0018). Mean levels shown from three independent experiments, each using triplicate wells. Bars denote standard deviation. Cfu denotes colony forming units.
The effect of GapA-1 mutation on meningococcal adhesion is capsule-independent
Our experiments demonstrate clearly that GapA-1 increased the ability of meningococci to adhere to host cells (Figure 4), suggesting a role in pathogenesis. However, flow cytometry indicated that GapA-1 is made inaccessible to antibodies on the surface of meningococci by capsule (Figure 3). In order to determine whether capsule expression influences the role of GapA-1 in adhesion to host cells we constructed a gapA-1 deficient derivative of MC58ΔsiaD, which does not express a capsule. After confirming that GapA-1 expression had been abolished in MC58ΔsiaD ΔgapA-1 (Figure 2, lanes 4 & 5), we determined the capacity of both strains to associate with HBME cells. GapA-1 deficient non-encapsulated meningococci had a significantly reduced capacity to adhere to monolayers of HBME cells compared to the parent strain (Figure 5), confirming our observation that GapA-1 is required for optimal host cell adhesion. However, this reduction was not enhanced in the non-encapsulated background, indicating that the role of gapA-1 in the adhesion process was not moderated by the production of capsule. In summary, these experiments show that GapA-1 plays a role in the adherence of N. meningitidis with human cells in a capsule-independent manner.
Figure 5.
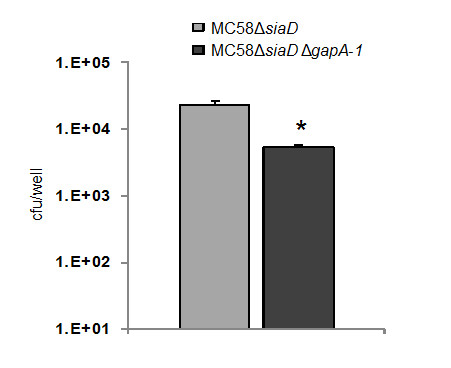
MC58ΔsiaD ΔgapA-1 has a reduced ability to associate with HBME cells compared to MC58ΔsiaD. The number of MC58ΔsiaD ΔgapA-1 cells associating was significantly lower than the capsule null (*P = 0.0008). Mean levels shown from three independent experiments, each using triplicate wells. Bars denote standard deviation. Cfu denotes colony forming units.
Discussion
It is now apparent that many of the classical cytoplasmic house-keeping enzymes, including enolase, FBA and GAPDH, are often localized to the surface of microbial pathogens, where they exhibit various functions, unrelated to their housekeeping roles [36-38]. Currently, there is considerable interest in identifying the additional roles of these bacterial glycolytic enzymes. In N. meningitidis, enolase was recently shown to be a surface-localized protein, where it acts to recruit plasminogen onto the bacterial surface [28]. In addition, we have recently demonstrated that FBA is also a partially surface-localized protein and is required for optimal adhesion to human cells through an unknown mechanism [29]. Furthermore, it is noteworthy that GAPDH is also a multi-functional protein in eukaryotic cells. For example, in addition to its role in central metabolic pathways, GAPDH is involved in controlling cell survival by delaying apoptosis via the inhibition of caspase-dependant proteolysis [39]. This raises the possibility that GAPDH on the surface of invasive bacterial pathogens such as N. meningitidis may influence intracellular processes of host cells to the advantage of the invading organism (including delaying apoptosis).
In our study, attempts to purify GapA-1 under native conditions were unsuccessful. Therefore, recombinant GapA-1 was purified under denaturing conditions in order to raise antiserum, but it was not possible to determine whether this protein had enzymatic activity. Rabbit polyclonal GapA-1 antiserum was used for immunoblot analysis of whole cell proteins from different clinical isolates of known MLST-type. These strains were representatives from lineages commonly causing invasive meningococcal disease. This showed that they all express GapA-1 suggesting that GapA-1 is constitutively-expressed in N. meningitidis. A GapA-1 knock-out mutant was created in N. meningitidis strain MC58 to facilitate studies of the potential role of GapA-1 in the pathogenesis of meningococcal disease. The GapA-1 mutant grew at the same rate (in broth culture and on solid media) as the wild-type and the complemented mutant strains, demonstrating that GapA-1 is not required for growth of the meningococcus under in vitro conditions. No differences in either colony or bacterial cell morphology (using light microscopy) were observed.
In a previous study, Grifantini et al. used microarrays to show that expression of gapA-1 was up-regulated in meningococcal strain MC58 (4.8-fold) following contact for 30 min with human 16HBE14 epithelial cells [27]. Subsequent flow cytometry experiments showed that GapA-1 could be detected on the cell surface of free grown and adherent meningococci [27]. However, the methodology used involved a pre-treatment of cells with 70% ethanol to permeabilize the capsule layer, thus making it unclear if GapA-1 is antibody-accessible in encapsulated meningococci. In our study, GapA-1 could only be detected on the meningococcal cell surface in mutants lacking capsule, suggesting that GapA-1 is usually masked by this structure.
In our adhesion experiments using siaD-knockout meningococci, the GapA-1 mutant strain exhibited a similarly significantly reduced capacity to adhere to host cells compared to the GapA-1 mutant in an encapsulated strain suggesting that the presence of capsule does not affect the role of GapA-1 in the adhesion process. It is not obvious why the influence of GapA-1 on adhesion is not itself modulated by the presence of masking capsule since the removal of capsule does increase the ability of meningococci to bind host cells via outer membrane adhesins [4]. In our adhesion experiments the binding of strains lacking capsule was approximately two-fold higher than the cognate encapulsulated strains (Figure 4 &5). This agrees with previous studies comparing the adherence of encapsulated and non-capsulated serogroup B meningococci to macrophages and buccal epithelial cells, where four-fold and less than two-fold increases, respectively, in adhesion were seen when capsule production was abolished [40,41]. Thus, it is possible that the influence of surface-localised GapA-1 on adhesion to host cells is indirect, possibly involving its enzymatic activity, and that a direct interaction of GapA-1 with the host cell surface is not required. Alternatively, capsule expression in meningococci is known to be down-regulated following the initial type IV pilus dependant-contact with host cells in order to facilitate intimate adherence [3]. Thus, we hypothesize that surface-localised GapA-1 may be unmasked following this change allowing it to influence subsequent steps in adhesion.
The observation that GapA-1 is detectable on the meningococcal cell surface suggests that GapA-1 is actively translocated to the outer membrane. An alternative hypothesis is that GapA-1 is released from lysed cells and recruited back onto the surface of intact meningococci. This maybe unlikely given the recent work on L. plantarum which showed that provoked cell lysis did not lead to re-association of GAPDH onto the cell surface [42]. Instead, it was suggested that changes in plasma membrane permeability during the growth cycle may be involved in the movement of GAPDH onto the external surface of the plasma membrane in this Gram-positive organism [42]. Clearly, such a mechanism could only account for periplasmic localization in a Gram-negative organism. We are currently investigating how GapA-1 is localized to the cell surface in N. meningitidis.
Conclusions
Meningococcal GapA-1 is a constitutively-expressed, highly-conserved surface-exposed protein which is antibody-accessible only in the absence of capsule. Mutation of GapA-1 does not affect the in vitro growth rate of N. meningitidis, but significantly affects the ability of the organism to adhere to human epithelial and endothelial cells in a capsule-independent process suggesting a role in the pathogenesis of meningococcal infection.
Authors' contributions
SAT carried out experiments and was involved in manuscript editing. NJO performed experiments and wrote the majority of the manuscript. DAAA, KGW and DPJT participated in the design of the experiments and wrote and edited portions of the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Isolates of N. meningitidis examined for the expression of GapA-1.
Contributor Information
Sarfraz A Tunio, Email: sarfraztunio@hotmail.com.
Neil J Oldfield, Email: neil.oldfield@nottingham.ac.uk.
Dlawer AA Ala'Aldeen, Email: daa@nottingham.ac.uk.
Karl G Wooldridge, Email: karl.wooldridge@nottingham.ac.uk.
David PJ Turner, Email: david.turner@nottingham.ac.uk.
Acknowledgements and Funding
We wish to thank Prof. Kim (John Hopkins University School of Medicine, Baltimore, US) for providing HBME cells and C. Tang (Imperial College, London, UK) for providing the MC58ΔsiaD strain. The work was funded by the University of Sindh, Pakistan. All authors have read and approved the final manuscript.
References
- Caugant DA, Maiden MCJ. Meningococcal carriage and disease - population biology and evolution. Vaccine. 2009;27(Suppl 2):B64–B70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine. 2009;27(Suppl 2):B71–77. doi: 10.1016/j.vaccine.2009.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deghmane AE, Giorgini D, Larribe M, Alonso JM, Taha MK. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol Microbiol. 2002;43:1555–1564. doi: 10.1046/j.1365-2958.2002.02838.x. [DOI] [PubMed] [Google Scholar]
- Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nature. 2009;7:274–286. doi: 10.1038/nrmicro2097. [DOI] [PubMed] [Google Scholar]
- Lottenberg R, Broder CC, Boyle MD, Kain SJ, Schroeder BL, Curtiss R. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J Bacteriol. 1992;174(16):5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176(2):415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winram SB, Lottenberg R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;142:2311–2320. doi: 10.1099/13500872-142-8-2311. [DOI] [PubMed] [Google Scholar]
- Jin H, Song YP, Boel G, Kochar J, Pancholi V. Group A streptococcal surface GAPDH, SDH, recognizes uPAR/CD87 as its receptor on the human pharyngeal cell and mediates bacterial adherence to host cells. J Mol Biol. 2005;350(1):27–41. doi: 10.1016/j.jmb.2005.04.063. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J Exp Med. 1997;186(10):1633–1643. doi: 10.1084/jem.186.10.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao Y, Yamaguchi M, Hamada S, Kawabata S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J Biol Chem. 2006;281(20):14215–14223. doi: 10.1074/jbc.M513408200. [DOI] [PubMed] [Google Scholar]
- Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67(3):1086–1092. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modun B, Morrissey J, Williams P. The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 2000;8:231–237. doi: 10.1016/S0966-842X(00)01728-5. [DOI] [PubMed] [Google Scholar]
- Seifert KN, McArthur WP, Bleiweis AS, Brady LJ. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. Can J Microbiol. 2003;49(5):350–356. doi: 10.1139/w03-042. [DOI] [PubMed] [Google Scholar]
- Ling E, Feldman G, Portnoi M, Dagan R, Overweg K, Mulholland F, Chalifa-Caspi V, Wells J, Mizrachi-Nebenzahl Y. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol. 2004;138(2):290–298. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg J, Diekmann O, Hagendorff P, Bergmann S, Rohde M, Hammerschmidt S, Jänsch L, Wehland J, Kärst U. The cell wall subproteome of Listeria monocytogenes. Proteomics. 2004;4(10):2991–3006. doi: 10.1002/pmic.200400928. [DOI] [PubMed] [Google Scholar]
- Egea L, Aguilera L, Gimenez R, Sorolla MA, Aguilar J, Badia J, Baldoma L. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int J Biochem Cell Biol. 2007;39(6):1190–1203. doi: 10.1016/j.biocel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Aguilera L, Giménez R, Badia J, Aguilar J, Baldoma L. NAD+-dependent post-translational modification of Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Int Microbiol. 2009;12:187–192. [PubMed] [Google Scholar]
- Alvarez RA, Blaylock MW, Baseman JB. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol Microbiol. 2003;48(5):1417–1425. doi: 10.1046/j.1365-2958.2003.03518.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita H, Uchida H, Kawai Y, Kawasaki T, Wakahara N, Matsuo H, Watanabe M, Kitazawa H, Ohnuma S, Miura K. et al. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J Appl Microbiol. 2008;104:1667–1674. doi: 10.1111/j.1365-2672.2007.03679.x. [DOI] [PubMed] [Google Scholar]
- Ramiah K, van Reenen CA, Dicks LM. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecalis. Res Microbiol. 2008;159:470–475. doi: 10.1016/j.resmic.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, Minamino N, Kuwahara H, Shizukuishi S. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun. 2009;77:5130–5138. doi: 10.1128/IAI.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Navarro I, Gil ML, Casanova M, O'Connor JE, Martinez JP, Gozalbo D. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J Bacteriol. 1997;179(16):4992–4999. doi: 10.1128/jb.179.16.4992-4999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalbo D, Gil-Navarro I, Azorin I, Renau-Piqueras J, Martinez JP, Gil ML. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun. 1998;66(5):2052–2059. doi: 10.1128/iai.66.5.2052-2059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonathan DC, Isla KS, Gillian CA, Norma RM, Neil ARG, Nuala AB. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol Microbiol. 2003;47(6):1637–1651. doi: 10.1046/j.1365-2958.2003.03390.x. [DOI] [PubMed] [Google Scholar]
- Lama A, Kucknoor A, Mundodi V, Alderete JF. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect Immun. 2009;77:2703–2711. doi: 10.1128/IAI.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ. et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287(5459):1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, Ratti G, Petracca R, Galli G, Agnusdei M. et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat Biotech. 2002;20(9):914–921. doi: 10.1038/nbt728. [DOI] [PubMed] [Google Scholar]
- Knaust A, Weber MV, Hammerschmidt S, Bergmann S, Frosch M, Kurzai O. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J Bacteriol. 2007;189(8):3246–3255. doi: 10.1128/JB.01966-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunio SA, Oldfield NJ, Berry A, Ala'Aldeen DAA, Wooldridge KG, Turner DPJ. The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol Microbiol. 2010;76:605–615. doi: 10.1111/j.1365-2958.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- Kizil G, Todd I, Atta M, Borriello SP, Ait-Tahar K, Ala'Aldeen DAA. Identification and characterization of TspA, a major CD4+ T-cell- and B-cell-stimulating Neisseria-specific antigen. Infect Immun. 1999;67:3533–3541. doi: 10.1128/iai.67.7.3533-3541.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi HA, Wooldridge KG, Robinson K, Ala'Aldeen DAA. Identification and characterization of App: an immunogenic autotransporter protein of Neisseria meningitidis. Mol Microbiol. 2001;41(3):611–623. doi: 10.1046/j.1365-2958.2001.02516.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T. et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404(6777):502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Vernikos GS, Snyder LAS, Churcher C, Arrowsmith C, Chillingworth T, Cronin A, Davis PH, Holroyd NE, Jagels K. et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genetics. 2007;3(2):e23. doi: 10.1371/journal.pgen.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yang L, Yang F, Yang J, Yan Y, Nie H, Zhang X, Xiong Z, Jiang Y, Cheng F. et al. Characterization of ST-4821 complex, a unique Neisseria meningitidis clone. Genomics. 2008;91(1):78–87. doi: 10.1016/j.ygeno.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Chhatwal G. Housekeeping enzymes as virulence factors for pathogens. Int J Med Microbiol. 2003;293:293–391. doi: 10.1078/1438-4221-00283. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Kulshreshtha P, Bambah Mukku D, Bhatnagar R. Alpha-enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim Biophys Acta. 2008;1784(7-8):986–994. doi: 10.1016/j.bbapap.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30(3):142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Jang M, Kang HJ, Lee SY, Chung SJ, Sunghyun K, Chi SW, Cho S, Lee S, Lee CK, Par BC. et al. Glyceraldehyde-3-phosphate, a glycolytic intermediate, plays a key role in controlling cell fate via inhibition of caspase activity. Mol Cells. 2009;28:559–563. doi: 10.1007/s10059-009-0151-7. [DOI] [PubMed] [Google Scholar]
- Read RC, Zimmerli S, Broaddus C, Sanan DA, Stephens DS, Ernst JD. The (alpha2-->8)-linked polysialic acid capsule of group B Neisseria meningitidis modifies multiple steps during interaction with human macrophages. Infect Immun. 1996;64(8):3210–3217. doi: 10.1128/iai.64.8.3210-3217.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, Spellman PA, Swartley JS. Effect of the (alpha 2-->8)-linked polysialic acid capsule on adherence of Neisseria meningitidis to human mucosal cells. J Infect Dis. 1993;167(2):475–479. doi: 10.1093/infdis/167.2.475. [DOI] [PubMed] [Google Scholar]
- Saad N, Urdaci M, Vignoles C, Chaignepain S, Tallon R, Schmitter JM, Bressollier P. Lactobacillus plantarum 299v surface-bound GAPDH: a new insight into enzyme cell walls location. J Microbiol Biotechnol. 2009;19:1635–1643. doi: 10.4014/jmb.0902.0102. [DOI] [PubMed] [Google Scholar]
- van Vliet AH, Wooldridge KG, Ketley JM. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolates of N. meningitidis examined for the expression of GapA-1.