Abstract
Background
The species status of two closely related Chinese oaks, Quercus liaotungensis and Q. mongolica, has been called into question. The objective of this study was to investigate the species status and to estimate the degree of introgression between the two taxa using different approaches.
Methodology/Principal Findings
Using SSR (simple sequence repeat) and AFLP (amplified fragment length polymorphism) markers, we found that interspecific genetic differentiation is significant and higher than the differentiation among populations within taxa. Bayesian clusters, principal coordinate analysis and population genetic distance trees all classified the oaks into two main groups consistent with the morphological differentiation of the two taxa rather than with geographic locations using both types of markers. Nevertheless, a few individuals in Northeast China and many individuals in North China have hybrid ancestry according to Bayesian assignment. One SSR locus and five AFLPs are significant outliers against neutral expectations in the interspecific F ST simulation analysis, suggesting a role for divergent selection in differentiating species.
Main Conclusions/Significance
All results based on SSRs and AFLPs reached the same conclusion: Q. liaotungensis and Q. mongolica maintain distinct gene pools in most areas of sympatry. They should therefore be considered as discrete taxonomic units. Yet, the degree of introgression varies between the two species in different contact zones, which might be caused by different population history or by local environmental factors.
Introduction
Natural hybridization occurs frequently in plants and animals [1]. Analysis of natural hybridization and hybrid zones provides insight into the processes of introgression, speciation and reproductive isolation [1]–[4]. While contemporary hybridization and introgression have long been thought to threaten species persistence, more recent work suggests that these processes are not necessarily a major impediment to effective species delimitation [5]. However, they can lead to species barriers of varying strength across different contact zones, a feature of great potential interest to understand the evolution of reproductive isolation as well as its breakdown [6], [7].
The oaks (Quercus) should be good models to evaluate the effects of hybridization and introgression on species delimitation. They have long been recognized as a challenge to the ideal standard of discrete biological species because of their propensity to intercross [8]–[10]. Morphologically intermediate forms are frequently observed [11], [12]. Such intermediate forms can be especially abundant locally [e.g. 13]. These populations are then called hybrid swarms, which are defined as “an extremely variable mixture of species, hybrids, backcrosses, and later-generation recombination types” [10].
Earlier studies on oaks have shown that chloroplast (cp) DNA haplotypes are often shared in areas of sympatry [14]–[16]. They also established that sibling species pairs are more distinctly discriminated by morphological or ecological (i.e., adaptive) traits than by isozyme or nuclear DNA markers [17]–[20]. However, in contrast to cpDNA markers, some genetic differentiation generally exists at nuclear DNA markers [e.g. 17], [ 18], [ 20], [ 21]. These studies also provided the first quantitative evidence that the strength of species barriers can vary geographically [22], [23]. More recent studies relying on a combination of powerful markers such as SSRs (in sufficient number) or AFLPs in combination with effective assignment methods have successfully distinguished among closely related oak species [24]–[28]. Individuals can generally be assigned to their respective species using their multilocus genotypes irrespective of their physical appearance, providing evidence for bimodal distribution of characters, with few individuals with intermediate characters and many parental types [25], [29]. For instance, Muir and Schlötterer [30] found that all studied individuals of Q. petraea and Q. robur, two closely related European oak species that have been extensively studied, can be assigned to either of the two species. This led them to call into question the importance of hybridization in these species, thereby resurrecting an old controversy [31]. However, subsequent studies relying on SSR markers confirmed that there are evolutionary significant rates of hybridization between Q. robur and Q. petraea and between other oak species pairs [32]–[37], confirming earlier work [12].
In China, problems of taxonomic discrimination occur between the Liaotung oak (Q. liaotungensis Koidz.) and the Mongolian oak (Q. mongolica Fisch. ex Lede). Taxonomists distinguish the two species by subtle morphological differences: Q. liaotungensis has smooth-cupule acorns and 5–7 pairs of lateral veins per leaf, whereas in Q. mongolica the acorn cupule is rough and there are 8–12 pairs of lateral veins per leaf [38]. However, due to the morphological plasticity of oaks and the potential for hybridization in sympatric populations, these characters can be confusing. For instance, different local floras record different ranges of variation in number of lateral veins within each species, such as 7–9, 7–10, 7–13 and 8–10 for Q. mongolica and 5–7, 5–8 and 5–9 for Q. liaotungensis [39]. The Chinese version of Flora of China [40], Higher Plants of China [38] and many local floras discriminate the two species, but the English version of Flora of China [41] considers them as belonging to the same species.
In an attempt to elucidate whether the morphologically identified Q. liaotungensis and Q. mongolica are genetically distinct from each other, we examine genetic variation in 419 individuals from 15 oak populations with both SSR and AFLP markers. We also investigate if the markers are equally powerful at delimiting taxa or if there is some heterogeneity among loci or marker type. Finally, we evaluate to what extent species boundaries are homogeneous across the sympatric range or instead vary in strength geographically. To answer these questions, we rely on conventional population genetic analysis as well as on a Bayesian clustering approach without consideration of sampling locations and taxonomic status.
Materials and Methods
Study species
According to the description of Higher Plants of China [38], Quercus mongolica Fisch. ex Ledeb. is a common tree species of temperate, low-elevation broadleaved woodlands, with a widespread distribution in North China, Northeast China and in parts of Russia, North Korea and Japan; Q. liaotungensis Koidz. ( = Q. wutaishanica Mayr) is another dominant broadleaved tree in the warm temperate zone, with a main distribution in northern China and partially in North Korea [38]. Recently, Q. liaotungensis has also been reported in the Russian Far East [42]. In accordance with their ecological amplitudes, the two species can occur in the same locality. They have similar reproductive biology, characterized by monoecy, anemophily and seed dispersal by gravity and animals.
Sampling design
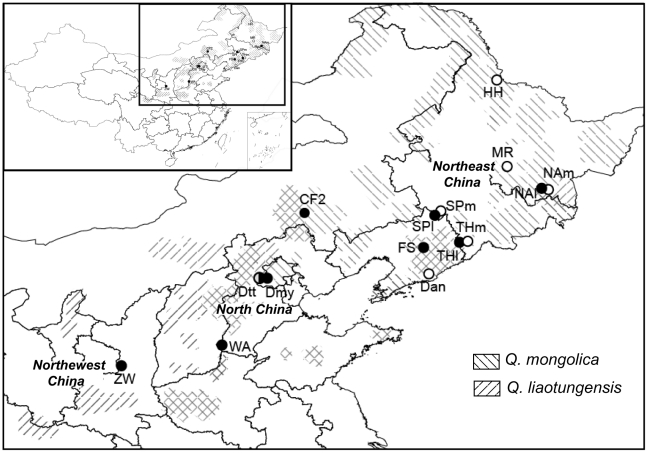
Leaf samples were collected from 419 individuals from a total of 15 oak populations across their distribution range in China (Table 1 and Fig. 1). Individuals were identified in the field as Q. liaotungensis (smooth acorn cupule), or Q. mongolica (rough acorn cupule), following Higher Plants of China [38]. Our observations suggest that Q. mongolica has smoother trunk bark than Q. liaotungensis. Therefore, the character of trunk bark was also considered where acorn cupule morphology alone was not sufficient to differentiate oak individuals (population THl and THm from Tonghua, and population SPl and SPm from Siping). According to above rules, four ‘pure’ Q. liaotungensis populations were sampled. Populations ZW and WA are located in areas where Q. mongolica is completely absent (allopatric range); CF2 and FS are located in areas from the sympatric range where Q. mongolica is locally absent. Three pure Q. mongolica populations were also collected. Populations HH and MR are located in areas where Q. liatungensis is completely absent (allopatric range); Dan is within the sympatric range in an area where Q. mongolica is locally absent. At three locations both species were found within mixed forests. Individuals were randomly sampled and then categorized into different species as far as possible, thus resulting in three pairs of populations: SPl and SPm, THl and THm, and NAl and NAm, where the lower-case letter ‘l’ represents Q. liaotungensis and ‘m’ Q. mongolica. In addition, one Q. liaotungensis-type population (Dmy) and one morphologically intermediate population (Dtt) were sampled from the Dongling Mountain region where controversy has arisen among Chinese taxonomists because of the existence of a broad array of morphologically intermediate trees [43]. These last two populations were excluded from the diversity and differentiation analyses due to the dominance of intermediates in Dtt and too small population sizes for Dmy. Leaf tissues (1–3 leaves per tree) were collected from each sampled tree, dried with silica gel and taken back to the laboratory.
Table 1. Description of 15 Quercus populations analyzed.
| Population type | Sampling Location | Population ID | Species | Longitude (E) | Latitude (N) | Sample size | HE | Hj |
| Pure site | Ziwuling, Shaanxi | ZW | Q. liaotungensis | 108°59′ | 35°30′ | 31 | 0.771 | 0.207 |
| Wu'an, Hebei | WA | Q. liaotungensis | 113°47′ | 36°55′ | 30 | 0.769 | 0.213 | |
| Chifeng, Neimenggu | CF2 | Q. liaotungensis | 117°58′ | 43°18′ | 30 | 0.764 | 0.229 | |
| Fushun, Liaoning | FS | Q. liaotungensis | 124°15′ | 41°50′ | 29 | 0.772 | 0.204 | |
| Heihe, Heilongjiang | HH | Q. mongolica | 127°19′ | 50°11′ | 29 | 0.713 | 0.214 | |
| Mao'ershan, Heilongjiang | MR | Q. mongolica | 127°40′ | 45°24′ | 32 | 0.724 | 0.205 | |
| Dandong, Liaoning | Dan | Q. mongolica | 124°10′ | 40°18′ | 28 | 0.760 | 0.211 | |
| Mixed site | Siping, Jilin | SPl | Q. liaotungensis | 124°15′ | 43°13′ | 26 | 0.797 | 0.219 |
| SPm | Q. mongolica | 124°15′ | 43°13′ | 29 | 0.793 | 0.223 | ||
| Tonghua, Jilin | THl | Q. liaotungensis | 125°55′ | 41°38′ | 30 | 0.730 | 0.212 | |
| THm | Q. mongolica | 125°55′ | 41°38′ | 30 | 0.757 | 0.211 | ||
| Ning'an, Heilongjiang | NAl | Q. liaotungensis | 129°32′ | 44°21′ | 25 | 0.785 | 0.234 | |
| NAm | Q. mongolica | 129°32′ | 44°21′ | 26 | 0.765 | 0.213 | ||
| Intermediate rich site | Dongling Mountain, Beijing | *Dmy | Q. liaotungensis | 115°27′ | 39°58′ | 12 | 0.776 | 0.232 |
| *Dtt | Intermediate | 115°26′ | 39°57′ | 32 | 0.772 | 0.225 |
*Populations that were eliminated from the diversity and differentiation comparison between species due to small population sizes or intermediate morphology; HE: mean expected heterozygosity across the 19 SSR loci; Hj: unbiased estimates of genetic diversity (analogous to HE) based on the 194 AFLP markers.
Figure 1. Geographic distribution of Quercus liaotungensis and Q. mongolica in China and location of populations sampled.
Shadows show the distribution of Q. liaotungensis and Q. mongolica, respectively, according to Zhang [38]. Filled circles: Q. liaotungensis; open circles: Q. mongolica; half filled circles: intermediate populations.
Marker analysis and scoring
Total genomic DNA was extracted from 25 mg of leaf tissue from each individual oak and purified using a Plant Genomic DNA Extraction Kit (Tiangen, Beijing, China). The oak samples were screened for variation at 19 nuclear SSR loci that had been developed for other oak species [44]–[49] (see Table 2 for primer details). PCR amplification with each primer pair was performed separately with a PTC-200 thermal cycler (MJ Research Inc.) in a 15-µL reaction volume. The PCR reaction mixture contained 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCL2, 200 µM dNTP, 0.3 µM (each) primer, 20 ng of DNA template, and 0.5 U Taq polymerase (TaKaRa Company, Tokyo, Japan). PCR amplifications were performed as follows: an initial denaturation step at 94°C for 4 min followed by 31 cycles of 45 s at 94°C, 45 s at an annealing temperature and 45 s at 72°C, and a final extension step at 72°C for 8 min.
Table 2. Description and reference of the 19 SSR loci analyzed in current study.
| Locus | Motif | Ta (°C) | Allele range (bp) | Labela | Reference |
| ssrQpZAG36 | (AG)19 | 50 | 206–246 | HEX | [46] |
| ssrQpZAG16 | (AG)21 | 58 | 137–177 | HEX | [46] |
| ssrQpZAG15 | (AG)23 | 50 | 102–150 | FAM | [46] |
| ssrQpZAG110 | (GA)14 | 50 | 190–244 | FAM | [46] |
| quru-GA-0C11 | (GA)11 | 53 | 190–248 | TAMRA | [48] |
| quru-GA-0C19 | (CT)7 | 53 | 193–227 | FAM | [48] |
| quru-GA-0M05 | (GA)15 | 53 | 182–240 | TAMRA | [48] |
| quru-GA-0M07 | (GA)19 | 48 | 180–252 | TAMRA | [48] |
| quru-GA-1C08 | (AG)10 | 52 | 251–289 | HEX | [48] |
| MSQ4 | (AG)12 | 48 | 191–269 | FAM | [44] |
| MSQ13 | (GA)29 | 50 | 190–246 | HEX | [44] |
| MSQ16 | (GA)7 | 55 | 178–232 | TAMRA | [45] |
| ssrQrZAG7 | (TC)17 | 50 | 117–169 | TAMRA | [47] |
| ssrQrZAG87 | (TC)20 | 50 | 97–185 | HEX | [47] |
| ssrQrZAG96 | (TC)20 | 53 | 133–195 | FAM | [47] |
| ssrQrZAG101 | (TC)20(AG)15 | 52 | 127–181 | FAM | [47] |
| ssrQrZAG102 | (GA)29 | 52 | 193–309 | FAM | [47] |
| ssrQrZAG112 | (GA)32 | 52 | 73–115 | TAMRA | [47] |
| bcqm42 | (GT)11 | 52 | 104–146 | HEX | [49] |
Forward primers were modified at the 5′ end with a fluorescent label: HEX (green), 6-FAM (blue), or TAMRA (yellow) (see the Materials and Methods, PCR amplification).
Ta: annealing temperature.
AFLP fingerprinting was performed according to the original protocol of Vos et al. [50] except that digestion and ligation were carried out simultaneously. In brief, 50–500 ng of DNA was digested for 3 h at 37°C with 8 U EcoRI and 2 U MseI in 20 µL 1×NEB buffer (10 mM Tris-HCl pH 7.5, 1 mM MgCL2, 5 mM NaCL, 0.0025% Triton X-100) with 2 µg/ml BSA. Simultaneously, two adaptors, one for the EcoRI ends and one for the MseI ends, were ligated to cutting sites by adding 0.2 µM adapters, 80 U T4 DNA ligase and 1 mM ATP within the digesting mixture. Selective pre-amplification was performed with primers (E01 = 5′-GACTGCGTACCAATTCA-3′ and M02 = 5′-GATGAGTCCTGAGTAAC-3′) complementary to adapters, but with one base extension. Each PCR was performed in a 20-µL reaction volume using 2 µL ligated product, primer concentration 0.25 µM, 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, 0.2 mM dNTP each and 0.5 U Taq DNA polymerase (TaKaRa Company, Tokyo, Japan). The selective pre-amplification was carried out in a PTC-200 thermal cycler (MJ Research Inc.) with the following thermo-cycling parameters: an initial denaturation step at 94°C for 2 min followed by 28 cycles of denaturation at 94°C for 45 s, annealing at 56°C for 45 s, extension at 72°C for 1 min, and a final extension step at 72°C for 10 min. The quality and quantity of the pre-amplified products obtained were determined on 1.0% agarose gels and diluted (1∶19) with ddH2O. The selective amplification was performed with four primer combinations (AGA/CTC, AGT/CTC, AAC/CTC, AAC/CAG). Each PCR was performed in a 20-µL reaction volume using 3 µL diluted pre-amplification product, 0.25 µM E-primer labeled fluorescence with 6-FAM (Sangon, Shanghai, China), 0.3 µM M-primer, 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, 0.2 mM dNTP and 0.5 U Taq DNA polymerase (TaKaRa Company, Tokyo, Japan). Amplification was performed with a touch down cycling process: an initial denaturation step at 94°C for 2 min, then 1 cycle of 30 s at 94°C, 30 s at 65°C, 1 min at 72°C, followed by 11 cycles in which the annealing temperature decreases 0.7°C per cycle, followed by 22 cycles of 30 s at 94°C, 30 s at 56°C and 1 min at 72°C, and a final extension step at 72°C for 5 min.
The SSR genotyping of all individuals was performed by assessing allele size on an ABI 3100 automated Genetic Analyzer (Applied Biosystems), using forward primers labeled with 6-FAM, HEX, or TAMRA (Sangon, Shanghai, China) and the ROX 500 (Applied Biosystems) as an internal standard. Allele sizing was performed using the GENEMAPPER software version 3.7 (Applied Biosystems). In the AFLP analysis, selective PCR products were also separated on an ABI 3100 automated sequencer (Applied Biosystems) with a genescan ROX 500 internal size standard. Electropherograms were subsequently analyzed using GENEMAPPER software version 3.7 (Applied Biosystems). The intensity of each individual peak was normalized on the basis of the total signal intensity and the peak was considered only if its intensity exceeded a fixed threshold. The multilocus AFLP profiles were scored as present (1), absent (0) or ambiguous (?) to create binary matrices. Each set of 48 reactions included a positive (known genotype) and a negative (water) control carried from restriction digest through to the final automated sequencer analysis for AFLP and from PCR through to the final automated sequencer analysis for SSR. Allele size determinations were performed twice manually to reduce scoring errors.
Genetic diversity analysis
Descriptive statistics for SSR such as the number of alleles, allele frequencies, observed and expected heterozygosities (HO and HE) were calculated using the program FSTAT 2.9.3 [51]. We used the software GENEPOP 4.0 for Windows [52] to test for homogeneity of allele distributions between species. We also counted the number of private alleles for each species.
For AFLP, percentage of polymorphic loci (5% level), unbiased estimates of genetic diversity (Hj, analogous to HE) and differentiation statistics were calculated using the AFLP-SURV 1.0 software [53]. With this software, allelic frequencies at AFLP loci were calculated from the observed frequencies of fragments using the Bayesian approach proposed by Zhivotovsky [54] for diploid species. A non-uniform prior distribution of allelic frequencies was assumed with its parameters derived from the observed distribution of fragment frequencies among loci [54]. These allelic frequencies were used as the input for the analysis of genetic diversity within and between samples following the method described in Lynch and Milligan [55].
Genetic differentiation
The significance of the genetic differentiation between species or among populations within species was tested by comparison of the observed F ST with a distribution of F ST under the hypothesis of no genetic structure, obtained by means of 5,000 random permutations of individuals between species or among populations. The F ST analogue θ of Weir and Cockerham [56] was calculated for the 19 SSR loci using the program FSTAT 2.9.3 [51]. For AFLP, F ST was calculated using the AFLP-SURV 1.0 software [53]. This program uses the approach of Lynch and Milligan [55] to calculate population genetic parameters on the basis of the expected heterozygosity of dominant marker loci.
Genetic differentiation of population pairs was estimated with θ of Weir and Cockerham [56] for the SSR loci and Φ PT (a statistic analogous to F ST [57]) for AFLPs. θ-values with significance level (obtained by bootstrapping loci 10,000 times) were calculated in FSTAT 2.9.3 [51] and Φ PT-values with significance level (obtained by 999 times permutation) were calculated in GenAlEx6 [58].
Genetic distances, DS [59] for SSR and D [60] modified for dominant markers by Lynch and Milligan [55] for AFLP, were calculated for each population pair using the programs GENDIST in the PHYLIP package [61] for SSR and AFLP-SURV 1.0 [53] for AFLP. The UPGMA (Unweighted Pair Group Method with Arithmetic mean) trees were generated using the program NEIGHBOR in the PHYLIP package [61] and bootstrapped 1000 times.
A principal coordinate analysis (PCo) was performed for both AFLP and SSR data sets to calculate principal co-ordinates from pairwise Euclidian distance estimates between individual genotypes. Analyses were executed in GenAlEx6 [58]. The first two axes were plotted graphically with Origin 7.5 (OriginLab, Northampton, MA).
Bayesian admixture analysis
A model-based clustering method implemented in the program STRUCTURE version 2.2 [62]–[64] was used to determine the probability of each individual (non-admixture models) or the proportion of each individual's genome (admixture model) from homogenous clusters without consideration of sampling locations for both the SSR and the AFLP data sets. Estimated posterior probabilities for the simulated model fitting the data were calculated for all samples assuming a uniform prior for K (number of possible clusters), and every cluster pattern from 1 to 10 was simulated. After fitting both the admixture and non-admixture models and an initial test of varying the burn-in and run length, ten replicates for each K were analyzed using the following parameters: we assumed correlated allele frequencies and an admixed origin of populations; burn-in was set to 100,000 with 1,000,000 additional cycles. STRUCTURE output, Pr(X|K), can be used as an indication of the most likely number of groups. In addition, following Evanno et al. [65], ΔK, where the modal value of the distribution is located at the real K, was also calculated using the software Structure2[1].2-sum (supplied by Dorothée Ehrich). For graphic visualization of the STRUCTURE results, we used DISTRUCT [66].
For the most likely STRUCTURE run (K = 2; see Results), the approximate Bayes factors (ratio of the estimated marginal likelihood of the admixture model to that of the non-admixture model) favoring admixture were 1.1×1036 (−33651.2 vs −33734.2) and 7.7×10131 (−41280.9 vs −41584.5) for SSR and AFLP, respectively. Under the admixture model, the posterior probability approximates the proportion of each individual's genome that is derived from each species [62]. The individuals were assigned to pure species or hybrid categories according to the estimated posterior probability at K = 2. A threshold value 0.8 was used as a compromise between efficiency and accuracy [67]. The percentage of hybrid oaks was compared among Siping, Tonghua, Ning'an and Dongling Mountain region where the two species and hybrids co-occur, using Chi-square tests with the crosstabs analysis in SPSS (SPSS Inc., Chicago IL).
Identification of loci under selection
To address the question of whether adaptive divergence occurs between Q. liaotungensis and Q. mongolica, we used a coalescent simulation outlined in Beaumont and Nichols [68] to identify ‘outlier loci’ whose empirically derived levels of differentiation place them at the upper extreme of simulated distributions of differentiation based on neutral loci. The program FDIST2 was used to simulate a null distribution of F ST values (conditional on heterozygosity) for the SSR data set under an infinite-alleles model and a symmetrical two-island model of population structure. Simulations employed the median of observed F ST and the same sample sizes as used in the empirical study. Outlier loci were detected by comparing the empirical distribution of F ST's with a simulated distribution derived from 50,000 paired values of F ST and heterozygosity at 99th quantiles. The significant AFLP outlier loci were identified by plotting F ST against heterozygosity under the assumption of Hardy-Weinberg equilibrium using the program Dfdist [68], [69]. Loci with F ST values that fell outside the 99th quantiles threshold were obtained by generating a null distribution of F ST-values based on 50,000 simulated loci with a mean F ST equivalent to ‘neutral’ mean F ST of empirical distribution, which was obtained by trimming the 30% highest and lowest F ST values.
Results
Genetic diversity and differentiation
All SSR loci studied were highly variable, with 5–45 alleles per locus (21 on average) and a total of 406 alleles found across all loci in our sample of 419 individuals. The mean expected heterozygosity (HE) across all loci for each population varied from 0.713 to 0.797 (Table 1). The number of alleles, observed heterozygosity (HO), expected heterozygosity (HE) across all loci were similar between Q. liaotungensis and Q. mongolica. However, there were some differences between species at some loci, e.g., ssrQpZAG15, ssrQpZAG110, quru-GA-0C11, ssrQrZAG87, and ssrQrZAG112 (Table 3). Q. liaotungensis and Q. mongolica shared most frequent alleles. Nevertheless, species-specific alleles were found at several loci, especially rare alleles restricted to Q. mongolica (Table 3). In fact, none of these private alleles had a frequency higher than 4%. Tests for heterogeneity of allele distributions were highly significant for all 19 SSR loci (P<0.0001).
Table 3. Comparison of genetic diversity and differentiation between Quercus liaotungensis and Q. mongolica based on the 19 SSR loci.
| N | An | AP | HO | HE | θ | ||||||||
| Locus | QL | QM | QL | QM | QL | QM | QL | QM | QL | QM | Between species | QL | QM |
| ssrQpZAG36 | 201 | 174 | 13 | 14 | 1 | 2 | 0.677 | 0.743 | 0.746 | 0.823 | 0.158 | 0.074 | 0.030 |
| ssrQpZAG16 | 201 | 174 | 19 | 18 | 1 | 0 | 0.851 | 0.878 | 0.903 | 0.914 | 0.008 | 0.041 | 0.024 |
| ssrQpZAG15 | 201 | 174 | 18 | 13 | 7 | 2 | 0.555 | 0.242 | 0.605 | 0.240 | 0.081 | 0.043 | 0.031 |
| ssrQpZAG110 | 201 | 173 | 15 | 21 | 1 | 7 | 0.680 | 0.542 | 0.646 | 0.595 | 0.103 | 0.055 | 0.022 |
| quru-GA-0C11 | 201 | 174 | 20 | 20 | 1 | 1 | 0.832 | 0.615 | 0.895 | 0.634 | 0.132 | 0.066 | 0.017 |
| quru-GA-0C19 | 201 | 173 | 5 | 5 | 0 | 0 | 0.405 | 0.287 | 0.395 | 0.290 | 0.008 | 0.049 | 0.001 |
| quru-GA-0M05 | 201 | 174 | 21 | 18 | 6 | 3 | 0.766 | 0.754 | 0.798 | 0.808 | 0.008 | 0.026 | 0.013 |
| quru-GA-0M07 | 201 | 174 | 9 | 12 | 0 | 3 | 0.732 | 0.766 | 0.770 | 0.829 | 0.043 | 0.040 | 0.018 |
| quru-GA-1C08 | 200 | 173 | 16 | 15 | 1 | 0 | 0.723 | 0.795 | 0.800 | 0.819 | 0.019 | 0.110 | 0.017 |
| MSQ4 | 201 | 174 | 18 | 27 | 1 | 10 | 0.807 | 0.780 | 0.899 | 0.919 | 0.008 | 0.017 | 0.031 |
| MSQ13 | 200 | 172 | 12 | 12 | 2 | 2 | 0.721 | 0.703 | 0.808 | 0.798 | 0.018 | 0.055 | 0.044 |
| MSQ16 | 199 | 171 | 15 | 19 | 0 | 4 | 0.855 | 0.799 | 0.894 | 0.809 | 0.051 | 0.025 | 0.015 |
| ssrQrZAG7 | 201 | 174 | 20 | 22 | 0 | 2 | 0.915 | 0.954 | 0.911 | 0.933 | 0.019 | 0.023 | 0.013 |
| ssrQrZAG87 | 200 | 173 | 23 | 36 | 0 | 13 | 0.770 | 0.919 | 0.818 | 0.951 | 0.061 | 0.044 | 0.007 |
| ssrQrZAG96 | 201 | 174 | 24 | 22 | 5 | 3 | 0.852 | 0.874 | 0.928 | 0.884 | 0.025 | 0.018 | 0.020 |
| ssrQrZAG101 | 201 | 172 | 21 | 24 | 2 | 5 | 0.794 | 0.930 | 0.842 | 0.918 | 0.046 | 0.110 | 0.025 |
| ssrQrZAG102 | 201 | 173 | 38 | 42 | 2 | 6 | 0.937 | 0.954 | 0.967 | 0.963 | 0.004 | 0.021 | 0.011 |
| ssrQrZAG112 | 201 | 174 | 16 | 17 | 2 | 3 | 0.825 | 0.711 | 0.841 | 0.759 | 0.097 | 0.039 | 0.025 |
| bcqm42F | 201 | 172 | 11 | 9 | 2 | 0 | 0.758 | 0.596 | 0.745 | 0.623 | 0.031 | 0.043 | −0.001 |
| All | 201 | 174 | 323 | 357 | 32 | 66 | 0.761 | 0.729 | 0.801 | 0.764 | 0.049** | 0.046** | 0.019** |
QL: seven populations of Q. liaotungensis; QM: six populations of Q. mongolica; N: number of individual analyzed; An: number of alleles over all populations for each species; AP: number of private alleles; HO: observed heterozygosity; HE: expected heterozygosity;
**: p<0.01.
The application of the four AFLP primer combinations to 419 oak individuals resulted in 194 non-monomorphic markers (1%<frequency<99%), of which 133 (69%) were polymorphic at the 5% level in Q. liaotungensis and 124 (64%) in Q. mongolica. The unbiased estimate of genetic diversity (Hj) for each population varied from 0.204 to 0.234 (Table 1). The vast majority of the diversity is partitioned within-population in both species. The levels of diversity within each species, either at the population (Hw) or the whole-sample level (Ht), were strikingly similar (Table 4).
Table 4. Comparison of genetic diversity and differentiation between Quercus liaotungensis and Q. mongolica based on 194 AFLP markers.
| Populations | N | Ht | Hw | SE(Hw) | Hb | SE(Hb) | F ST | lower 99% F ST | upper 99% F ST |
| Between species | 2 | 0.234 | 0.212 | 0.005 | 0.022 | <0.001 | 0.093 | −0.001 | 0.003 |
| Q. liaotungensis | 7 | 0.227 | 0.217 | 0.004 | 0.010 | 0.001 | 0.044 | −0.005 | 0.003 |
| Q. mongolica | 6 | 0.216 | 0.213 | 0.002 | 0.003 | <0.001 | 0.014 | −0.006 | 0.002 |
N: number of populations; Ht: total diversity; Hw: average diversity within population; Hb: average diversity between populations; F ST: differentiation between populations.
The Weir and Cockerham's (1984) estimator of genetic differentiation between Q. liaotungensis and Q. mongolica was low but significant across all SSR loci (θ_inter = 0.049, p<0.01). At most SSR loci, the interspecific θ was low, with 8 of the 19 loci displaying values <0.02 and only 3 having values >0.10 (Table 3). The differentiation among populations within species was lower though also significant for both species [θ _intra = 0.046 (p<0.01) for Q. liaotungensis; θ _intra = 0.019 (p<0.01) for Q. mongolica]. When the analysis was restricted to ‘pure’ individuals (i.e. individuals assigned at >0.8 to one of the two species), the interspecific differentiation across all SSR loci was larger (θ_pure inter = 0.067, p<0.01). The differentiation among populations within species was also higher [θ _pure intra = 0.049 (p<0.01) for Q. liaotungensis; θ _pure intra = 0.020 (p<0.01) for Q. mongolica]. The genetic differentiation between Q. liaotungensis and Q. mongolica across the 194 AFLP markers was low but significant (F ST_inter = 0.093, p<0.01; Table 4). The differentiation among populations within species was lower but significant for both species [F ST_intra = 0.044 (p<0.01) for Q. liaotungensis; F ST_intra = 0.014 (p<0.01) for Q. mongolica; Table 4]. When the analysis was restricted to ‘pure’ individuals, the interspecific differentiation across the 194 AFLP markers became more pronounced (F ST_pure inter = 0.131, p<0.01) and was much higher than the differentiation among populations within species [F ST_pure intra = 0.042 (p<0.01) for Q. liaotungensis; F ST_pure intra = 0.010 (p<0.01) for Q. mongolica]. The higher differentiation within Q. liaotungensis than within Q. mongolica was mainly caused by the divergence of THl and of ZW and WA, the two allopatric Q. liaotungensis populations (see Table 5).
Table 5. Pairwise θ (Weir and Cockerham, 1984) based on 19 SSRs (above the diagonal) and pairwise Φ PT (a statistic analogous to F ST, Peakall et al., 1995) based on 194 AFLPs (below the diagonal).
| ZW | WA | CF2 | FS | SPl | THl | NAl | HH | MR | Dan | SPm | THm | NAm | Dmy | Dtt | |
| ZW | 0.036 ** | 0.050 ** | 0.052 ** | 0.036 ** | 0.078 ** | 0.055 ** | 0.112 ** | 0.120 ** | 0.096 ** | 0.074 ** | 0.097 ** | 0.095 ** | 0.029 ** | 0.066 ** | |
| WA | 0.022 ** | 0.061 ** | 0.041 ** | 0.032 ** | 0.086 ** | 0.059 ** | 0.124 ** | 0.120 ** | 0.109 ** | 0.077 ** | 0.103 ** | 0.100 ** | 0.047 ** | 0.074 ** | |
| CF2 | 0.111 ** | 0.097 ** | 0.039 ** | 0.025 ** | 0.070 ** | 0.029 ** | 0.067 ** | 0.077 ** | 0.059 ** | 0.046 ** | 0.065 ** | 0.056 ** | 0.045 ** | 0.034 ** | |
| FS | 0.089 ** | 0.070 ** | 0.050 ** | 0.007 * | 0.052 ** | 0.023 ** | 0.087 ** | 0.100 ** | 0.076 ** | 0.043 ** | 0.071 ** | 0.069 ** | 0.055 ** | 0.052 ** | |
| SPl | 0.094 ** | 0.070 ** | 0.049 ** | 0.016 ** | 0.047 ** | 0.011 NS | 0.063 ** | 0.074 ** | 0.052 ** | 0.027 ** | 0.055 ** | 0.044 ** | 0.034 * | 0.032 ** | |
| THl | 0.142 ** | 0.118 ** | 0.112 ** | 0.091 ** | 0.086 ** | 0.044 ** | 0.105 ** | 0.110 ** | 0.087 ** | 0.061 ** | 0.073 ** | 0.086 ** | 0.076 ** | 0.067 ** | |
| NAl | 0.108 ** | 0.097 ** | 0.038 ** | 0.038 ** | 0.026 ** | 0.092 ** | 0.043 ** | 0.051 ** | 0.037 ** | 0.017 ** | 0.032 ** | 0.027 ** | 0.044 ** | 0.026 ** | |
| HH | 0.233 ** | 0.243 ** | 0.151 ** | 0.165 ** | 0.137 ** | 0.188 ** | 0.106 ** | 0.035 ** | 0.021 ** | 0.017 ** | 0.021 ** | 0.014 ** | 0.079 ** | 0.031 ** | |
| MR | 0.260 ** | 0.270 ** | 0.166 ** | 0.192 ** | 0.165 ** | 0.218 ** | 0.119 ** | 0.043 ** | 0.032 ** | 0.036 ** | 0.021 ** | 0.022 ** | 0.091 ** | 0.037 ** | |
| Dan | 0.238 ** | 0.238 ** | 0.144 ** | 0.166 ** | 0.135 ** | 0.182 ** | 0.091 ** | 0.035 ** | 0.046 ** | 0.010 ** | 0.013 ** | 0.013 ** | 0.065 ** | 0.019 ** | |
| SPm | 0.168 ** | 0.169 ** | 0.081 ** | 0.092 ** | 0.075 ** | 0.133 ** | 0.042 ** | 0.036 ** | 0.043 ** | 0.017 ** | 0.010 ** | 0.009 ** | 0.044 ** | 0.013 ** | |
| THm | 0.234 ** | 0.236 ** | 0.147 ** | 0.161 ** | 0.132 ** | 0.170 ** | 0.094 ** | 0.029 ** | 0.044 ** | 0.018 ** | 0.012 ** | 0.009 NS | 0.065 ** | 0.017 ** | |
| NAm | 0.217 ** | 0.225 ** | 0.121 ** | 0.148 ** | 0.113 ** | 0.170 ** | 0.072 ** | 0.022 ** | 0.041 ** | 0.025 ** | 0.012 * | 0.012 * | 0.066 ** | 0.022 ** | |
| Dmy | 0.084 ** | 0.081 ** | 0.044 ** | 0.051 ** | 0.037 ** | 0.114 ** | 0.026 ** | 0.137 ** | 0.152 ** | 0.110 ** | 0.053 ** | 0.106 ** | 0.096 ** | 0.035 ** | |
| Dtt | 0.129 ** | 0.128 ** | 0.070 ** | 0.072 ** | 0.043 ** | 0.102 ** | 0.036 ** | 0.062 ** | 0.080 ** | 0.053 ** | 0.017 ** | 0.045 ** | 0.044 ** | 0.036 ** |
*: 0.01<p<0.05,
**: p<0.01,
NS: Non-Significant. Pairwise genetic differentiation within species is indicated in bold.
Pairwise population θ and Φ PT values with corresponding significant levels are presented in Table 5. Significant genetic differentiation was found among all population pairs in the AFLP data set and all but two (SPl and NAl, THm and NAm) in the SSR data set. For both AFLP and SSR data sets, genetic differentiation was low between all Q. mongolica population pairs; whereas among Q. liaotungensis populations, the THl, ZW and WA population were significantly differentiated from other Q. liaotungensis populations.
Genetic distance-based analysis
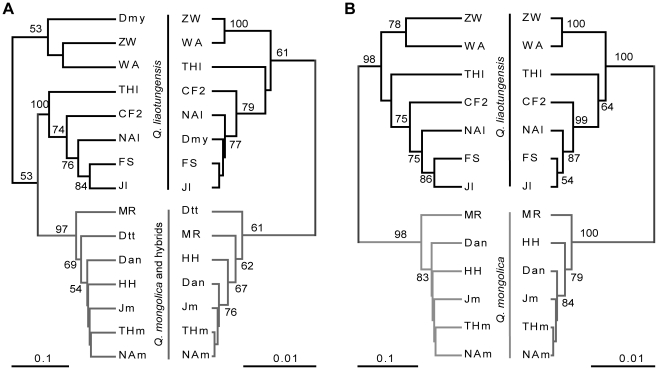
Overall, the two UPGMA trees based on SSR and AFLP data sets were highly congruent (Fig. 2A). All typical Q. mongolica populations and the Dtt population shared a common node with a bootstrap value over 60% in both trees; all Q. liaotungensis populations shared the other common node in the AFLP tree. In the SSR tree, Q. liaotungensis populations were separated in two groups: the sympatric group vs the allopatric group; the sympatric Q. liaotungensis group, which is comprised of populations from northeast China and Chifeng, was more closely related to Q. mongolica populations than to the allopatric Q. liaotungensis group comprised of ZW, WA and Dmy populations. When the Dtt and Dmy populations were excluded from the analysis, the SSR and AFLP trees had exactly the same topology, with the two species separated at very strong bootstrap support (98% for SSR and 100% for AFLP; Fig. 2B).
Figure 2. Dendrograms of oak populations based on genetic distance.
(A) With all populations. (B) After excluding the two populations from the Dongling Mountain region. In both panels, left dendrograms are based on genetic distance of 19 SSR loci and right dendrograms are based on genetic distance at 194 AFLP bands. The dendrograms were computed using a UPGMA approach implemented in PHYLIP. Numbers are bootstrap support values.
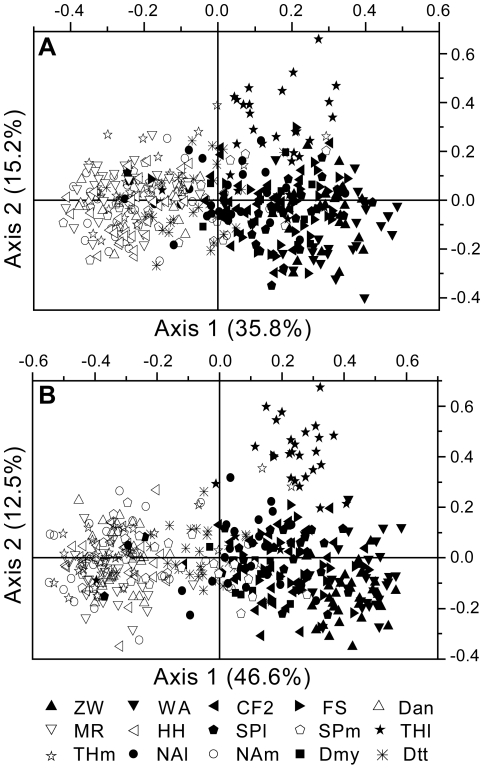
The results of the PCo for the pairwise individual genetic distances are presented in Fig. 3. The two first PCo-axes of the SSR (Fig. 3A) and the AFLP (Fig. 3B) plot accounted for about 51% and 59% of the variation, respectively. Both SSR and AFLP data grouped individuals from Q. liaotungensis and Q. mongolica populations separately, but AFLP distinguished the individuals of the two groups slightly better. Most individuals from the Dmy population had a close relationship with Q. liaotungensis, and most individuals from Dtt had intermediate positions. The AFLP plot generally showed a higher resolution, and species groups appeared more distinct than in the SSR plot.
Figure 3. PCo plots of pairwise individual genetic distances Φ at SSR (A) and AFLP (B) markers.
Filled symbols: Quercus liaotungensis; open symbols: Q. mongolica; radial symbols: intermediate individuals from population Dtt.
Bayesian cluster results
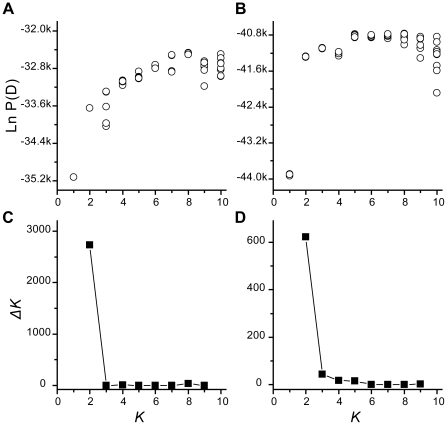
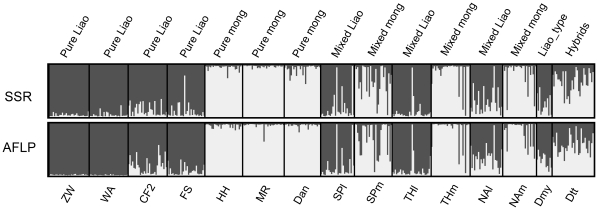
The STRUCTURE output for both SSR and AFLP data sets suggested the existence of two clusters. The AFLP and SSR data sets gave similar results, with K = 2 being considerably more likely than K = 1, while K≥3 being only slightly more likely (Fig. 4A and B). ΔK distribution further supported the choice of K = 2, showing distinct modal distribution at K = 2 (Fig. 4C and D). The cluster patterns for the two data sets were also very similar; moreover, the two genetically distinct clusters corresponded well to our morphological assignment of populations to Q. liaotungensis and Q. mongolica (Fig. 5 and Fig. 6).
Figure 4. Indication of the most likely number of clusters in the STRUCTURE analysis.
Both the estimated logarithmic probability (panels A and B) and magnitude of ΔK (panels C and D) as a function of K suggested the existence of two clusters. Results are from 10 replicates for each of 1≤K≤10 with both SSR (panels A and C) and AFLP markers (panels B and D).
Figure 5. STRUCTURE results for two clusters with no prior population knowledge.
Results are based on 19 SSR loci (above) or 194 AFLP markers (below). Each individual is represented by a thin vertical line. Black lines separate individuals of different populations. Populations are labelled below the figure with their regional affiliations. Previous population morphological information is provided above the figure. (Pure Liao: pure Quercus liaotungensis; Pure Mong: pure Q. mongolica; Mixed Liao and Mixed Mong: Q. liaotungensis and Q. mongolica from mixed forests, respectively.
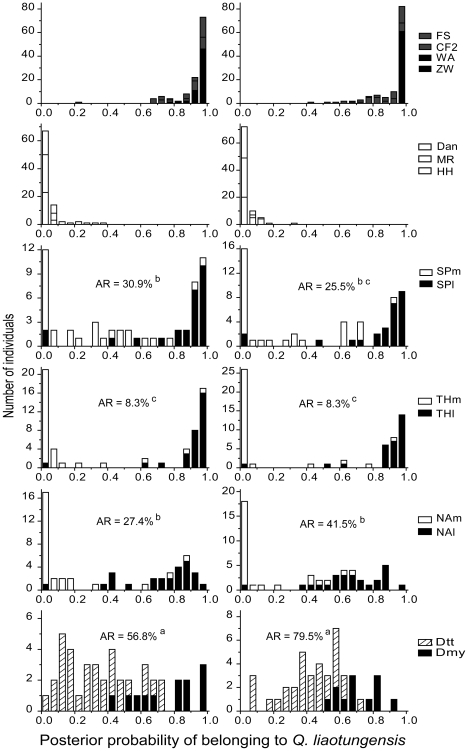
Figure 6. Distribution of ancestry estimates in each study site.
The ancestry is based on posterior probability of belonging to Quercus liaotungensis at K = 2 in the STRUCTURE analysis using 19 SSR loci (left) or 194 AFLP markers (right). AR = admixture rate according to the STRUCTURE assignment analysis, with the superscript a, b and c indicating statistically significant differences.
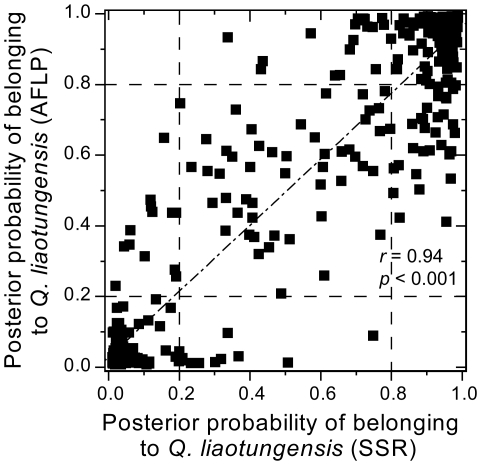
More specifically, SSRs results indicated that 42% of all individuals had a posterior probability >0.8 of belonging to Q. liaotungensis, and 38% to Q. mongolica; the corresponding values based on AFLPs are 40% and 38% (Table 6). The assignment results based on the two types of markers were highly congruent. The mean posterior probability of belonging to Q. liaotungensis is 0.52 for SSRs and 0.51 for AFLPs. The Pearson correlation coefficient between posterior probabilities for SSRs and for AFLPs was high and significant (r = 0.94, p<0.001, Fig. 7). The correlation remained significant but dropped sharply after excluding genotypes assigned to pure species with both approaches (r = 0.52, p<0.001). With both SSR and AFLP datasets, the majority of individuals from pure sites were assigned to their respective clusters, except in Chifeng, where the percentage of hybrids reached 30% (according to SSR) and 37% (according to AFLP). Most individuals from the three mixed sites where both species are present were also successfully assigned to their respective clusters, with only a few individuals being assigned to the alternative cluster (Siping: 7.3% and 5.5%; Tonghua: 5.0% and 3.3%; Ning'an: 3.9% and 2.0%, for SSR and AFLP respectively). The percentage of hybrids in the three mixed sites varied among different sites (Table 6). Tonghua had the fewest hybrids (8.3% and 8.3% for SSR and AFLP, respectively) as compared to Siping (SSR: 30.9%, χ 2 = 9.5, p = 0.002; AFLP: 25.5%, χ 2 = 6.1, p = 0.014) and Ning'an (SSR: 27.4%, χ 2 = 7.1, p = 0.008; AFLP: 45.1%, χ 2 = 17.8, p<0.001), whereas the percentage of hybrids in Dongling Mountain (56.8% for SSR and 79.5% for AFLP) was significantly higher than at any of the three mixed sites (SSR: χ 2>6.7, p<0.01 for all comparisons; AFLP: χ 2>11.8, p≤0.001 for all comparisons). Most hybrids there were from Dtt population. In contrast, a total of 7 (according to SSR) and 4 (according to AFLP) of the 12 Dmy individuals had a posterior probability >0.8 of belonging to the Q. liaotungensis cluster.
Table 6. Number (and percentage) of pure species and hybrid oaks as assigned by the STRUCTURE software in the different studied sites.
| SSR | AFLP | |||||||
| Study site | Population ID | N | QL | QM | Hybrids | QL | QM | Hybrids |
| Ziwu | ZW | 31 | 31 (100.0%) | - | - | 31 (100.0%) | - | - |
| Wuan | WA | 30 | 29 (96.7%) | - | 1(3.3%) | 30 (100.0%) | - | - |
| Chifeng | CF2 | 30 | 21 (70.0%) | - | 9 (30.0%) | 19 (63.3%) | - | 11 (36.7%) |
| Fushun | FS | 29 | 24 (82.8%) | - | 5(17.2%) | 25 (86.2%) | - | 4 (13.8%) |
| Heihe | HH | 29 | - | 28 (96.6%) | 1 (3.4%) | - | 29 (100.0%) | - |
| Mao'ershan | MR | 32 | - | 32 (100.0%) | - | - | 31 (96.8%) | 1 (3.2%) |
| Dandong | Dan | 28 | - | 24 (85.7%) | 4 (14.3%) | - | 28 (100.0%) | - |
| Siping | SPl+SPm | 55 | 22 (40.0%) | 16 (29.1%) | 17 (30.9%) b | 22 (4.0%) | 19 (34.5%) | 14 (25.5%) bc |
| Tonghua | THl+THm | 60 | 29 (48.3%) | 26 (43.3%) | 5 (8.3%) c | 28 (46.7%) | 27 (45.0%) | 5 (8.3%) c |
| Ning'an | NAl+NAm | 51 | 14 (27.4%) | 23 (45.1%) | 14 (27.4%) b | 8 (15.7%) | 20 (39.2%) | 23 (45.1%) b |
| Dongling Mountain | Dtt+Dmy | 44 | 7 (13.7%) | 12 (25.0%) | 25 (56.8%) a | 4 (9.1%) | 5 (11.4%) | 35 (79.5%) a |
| Total | 419 | 177 (42.2%) | 161 (38.4%) | 81(19.3%) | 167 (39.9%) | 159 (37.9%) | 93 (22.2%) | |
N: number of sampled oaks; QL: Quercus liaotungensis; QM: Q. mongolica;
indicated statistic significant (p<0.01) differences for the hybridization rate comparison between different sites.
Figure 7. Correspondence between the SSRs and AFLPs assignment results in STRUCTURE analysis.
Each black square represents a single individual. Dashed lines denote the threshold of the successful assignment to either cluster (posterior probability <0.2: Quercus mongolica; posterior probability >0.8: Q. liaotungensis; 0.2< posterior probability <0.8: hybrids).
Loci under disruptive selection
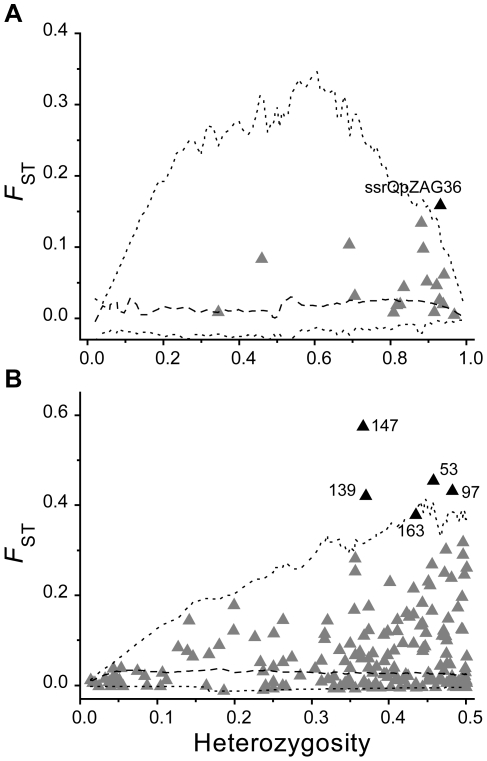
Simulation analysis revealed that one SSR locus (ssrQpZAG36) and five AFLPs are significant outliers on a plot of interspecific F ST against heterozygosity (Fig. 8). Therefore they are likely to be either under disruptive selection or linked to a locus under selection [68], [69]. Most of these outlier loci were also identified as outliers in the separate pairwise interspecific comparisons within the three mixed populations (Fig. S1). The AFLP outlier locus with the highest F ST value (0.57) had a large difference in allelic frequency between Q. liaotungensis (95%) and Q. mongolica populations (14%). However, the interspecific F ST of most SSR and AFLP loci exhibited very low values, suggesting substantial genetic exchanges at loci unlikely to be under disruptive selection (Fig. 8).
Figure 8. Distribution of per-locus F ST values (differentiation between Quercus liaotungensis and Q. mongolica) against heterozygosity.
Each triangle represents a SSR (panel A) or AFLP (panel B) marker. The black triangles above the upper line are classified as outliers potentially under divergent selection. Dotted lines denote 99th and 1th quantiles estimated from simulation, and dashed lines denote the medians.
Discussion
Species status
The taxonomic status of closely related oak species has long been an issue of controversy because of continuous variation in morphological, ecological, and genetic traits due to interspecific hybridization and/or shared ancient polymorphisms [11], [12], [18], [31], [70]. This is the case for Q. liaotungensis and Q. mongolica, dominant members of warm temperate forests in northern China and the surrounding regions. A previous molecular study of the two oaks inferred strong gene flow between species and did not resolve the species status issue [43]. The present study revealed a clear differentiation of the Chinese oak gene pool into two entities corresponding to Q. liaotungensis and Q. mongolica. Acorn cupule and trunk bark characteristics were helpful to discriminate the species; in contrast, the number of lateral veins did not appear helpful for taxonomic purposes.
Although clustering based on population genetic distance relies on a priori classification of individuals or populations, it can provide useful insights. In our study, high bootstrap values and consistent topology between SSR and AFLP trees (Fig. 2) indicated that the differentiation based on the acorn cupule and bark morphology was associated with a clear and stable genetic differentiation at molecular markers. In particular, at the three mixed sites, populations were clustered into two groups according to species rather than to geographic origin. Two analyses that do not rely on a priori classifications (Bayesian clustering analysis using the software STRUCTURE and principal coordinate analysis based on individual pairwise distances) were then carried out. Both revealed a separation of the individuals into two groups that correspond well with the taxonomic species. Comparable results in the European white oak complexes have been obtained in both SSR [24] and AFLP surveys [71]. These analyses established the species status of sessile (Q. petraea) and pedunculate oaks (Q. robur). However, the bootstrap support for each group in our SSR study was not 100%, as in Muir et al. [24], for the following reasons. First, we did not get detailed morphological data for each individual. The identification based on one or two morphological characteristics might have led to false classification, thus ultimately reducing the support values. Second, genotypic and phenotypic mismatch might exist for a few individual trees, especially for hybrids [25], which could reduce the genetic distance between species.
The most likely clustering in STRUCTURE (with K = 2) also revealed that the taxonomical signal was much stronger than the geographic signal. The Bayesian clustering approach that was developed to identify genetic structure in the mixed populations has often been applied to test for the presence of a taxon without assigning individuals to a predefined group [25], [27], [29], [72]. This approach allows evaluation of the statistical significance of clusters. Moreover, the origin of individuals can be inferred by calculating the probability that individual multilocus genotypes belong to different genetic clusters, or alternatively, are hybrid in origin [62], [63]. Without detailed individual morphological data, our multilocus analysis assigned most individuals from both pure and mixed sites into groups recognizable as the two species, Q. liaotungensis and Q. mongolica, with only a few mismatched individuals and hybrids. When each of three pairs of mixed populations was analyzed separately, AFLP could also categorize individuals into the respective species (Fig. 10 in [73]). These results suggest that Q. liaotungensis and Q. mongolica occur as distinct clusters of genotypes even where they co-occur locally. One population that we have to mention is Dmy. Its nearest population, Dtt, consisted mostly of hybrid individuals; however, STRUCTURE analysis suggested that most individuals in the Dmy population were Q. liaotungensis, consistent with their morphology (smooth-cupules acorns).
The existence of outlier loci indicated that, although Q. liaotungensis and Q. mongolica form hybrids, they remain generally distinct at some genomic regions when in sympatry. Q. robur and Q. petraea have also been shown to represent separate species using both morphological data [74] and molecular markers [24], [71]; however, until now, no single marker has been identified that can differentiate between them at the individual tree level. With 176 polymorphic AFLP markers, Coart et al. [71] classify Q. petraea and Q. robur populations into two main groups with very high bootstrap support (100%), in agreement with their taxonomic status. However, in their study, only one marker displays a difference in frequency up to 71%. With comparable polymorphic markers, we identified one fragment with a difference in frequency between Q. liaotungensis and Q. mongolica of 81% (95% vs 14%); this difference would be even higher if the individuals suggested by STRUCTURE as having been misidentified had been corrected, suggesting that the two species studied are at least as valid as the well-investigated Q. robur and Q. petraea.
Comparison between AFLP markers and SSR loci
Although AFLP and SSR markers have been widely used in describing interspecific differentiation in oaks, direct comparisons of genotype assignment using AFLP and SSR are still lacking. In our study, the interspecific differentiation estimated by F ST between Q. liaotungensis and Q. mongolica was slightly higher for AFLP (F ST_inter = 0.093) than for SSR (θ_inter = 0.049), in line with findings for Q. robur and Q. petraea by Mariette et al. [75] (but see [76]). Our assignment of all individuals into groups with the STRUCTURE program gave very similar results with the AFLP and the SSR data sets, as did the PCo plot based on individual distances and the topology identified in the UPGMA trees. However, the AFLP markers were more powerful than the SSR loci in discriminating the origin of individuals from different species. Indeed, the AFLP multilocus Bayesian cluster analysis showed higher assignment success than SSR loci: (1) For individuals from allopatric pure sites, the posterior probability of belonging to their respective species measured with AFLP markers was generally higher than the probability measured with SSR loci; (2) when we did Bayesian cluster analysis separately for each of the three pairs of mixed populations (THl vs THm, SPl vs SPm, and NAl vs NAm), AFLP markers could successfully assign individuals into distinct species for all three pairs of mixed populations, whereas SSR loci succeeded only in Tonghua location (THl vs THm). Furthermore, a larger proportion of variation was explained by the two first axes of the PCo with the AFLPs than with the SSRs, and the AFLP plots gave higher resolution and distinguished the individuals of different species slightly better (Fig. 3). Better genome coverage due to the larger number of loci is likely responsible for the higher resolution with the AFLP dataset compared to the SSR dataset.
Although the AFLP dataset provided higher resolution than the SSR dataset, both types of markers provided comparable and unbiased results. The overall mean species assignments were very close with each type of markers and the correlation between individual assignment values for the two types of data was high, although it was much lower for admixed individuals. These results indicate that similar conclusions regarding species delimitation can be arrived at using independent marker sets but also show that precise estimates of individual introgression rates require sizeable genome-wide datasets.
Hybridization and hybrid zones
Our investigation of Q. liaotungensis and Q. mongolica suggested that hybridization occurs between the two species in their sympatric range. First, the approximate Bayes factors (ratio of the estimated marginal likelihood of the admixture model to that of the non-admixture model) was greater than 100:1 for both the SSR and AFLP data sets, which could be considered as ‘decisive’ [77]. Second, the analyses of the AFLP and SSR datasets independently pointed to the existence of a subset of admixed individuals (with a posterior probability <0.8 of belonging to either cluster). Third, the interspecific F ST values were remarkably variable across markers, with many loci displaying low F ST values (<0.02). Fourth, past hybridization between the two species is suggested by the extensive sharing of cpDNA in North China and Northeast China [73].
Bayesian assignment using multilocus genotype suggests that the proportion of hybrids varies in different geographic contact sites of Q. liaotungensis and Q. mongolica (Table 6 and Fig. 6). The hybrid zones from Northeast China consisted largely of genotypes resembling the parental forms, and thus constitute bimodal hybrid zones, suggestive of well-developed (although incomplete) reproductive isolation [3], [78]. In contrast, the Dtt hybrid zone in North China was composed largely of recombinant individuals, corresponding to a unimodal hybrid zone or hybrid swarm, suggestive of incomplete reproductive isolation.
The variation in hybridization patterns between these two species in different parts of the range is of great interest. Such patterns have been reported in several plant species and many possible causes for this variation have been discussed. First, the degree of actual intermixing might not be uniform in different contact zones, resulting in differential hybridization and introgression [e.g. 27]. Second, differences in relative abundance of each species locally could affect rates and direction of introgression [34]. Third, the presence of open, intermediate, marginal or disturbed habitats could promote hybridization, as reported previously in oaks [12], [79]–[84]. In fact, during range expansions, both asymmetric population size and the presence of open habitats could result in a short-term increase of the proportion of hybrids [85], [86]. Another possibility that has been much less explored is reproductive character displacement, also called reinforcement of reproductive barriers, a pattern of greater divergence of a trait between closely related taxa in areas of sympatry than in areas of recent contact following allopatric divergence [87].
STRUCTURE assignment results suggest that hybrid frequency in Northeast China varies among the three mixed sites, ranging from 8% to 31–45%, depending on marker type. This proportion of hybrids is comparable to that of the European white oak complex (11–30%, see [34]). Q. liaotungensis and Q. mongolica shared most nuclear alleles, with only a few low-frequency private alleles being identified in our studied populations. The introgression direction differed depending on the location of hybrid zones. The hybrid zone in Siping suggests bidirectional introgression, whereas in Ning'an hybrids had a genetic composition closer to Q. liaotungensis (Fig. 6), indicating directional introgression. The directional introgression in Ning'an mixed forest might be due to the relatively low abundance of Q. liaotungensis in this region (see [34]). According to the Higher Plants of China ([38], see Fig. 1), and as confirmed by our own field observation, Ning'an represents the northeastern edge of the present distribution of Q. liaotungensis. The heterogeneous level of admixture and introgression direction in Northeast China might also relate to the precise location of oaks' glacial refugium and subsequent recolonization processes. Further studies are needed to investigate the role of hybridization and introgression during recolonization processes of Chinese oaks since the last glacial maximum [88].
Alternatively, the differences between contact zones might be explained by geographically variable natural selection against hybrids. The infrequency of hybrids in Northeast China might result from their having lower fitness than the parental species. In fact, in Tonghua, where rates of introgression were very low, more loci under disruptive selection were identified in comparison with the two other mixed sites (Fig. S1), even after accounting for misclassified individuals (analysis not shown). The existence of loci potentially under disruptive selection suggests directional selection on a subset of loci between Q. liaotungensis and Q. mongolica genomes in mixed sites of Northeast China. However, hybrids might exhibit an increased fitness in an intermediate or altered (mostly anthropogenic) environment uncharacteristic of either parental species, such as in Dtt, a touristic place located in the suburb of Beijing. Further artificial pollination and transplant experiments are needed to test the mechanism by which reproductive isolation and habitat selection affects species delimitation of Q. liaotungensis and Q. mongolica.
Finally, to evaluate the hypothesis of reproductive character displacement, it will be necessary to reconstruct the history of the two species in their different contact zones. For instance, long time co-occurrence of Q. liaotungensis and Q. mongolica in Northeast China could have reinforced their reproductive isolation barrier, resulting in stronger barriers than in the new contact zone between Q. liaotungensis and Q. mongolica in northern China.
Conclusions
Our molecular analysis led to the conclusion that Q. liaotungensis and Q. mongolica have maintained distinct gene pools, even in mixed stands, and should be considered as discrete taxonomic units, despite the existence of interspecific hybridization. Results based on SSRs and AFLPs were highly congruent, indicating that the conclusions reached regarding species delimitation and hybridization are of general value. Interestingly, hybridization rates were not uniform in different contact zones. More work is needed to tease apart the mechanisms underlying this heterogeneity of interspecific genetic differentiation across the species' ranges.
Supporting Information
Distribution of per-locus interspecific FST values against heterozygosity in each study site.
(TIF)
Acknowledgments
We would like to thank Quan-Guo Zhang, Meng Wang and many other people for helping with the sampling, Wei-Ning Bai, Yuan-Ye Zhang and Chuan Ni for technical assistance, Yan Hou and Peng-Cheng Yan for helping with STRUCTURE analysis, Shou-Hsien Li and Jian-Lin Han for guidance in the laboratory, Dorothée Ehrich for supplying the unpublished Structure2[1].2-sum software scripts, and the Scientific Computing Center of BNU for supplying high-performance computing for our STRUCTURE analysis. RJP is indebted to the organizers of the Conference New Frontiers in Plant Systematics and Evolution (Beijing, 2010, July 7-9) and to Prof. J. Liu from Lanzhou University for inviting him to China.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was financially supported by the National Natural Science Foundation of China (30430160) and Beijing Normal University. RJP's research is supported by the European Union-sponsored Network of Excellence EVOLTREE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Harrison RG. Hybrids and hybrid zones: historical perspective. In: Harrison RG, editor. Hybrid zones and the evolutionary process. Oxford, UK: Oxford University Press; 1993. pp. 3–12. [Google Scholar]
- 2.Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- 3.Jiggins CD, Mallet J. Biomodal hybrid zones and speciation. Trends Ecol Evol. 2000;15:250–255. doi: 10.1016/s0169-5347(00)01873-5. [DOI] [PubMed] [Google Scholar]
- 4.Coyne JA, Orr HA. Sunderland, MA: Sinauer Associates, Inc; 2004. Speciation.545 [Google Scholar]
- 5.Rieseberg LH, Wood TE, Baack EJ. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aldridge G, Campbell DR. Genetic and morphological patterns show variation in frequency of hybrids between Ipomopsis (Polemoniaceae) zones of sympatry. Heredity. 2009;102:257–265. doi: 10.1038/hdy.2008.112. [DOI] [PubMed] [Google Scholar]
- 7.Harrison RG. Hybrid zones: Windows on evolutionary process. Oxford Surveys in Evolutionary Biology. 1990;7:69–128. [Google Scholar]
- 8.Burger WC. The species concept in Quercus. Taxon. 1975;24:45–50. [Google Scholar]
- 9.Van Valen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239. [Google Scholar]
- 10.Grant V. New York: Columbia University Press; 1981. Plant speciation.563 [Google Scholar]
- 11.Jensen RJ, Hokanson SC, Isebrands JG, Hancock JF. Morphometric variation in oaks of the Apostle Islands in Wisconsin: evidence of hybridization between Quercus rubra and Q. ellipsoidalis (Fagaceae). Am J Bot. 1993;80:1358–1366. [Google Scholar]
- 12.Rushton BS. Natural hybridization within the genus Quercus L. Ann Sci For. 1993;50:73s–90s. [Google Scholar]
- 13.Benson L, Phillips EA, Wilder PA. Evolutionary sorting of characters in a hybrid swarm. I: Direction of slope. Am J Bot. 1967;54:1017–1026. [Google Scholar]
- 14.Whittemore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumolin-Lapègue S, Kremer A, Petit RJ. Are chloroplast and mitochondrial DNA variation species independent in oaks? Evolution. 1999;53:1406–1413. doi: 10.1111/j.1558-5646.1999.tb05405.x. [DOI] [PubMed] [Google Scholar]
- 16.Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, et al. Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecol Manag. 2002;156:5–26. [Google Scholar]
- 17.Bodénès C, Joandet S, Laigret F, Kremer A. Detection of genomic regions differentiating two closely related oak species Quercus petraea (Matt.) Liebl. and Quercus robur L. Heredity. 1997;78:433–444. [Google Scholar]
- 18.Bruschi P, Vendramin GG, Bussotti F, Grossoni P. Morphological and molecular differentiation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. (Fagaceae) in Northern and Central Italy. Ann Bot. 2000;85:325–333. [Google Scholar]
- 19.Muir G, Fleming CC, Schlötterer C. Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Mol Biol Evol. 2001;18:112–119. doi: 10.1093/oxfordjournals.molbev.a003785. [DOI] [PubMed] [Google Scholar]
- 20.González-Rodríguez A, Arias DM, Valencia S, Oyama K. Morphological and RAPD analysis of hybridization between Quercus affinis and Q. laurina (Fagaceae), two Mexican red oaks. Am J Bot. 2004;91:401–409. doi: 10.3732/ajb.91.3.401. [DOI] [PubMed] [Google Scholar]
- 21.Howard DJ, Preszler RW, Williams J, Fenchel S, Boecklen WJ. How discrete are oak species? Insights from a hybrid zone between Quercus grisea and Quercus gambelii. Evolution. 1997;51:747–755. doi: 10.1111/j.1558-5646.1997.tb03658.x. [DOI] [PubMed] [Google Scholar]
- 22.Zanetto A, Roussel G, Kremer A. Geographic variation of inter-specific differentiation between Quercus robur L. and Quercus petraea (Matt.) Liebl. Forest Genet. 1994;1:111–123. [Google Scholar]
- 23.Bodénès C, Labbé T, Pradère S, Kremer A. General vs. local differentiation between two closely related white oak species. Mol Ecol. 1997;6:713–724. [Google Scholar]
- 24.Muir G, Fleming CC, Schlötterer C. Species status of hybridizing oaks. Nature. 2000;405:1016–1016. doi: 10.1038/35016640. [DOI] [PubMed] [Google Scholar]
- 25.Craft KJ, Ashley MV, Koenig WD. Limited hybridization between Quercus lobata and Quercus douglasii (Fagaceae) in a mixed stand in central coastal California. Am J Bot. 2002;89:1792–1798. doi: 10.3732/ajb.89.11.1792. [DOI] [PubMed] [Google Scholar]
- 26.Ishida TA, Hattori K, Sato H, Kimura MT. Differentiation and hybridization between Quercus crispula and Q. dentata (Fagaceae): insights from morphological traits, amplified fragment length polymorphism markers, and leafminer composition. Am J Bot. 2003;90:769–776. doi: 10.3732/ajb.90.5.769. [DOI] [PubMed] [Google Scholar]
- 27.Valbuena-Carabaña M, González-Martínez SC, Sork VL, Collada C, Soto A, et al. Gene flow and hybridisation in a mixed oak forest (Quercus pyrenaica Willd. and Quercus petraea (Matts.) Liebl.) in central Spain. Heredity. 2005;95:457–465. doi: 10.1038/sj.hdy.6800752. [DOI] [PubMed] [Google Scholar]
- 28.Dodd RS, Afzal-Rafii Z. Selection and dispersal in a multispecies oak hybrid zone. Evolution. 2004;58:261–269. [PubMed] [Google Scholar]
- 29.Gugerli F, Brodbeck S, Holderegger R. Utility of multilocus genotypes for taxon assignment in stands of closely related European white oaks from Switzerland. Ann Bot. 2008;102:855–863. doi: 10.1093/aob/mcn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir G, Schlötterer C. Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.). Mol Ecol. 2005;14:549–561. doi: 10.1111/j.1365-294X.2004.02418.x. [DOI] [PubMed] [Google Scholar]
- 31.Gardiner AS. Pedunculate and sessile oak (Quercus robur L. and Quercus petraea (Mattuschka) Liebl.): A review of the hybrid controversy. Forestry. 1970;43:151–160. [Google Scholar]
- 32.Curtu AL, Gailing O, Finkeldey R. Patterns of contemporary hybridization inferred from paternity analysis in a four-oak-species forest. BMC Evol Biol. 2009;9:284. doi: 10.1186/1471-2148-9-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen J, Larsen A, Nielsen LR, Cottrell J. Hybridization between Quercus robur and Q. petraea in a mixed oak stand in Denmark. Ann For Sci. 2009;66:706–717. [Google Scholar]
- 34.Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, et al. Species relative abundance and direction of introgression in oaks. Mol Ecol. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- 35.Salvini D, Bruschi P, Fineschi S, Grossoni P, Kjaer ED, et al. Natural hybridisation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. within an Italian stand as revealed by microsatellite fingerprinting. Plant Biology. 2009;11:758–765. doi: 10.1111/j.1438-8677.2008.00158.x. [DOI] [PubMed] [Google Scholar]
- 36.Peñaloza-Ramírez JM, González-Rodríguez A, Mendoza-Cuenca L, Caron H, Kremer A, et al. Interspecific gene flow in a multispecies oak hybrid zone in the Sierra Tarahumara of Mexico. Ann Bot. 2010;105:389–399. doi: 10.1093/aob/mcp301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J, et al. Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q. petraea (Matt.) Liebl. Mol Ecol. 1999;8:831–841. [Google Scholar]
- 38.Zhang YT. Fagaceae. In: Fu LK, Chen TQ, Lang KY, Hong T, Lin Q, editors. Higher Plants of China. Qingdao: Qingdao Publishing House; 2000. pp. 177–254. [Google Scholar]
- 39.Wang HX, Hu ZA, Zhong M, Yun R, Qian YQ. Genetic Diversity of Liaodong Oak Populations. In: Hu ZA, Zhang YP, editors. Genetic Diversity of Animals and Plants in China. Hangzhou: Zhejiang Science and Technology Press; 1997. pp. 155–159. [Google Scholar]
- 40.Chen HY, Huang CJ. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; 1998. Quercus. pp. 213–332. [Google Scholar]
- 41.Huang CJ, Zhang YT, Bartholomew B Flora of China Editorial Committee. Flora of China. Beijing: Science Press and St. Louis: Missouri Botanical Garden Press; 1999. Fagaceae. pp. 314–402. [Google Scholar]
- 42.Beljaev EA. A new oak species for the flora of Russia Quercus wutaishanica (Fagaceae) from Primorskiy territory. Bot Zh. 2004;89:1665–1672. [Google Scholar]
- 43.Yun R, Wang HX, Hu ZA, Zhong M, Wei W, et al. Genetic differentiation of Quercus mongolica and Q. liaotungensis based on morphological observation, isozyme and DNA analysis. Acta Bot Sin. 1998;40:1040–1046. [Google Scholar]
- 44.Dow BD, Ashley MV, Howe HF. Characterization of highly variable (GA/CT)n microsatellites in the bur oak, Quercus macrocarpa. Theor Appl Genet. 1995;91:137–141. doi: 10.1007/BF00220870. [DOI] [PubMed] [Google Scholar]
- 45.Dow BD, Ashley MV. Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol. 1996;5:615–627. [Google Scholar]
- 46.Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, et al. Identification and characterization of (GA/CT)n - microsatellite loci from Quercus petraea. Plant Mol Biol. 1997;33:1093–1096. doi: 10.1023/a:1005736722794. [DOI] [PubMed] [Google Scholar]
- 47.Kampfer S, Lexer C, Glössl J, Steinkellner H. Characterization of (GA)n microsatellite loci from Quercus robur. Hereditas. 1998;129:183–186. [Google Scholar]
- 48.Aldrich PR, Michler CH, Sun WL, Romero-Severson J. Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes. 2002;2:472–474. [Google Scholar]
- 49.Mishima K, Watanabe A, Isoda K, Ubukata M, Takata K. Isolation and characterization of microsatellite loci from Quercus mongolica var. crispula. Mol Ecol Notes. 2006;6:695–697. [Google Scholar]
- 50.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, et al. AFLP: a new technique for DNA fingerpringting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goudet J. FSTAT, a program to estimate and test gene diverisities and fixation indices. 2001. Version 2.9.3 ed.
- 52.Rousset F. GENEPOP ' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 53.Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol. 2002;11:139–151. doi: 10.1046/j.0962-1083.2001.01415.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhivotovsky LA. Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol. 1999;8:907–913. doi: 10.1046/j.1365-294x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 55.Lynch M, Milligan BG. Analysis of population genetic structure with RAPD markers. Mol Ecol. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 56.Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 57.Peakall R, Smouse PE, Huff DR. Evolutionary implications of allozyme and RADP variation in diploid populations of dioecious buffalograss (Buchloe dactyloides). Mol Ecol. 1995;4:135–147. [Google Scholar]
- 58.Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. [Google Scholar]
- 60.Nei M. New York: Columbia University Press; 1987. Molecular evolutionary genetics.512 [Google Scholar]
- 61.Felsenstein J. Seattle, Washington: Department of Genome Sciences, University of Washington; 2004. PHYLIP (Phylogeny Inference Package). version 3.6. ed. [Google Scholar]
- 62.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 66.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 67.Vähä JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- 68.Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B. 1996;263:1619–1626. [Google Scholar]
- 69.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 70.Tomlinson PT, Jensen RJ, Hancock JF. Do whole tree silvic characters indicate hybridization in red oak (Quercus section Lobatae)? Am Midl Nat. 2000;143:154–168. [Google Scholar]
- 71.Coart E, Lamote V, De Loose M, Van Bockstaele E, Lootens P, et al. AFLP markers demonstrate local genetic differentiation between two indigenous oak species [Quercus robur L. and Quercus petraea (Matt.) Liebl.] in Flemish populations. Theor Appl Genet. 2002;105:431–439. doi: 10.1007/s00122-002-0920-6. [DOI] [PubMed] [Google Scholar]
- 72.Duminil J, Caron H, Scotti I, Cazal S, Petit RJ. Blind population genetics survey of tropical rainforest trees. Mol Ecol. 2006;15:3505–3513. doi: 10.1111/j.1365-294X.2006.03040.x. [DOI] [PubMed] [Google Scholar]
- 73.Zeng YF. Beijing: Beijing Normal University; 2008. The species status, phylogeography and population genetic structure of Quercus liaotungensis and Quercus mongolica.127 [Google Scholar]
- 74.Kremer A, Dupouey JL, Deans JD, Cottrell J, Csaikl U, et al. Leaf morphological differentiation between Quercus robur and Quercus petraea is stable across western European mixed oak stands. Ann For Sci. 2002;59:777–787. [Google Scholar]
- 75.Mariette S, Cottrell J, Csaikl UM, Goikoechea P, König A, et al. Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (Matt.) Liebl. and Q. robur L. stands. Silvae Genet. 2002;51:72–79. [Google Scholar]
- 76.Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, et al. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q. petraea (Matt.) Liebl.]. Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodman SN. Toward evidence-based medical statistics. 2: The Bayes factor. Ann Intern Med. 1999;130:1005–1013. doi: 10.7326/0003-4819-130-12-199906150-00019. [DOI] [PubMed] [Google Scholar]
- 78.Harrison RG, Bogdanowicz SM. Patterns of variation and linkage disequilibium in a field cricket hybrid zone. Evolution. 1997;51:493–505. doi: 10.1111/j.1558-5646.1997.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 79.Williams JH, Boecklen WJ, Howard DJ. Reproductive processes in two oak (Quercus) contact zones with different levels of hybridization. Heredity. 2001;87:680–690. doi: 10.1046/j.1365-2540.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- 80.Muller CH. Ecological control of hybridization in Quercus: a factor in the mechanism of evolution. Evolution. 1952;6:147–161. [Google Scholar]
- 81.Alberto F, Niort J, Derory J, Lepais O, Vitalis R, et al. Population differentiation of sessile oak at the altitudinal front of migration in the French Pyrenees. Mol Ecol. 2010;19:2626–2639. doi: 10.1111/j.1365-294X.2010.04631.x. [DOI] [PubMed] [Google Scholar]
- 82.Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, et al. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity. 2009;102:442–452. doi: 10.1038/hdy.2009.8. [DOI] [PubMed] [Google Scholar]
- 83.Nason JD, Ellstrand NC, Arnold ML. Patterns of hybridization and introgression in populations of oaks, manzanitas, and irises. Am J Bot. 1992;79:101–111. [Google Scholar]
- 84.Silliman FE, Leisner RS. An analysis of a colony of hybrid oaks. Am J Bot. 1958;45:730–736. [Google Scholar]
- 85.Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 87.Howard DJ. Reinforcement: origin, dynamics and fate of an evolutionary hypothesis. In: Harrison RG, editor. Hybrid zones and the evolutionary process. Oxford: Oxford University Press; 1993. pp. 46–69. [Google Scholar]
- 88.Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytol. 2004;161:151–164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of per-locus interspecific FST values against heterozygosity in each study site.
(TIF)