Abstract
Much of what we know about cytokinesis in bacteria has come from studies with Escherichia coli, and efforts to comprehensively understand this fundamental process in this organism continue to intensify. Major recent advances include in vitro assembly of a membrane-tethered version of FtsZ into contractile rings in lipid tubules, in vitro dynamic patterning of the Min proteins and a deeper understanding of how they direct assembly of the FtsZ-ring to midcell, the elucidation of structures, biochemical activities and interactions of other key components of the cell fission machinery, and the uncovering of additional components of this machinery with often redundant but important roles in invagination of the three cell envelope layers.
Introduction
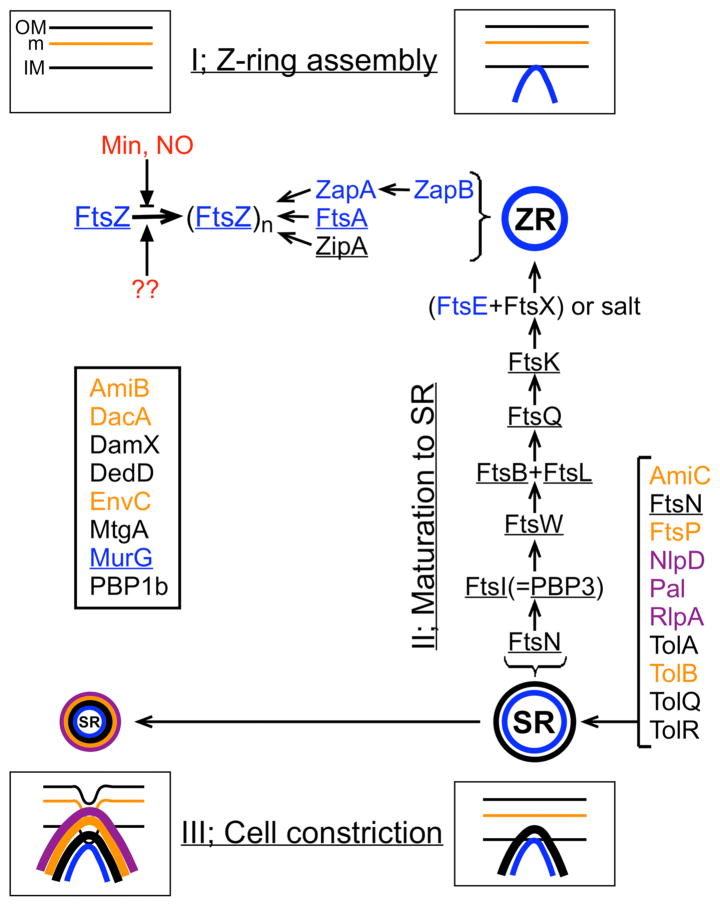
Cytokinesis in bacteria is driven by a complex ring-shaped organelle variously referred to as divisome, septasome, FtsZ ring, or septal ring (SR) [1–3]. For this review, I’ll use Z-ring (ZR) to indicate the mostly cytoplasmic intermediate structure that assembles first, and septal ring (SR) to indicate more mature forms of the organelle (Fig. 1).
Figure 1.
Schematic representation of septal ring assembly in E.coli. Indicated are three stages in development of the septal ring, known protein components, and the approximate step at which they become associated with the apparatus. Proteins that are essential for viability are underlined. Proteins assembling at the cytoplasmic face of the inner membrane are in blue, trans-membrane inner-membrane proteins are in black, periplasmic proteins in orange, and outer-membrane (lipo-) proteins are in purple. Regulators of Z-ring positioning are in red. Some septal ring components (listed in box) were left out of the recruitment pathway for clarity, or because pertinent information is missing.
The mature, constriction-competent, SR in E.coli is now known to contain over two dozen different protein components. Ten of these (FtsA, B, I, K, L, N, Q, W, Z, and ZipA) are essential to the cell constriction process, and can be considered to form the core of the apparatus. Cells lacking any of the core proteins fail to constrict and display the classical lethal division phenotype where they elongate into very long filaments with a commensurate number of evenly spaced nucleoids and non-functional ZR or SR structures before eventually dying. The number of known non-essential protein components of the SR has increased steadily in the last few years. Though none of these are individually required for cell fission and viability, many have overlapping roles in important aspects of cell constriction and their study is required for a satisfactory understanding of the whole process.
Though this review is focused on progress in our understanding of the cell fission process in E.coli in the last two years, I’ll freely refer to excellent recent progress with Bacillus subtilis, Caulobacter crescentus and other organisms, especially when particularly relevant to the E.coli system
Z-ring assembly
FtsZ plays a key role in cytokinesis of most prokaryotes and plastids, and is the likely ancestor of eukaryotic tubulin. Construction of the SR starts with the accumulation of FtsZ in a ring-like arrangement on the cytoplasmic face of the inner-membrane (IM) at the future site of cell constriction (Fig. 1, stage I). This involves the GTP-dependent homopolymerization of FtsZ into linear protofilaments and a means to tether these to the IM. FtsZ has no intrinsic affinity for phospholipid membrane and assembly of a Z-ring in E.coli cells minimally requires the presence of either FtsA or ZipA. Both FtsA and ZipA are membrane-associated and interact with a small domain (C-core) at the extreme C-terminus of FtsZ [1,4,5].
One profound development in the last two years was the recreation of Z-ring assembly in a minimal in vitro system by Erickson and coworkers [6**]. They smartly bypassed the requirement for a separate membrane-associated partner (e.g. FtsA or ZipA) by replacing the tail of FtsZ with YFP and the C-terminal amphiphatic membrane-targeting sequence (MTS) of MinD (see below). Remarkably, when mixed with liposomes and GTP, this FtsZ-YFP-MTS chimera somehow ended up in the lumen of some multilamellar phospholipid tubules where it then readily formed membrane-associated rings. Moreover, bright rings were associated with clear indentations of the liposome wall, implying they exerted a constrictive force [6**]. These results greatly clarified a number of issues, and showed that GTP, a membrane-tether, and a curved membrane surface are sufficient for FtsZ to spontaneously form ZRs that are at least partially functional.
One next hurdle will be to assemble Z-rings in vitro using native FtsZ and its natural partners, including FtsA, ZipA, and ZapA. Besides helping to tether FtsZ to the IM, FtsA and ZipA are normally both required for further maturation of the ZR to an SR as well (Fig. 1, stage II). ZipA is a bitopic membrane protein (N-out) with a large cytoplasmic domain that binds and bundles FtsZ protofilaments in vitro and helps to stabilize ZRs in vivo. While ZipA appears restricted to the γ-proteobacteria, FtsA is much better conserved and implicated in several important aspects of SR formation and function. FtsA belongs to the actin/Hsp70 superfamily of ATPases, and binds the IM peripherally via a C-terminal amphiphatic MTS [1–3]. Membrane binding is critical to FtsA function but its MTS can be replaced with a heterologous one or, remarkably, with a regular trans-membrane helix, indicating that any reversibility in membrane association is not a crucial property of FtsA [7,8*]. Studies on E.coli FtsA have long been frustrated by the purified native protein showing little biochemical activities. Significant progress, therefore, is the recent demonstration that a purified hypermorphic variant, FtsA* (R286W), stimulates curvature and de-polymerization of FtsZ protofilaments in an ATP-dependent fashion [9**]. In vivo, FtsA* stimulates cell fission at reduced cell mass, and also has the remarkable property of supporting efficient cell division in the complete absence of ZipA [10,11]. The FtsZ depolymerizing activity of FtsA* suggests a role in Z-ring constriction during active cell envelope invagination (Fig. 1, stage III), but seems less compatible with the role of FtsA in ZR assembly and with the ability of FtsA* to compensate for the absence of a protein (ZipA) that normally stabilizes Z-rings in vivo and FtsZ polymers in vitro. Evidently, FtsZ depolymerization by FtsA must be tightly regulated in the cell, and future work on FtsA* may well elucidate how this is accomplished [9**].
The ZapA and ZapB proteins also associate with early Z-ring assemblies. Unlike FtsA and ZipA, these proteins are not essential and E.coli single mutants show only modest phenotypes [12–15*]. ZapA is well conserved, and like ZipA, binds and bundles FtsZ polymers in vitro, and promotes the assembly and stability of the ZR in vivo [16–18*]. The recently discovered ZapB appears restricted to the γ-proteobacteria. It is a small and abundant protein that forms antiparallel coiled-coil dimers that readily polymerize into filaments in vitro [15*]. Initially suspected to interact with FtsZ directly, it primarily associates with the Z-ring via ZapA instead [14*]. Remarkably, fluorescent fusions to ZapB accumulate in a slightly smaller ring than ZapA or FtsZ itself, indicating that ZapB molecules form or decorate some structure that extends from the Z-ring far enough into the cytoplasm to be detectible by light microscopy [14*]. The purpose of such a structure is unclear, but it is possible that a cytoplasmic barrier forms before actual closure of the IM septal pore and that this may be advantageous.
Positioning of the Z-ring
Proper fission at midcell requires accurate placement of the Z-ring along the cell axes. One remarkable observation with the FtsZ-YFP-MTS chimera was that it spontaneously formed ‘closed’ rings in lipid tubules [6**], arguing that positioning of the ZR along the short axis can be largely determined by cell geometry and inherent properties of FtsZ itself.
Lateral positioning of the ZR is determined by two negative regulatory systems, Min and NO (nucleoid occlusion). NO counteracts assembly of Z-rings in the close vicinity of chromosomes and is mediated by DNA-binding proteins SlmA in E.coli [19] and by Noc in B.subtilis [20]. Noc binds to ~70 chromosomal sites that are conspicuously absent from the chromosomal terminus, and placing Noc binding sites in this region delays cell division, indicating Noc helps coordinate ZR assembly with progression of chromosome replication [21**]. Interestingly, it is likely only the DNA-bound form of Noc that is able to counteract ZR assembly, but how it does so still needs to be determined [21**]. E.coli SlmA interacts directly with FtsZ, but the precise mechanism by which it inhibits Z-ring assembly also needs clarification [19].
The Min system in E.coli is a dominant determinant of lateral positioning of the ZR, and its absence leads to frequent fission near a cell pole and the consequent release of chromosome-less minicells. It comprises the MinC, D, and E proteins and all three rapidly oscillate from one cell end to the other in a coordinated and interdependent fashion. Typically MinC, MinD, and a portion of MinE accumulate along the membrane at one cell end as another portion of MinE accumulates more sharply in a MinE-ring that appears to cap the Min polar zone and prevent it from extending past midcell. The MinE ring then moves poleward as the Min polar zone shrinks until most protein is released from that cell end and all three proteins reassemble on the membrane at the opposite end. MinD is a founding member of the MinD/ParA/Soj Walker A cytoskeletal ATPases (WACA) and is central to the Min system. Upon ATP binding, MinD forms a sandwich dimer and engages the IM via its C-terminal amphiphatic MTS. It then recruits additional MinD.ATP, forming larger oligomers/polymers on the membrane surface, as well as MinC and MinE homodimers from cytoplasmic pools [4,5,22,23].
MinC/MinD.ATP complexes (MinC/MinD) interfere with Z-ring assembly, and the mechanism has recently been further clarified. MinD binds to the C-terminal domain of MinC (MinCc) and the MinCc/MinD moiety of the complex in turn binds the C-core domain of FtsZ. This interaction itself interferes with Z ring assembly to some extent as: i) MinCc/MinD competes with FtsA and ZipA for binding FtsZ polymers, and ii) MinCc reduces lateral interactions between the polymers [24**,25*]. Importantly, binding of MinC/MinD to the C-core of FtsZ also positions the N-terminal domain of MinC (MinCN) near its substrate. MinCN is sufficient to inhibit Z ring assembly when overexpressed by itself in vivo, and it shortens FtsZ polymers in vitro [24**]. Unlike the SOS-induced division inhibitor SulA [26*], MinCN does not interfere with polymerization per se, but is now proposed to specifically weaken those longitudinal bonds in FtsZ filaments that harbor GDP in the FtsZ subunit interface via an interaction with helix-10 of FtsZ [24**,27*].
Pole-to-pole oscillation of the Min proteins is critical as it ensures that the time-integrated concentration of membrane-associated MinC/MinD is highest at the cell ends and lowest at the desired site for SR assembly and fission (midcell). MinE promotes release of MinC/MinD complexes from the membrane both by competition with MinC for binding MinD and, importantly, by stimulating MinD to hydrolyze bound ATP. Oscillation results from the interplay of the interactions between MinD with ATP, itself, membrane, and MinE, and this has been extensively modeled and reviewed [4,22,28]. Most models predicted it would be possible to recapitulate MinD/MinE oscillation in minimal in vitro systems, and two groups reported important strides towards that end [29**,30**]. Both demonstrate spontaneous ATP-dependent dynamic patterning of MinD and MinE on the surface of planar lipid bilayers and that, as predicted by most models, pattern dynamics involves rapid cycling of the proteins on/off the membrane. Traveling waves of MinD/E as seen in the first study [29**] are in the second study [30**] often preceded by spatially homogenous oscillations and followed by the formation of dynamic ‘amoebas’ of a MinD-rich zone ringed by a MinE-rich border. The dimensions and dynamics of ‘amoebas’ most closely resemble a surface projection of MinD/E patterns seen in vivo [30**]. Notably, the latter study also argues against bulk diffusion coefficients of MinD and MinE as critical determinants in generating surface patterns as is assumed in many current models for MinD/E oscillation. Obviously, our understanding of Min oscillation is still too limited, and more refined models that consider direct interaction of MinE with phospholipids [31,32], effects of membrane potential on MinD-membrane binding [33*], effects of MinD/E on phospholipid surface properties [34–36], and possible biochemical timing mechanisms [30**] are needed. A solid understanding of MinD/E oscillation is important not only for understanding Z-ring placement, but also for our understanding of other WACA class cytomotive systems, and for our understanding of biological pattern formation in general.
In the absence of Min and NO, FtsZ still displays a modest bias for accumulation in-between segregated nucleoids, indicating the existence of a third system that contributes to positioning of the Z-ring [19,20]. Work with outgrown spores suggests that initiation of chromosome replication at oriC somehow helps control ZR positioning in B.subtilis in a Noc-independent and, possibly, positive fashion [37*]. This is an enticing prospect, but much needs to be learned before the effects can be fully understood.
Maturation of the Z-ring to a Septal ring
Temporally, there is a substantial delay between assembly of the Z-ring and recruitment of the later assembling components [38,39]. During this interval, the Z-ring directs cylindrical murein synthesis at midcell [2,40,41]. How the Z-ring does so, and what signals it to begin further maturation to an SR, are still pressing questions. Maturation involves recruitment of other SR proteins in an ordered fashion (Fig. 1, stage II). Structures of SR proteins [15*,42*–45*], and detailed information on the multitude of interactions between them [46–51*] continue to emerge, helping to construct a picture of how components fit together in the constriction-competent organelle. This goal is also getting more challenging, however, as the number of known components of the division apparatus continues to climb. Besides ZapB [15*], recently identified components in E.coli include MurG [41,52], MtgA [53], FtsP [45*,54], YmgF [55], NlpD [56*], DacA [57], the SPOR-domain proteins DamX, DedD, and RlpA [58*,59], and the Tol-Pal complex [60,61*]. How each of the SR proteins joins the complex is still unclear for many. Though it is likely that specific protein-protein contacts often contribute, local substrate availability [57,62] or transient forms of murein generated at constriction sites [58*,59,63*] are proposed to provide important topological cues for localization of some of the SR proteins.
One intriguing example of the latter is FtsN, a core component whose accumulation at the SR appears to coincide with initiation of the constriction process [63*,64]. A major localization determinant is its periplasmic C-terminal SPOR domain that is now thought to specifically recognize a transient form of septal murein [58*,59,63*,65]. However, initiation of constriction and generation of the SPOR localization substrate depends on a small periplasmic juxta-membrane domain of FtsN, leading to a model in which self-enhanced accumulation of FtsN at the SR helps to trigger and sustain the constriction process (Fig. 1, stage III) [58*,63*]. Despite progress, the exact essential activity of FtsN is still unclear. One reasonable model is that the small essential domain of FtsN promotes the onset of constriction by allosterically modulating the periplasmic murein synthase activities of PBP3 (FtsI), MtgA, and/or PBP1A and PBP1B [47,53,58*,63*]. However, the fact that production of a mutant form of FtsA can rescue division in the complete absence of FtsN (albeit inefficiently) suggests that the signal for septal murein synthesis may more directly emanate from the Z-ring on the cytoplasmic side of the membrane [58*,66].
Cell constriction
Once constriction ensues (Fig. 1, stage III) the SR needs to accomplish a number of tasks in a coordinated manner: i) IM invagination and fission, ii) septal murein synthesis, iii) septal murein splitting, and iv) Outer membrane (OM) invagination and fission.
The ability of FtsZ assemblies to deform membrane surfaces in vitro [6**,67**] has provided strong support for the idea that IM invagination during cell constriction is one of the jobs of the Z-ring portion of the SR. How the ZR exerts force on the membrane is the subject of much modeling activity and debate, but I’ll defer to an excellent recent review on the topic [68]. One key experimental observation has been the in situ visualization of FtsZ polymers at the division site of Caulobacter crescentus cells by cryo-EM tomography, indicating they are unexpectedly sparse and solitary [69]. Whether this is true for other bacteria is unclear but, at least for C.crescentus, this result argues against lateral association of FtsZ polymers playing a decisive role in force generation. Bending of individual FtsZ polymers could provide a driving force [68,69]. Recent evidence that the bending direction of an FtsZ polymer is fixed relative to its long axis and also determines the direction of membrane deformation in vitro provides compelling support for this idea [67**].
Septal murein synthesis requires the transpeptidase PBP3 (FtsI) and at least one other synthase, likely PBP1A, PBP1B, and/or Mtg [47,53,70]. How septal murein is laid down is still poorly understood, but the simplest model is that it is added perpendicular to the cylindrical murein in a relatively thick layer that subsequently needs to be carefully split from the OM-proximal side in order to shape the two nascent cell poles and to allow OM invagination [71,72]. Splitting of septal murein is accomplished by murein hydrolases, most prominently the three murein amidases AmiA, B, and C [71,73,74**]. The LytM-domain proteins EnvC and NlpD are as critical for proper septal murein processing as the amidases, but how they act has been unclear [56*,75]. Recent in vitro and in vivo evidence convincingly shows that EnvC directly stimulates the murein amidase activities of AmiA and AmiB while NlpD stimulates that of AmiC [74**]. This work provides an important step in understanding how potentially lethal enzyme activities are controlled to perform precision surgery on the murein sacculus without compromising its structural integrity. The work also adds to other recent examples of murein synthase or hydrolase activities being controlled by protein regulators [47,76**] and increased efforts in this area can be expected.
Invagination of the OM was long thought to be a rather passive process as tethering of the OM to the murein sacculus by abundant murein-binding OM proteins such as Lpp and OmpA would inevitably cause the OM to move inwards as the septal murein was split into new poles during the constriction process. It is now clear, however, that efficient OM invagination requires the action of the Tol-Pal complex [60,61*], previously implicated in OM integrity and the uptake of colicins and ssDNA phages [77]. All components of the complex, comprising three IM proteins (TolQAR), one periplasmic protein (TolB) and the OM lipoprotein Pal, accumulate at the SR near the time of constriction initiation and mutants lacking the intact complex display a substantial delay in OM invagination [60,61*]. It is proposed that Tol-Pal forms a sub-complex of the SR that uses proton motive force to establish transient transenvelope connections that draw in the OM as the IM and murein layers invaginate [60].
Finally, the septal pore in the IM must close to compartmentalize the cytoplasm and a similar closure of the OM must occur to compartmentalize the periplasm and finalize the separation of daughter cells. We know preciously little about either membrane fission/sealing event. SpoIIIE is required for septal membrane fission during endospore formation in B.subtilis, but only if the pore entraps chromosomal DNA [78*]. This could also be true for the analogous DNA translocase FtsK during division in E.coli, but it is clear that FtsK-independent mechanisms of membrane fission must be operative as well [79*,80]. Whether OM septal pore closure is a spontaneous or controlled event is not known.
Conclusions
Progress in the last two years has been quite exhilarating and the pace of new discoveries is likely to accelerate further. The field is clearly benefiting from increased activities by both new laboratory leaders and by established labs across disciplines that bring valuable expertise in imaging, biophysical, biochemical, and theoretical approaches. Efforts to reconstitute parts of the division machinery and its regulators with purified components in vitro will increase, as will efforts to understand its precise molecular composition and molecular architecture. Much remains to be learned, not least about how all the activities of septal ring proteins are controlled to achieve an efficient and coordinated invagination of the three envelope layers, or how the whole constriction process is coordinated with other cell cycle events, such as chromosome replication and cell elongation/growth. Based on the last few years, answers to these and many other interesting questions will emerge sooner than one might think.
Acknowledgments
Work in the author’s laboratory is supported by NIH GM57059.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 2.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 3.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 4.Vats P, Yu J, Rothfield L. The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci. 2009;66:3353–3362. doi: 10.1007/s00018-009-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 6**.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. The authors show spontaneous assembly of a FtsZ-MTS chimera into contractile rings in lipid tubules. The work inspired much theoretical modeling on FtsZ assembly and force generation, and is an important step in reconstituting additional components of the division apparatus in in vitro systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55 :1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 8*.Shiomi D, Margolin W. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol Microbiol. 2008;67:558–569. doi: 10.1111/j.1365-2958.2007.06085.x. It is demonstrated that the native amphiphatic membrane tether of FtsA can be replaced with a transmembrane helix without compromising its essential activity in cell division. Thus, any reversibility in the interaction of FtsA with the membrane is not critical to its function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Beuria TK, Mullapudi S, Mileykovskaya E, Sadasivam M, Dowhan W, Margolin W. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem. 2009;284:14079–14086. doi: 10.1074/jbc.M808872200. E.coli FtsA has long been recalcitrant to biochemical characterization. This paper shows that a hypermorphic variant of FtsA can stimulate disassembly of FtsZ polymers in vitro, opening a door to more detailed biochemical studies on this important division protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissler B, Shiomi D, Margolin W. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology. 2007;153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JE, Lackner LL, Hale CA, De Boer PA. ZipA Is Required for Targeting of DMinC/DicB, but Not DMinC/MinD, Complexes to Septal Ring Assemblies in Escherichia coli. J Bacteriol. 2004;186:2418–2429. doi: 10.1128/JB.186.8.2418-2429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, Martos A, Alfonso C, van Marle J, Rivas G, den Blaauwen T. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry. 2009;48:11056–11066. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07183.x. The authors find that ZapB joins the Z-ring via an interaction with ZapA, and forms or decorates some unanticipated structure that emanates from the Z-ring into the cytoplasm. [DOI] [PubMed] [Google Scholar]
- 15*.Ebersbach G, Galli E, Moller-Jensen J, Lowe J, Gerdes K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol. 2008;68:720–735. doi: 10.1111/j.1365-2958.2008.06190.x. Describes the discovery of ZapB as a new Z-ring component, and presents its structure. [DOI] [PubMed] [Google Scholar]
- 16.Low HH, Moncrieffe MC, Lowe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Small E, Marrington R, Rodger A, Scott DJ, Sloan K, Roper D, Dafforn TR, Addinall SG. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J Mol Biol. 2007;369:210–221. doi: 10.1016/j.jmb.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 18*.Monahan LG, Robinson A, Harry EJ. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol Microbiol. 2009;74:1004–1017. doi: 10.1111/j.1365-2958.2009.06914.x. This paper provides additional evidence that Z-ring assembly in B.subtilis cells occurs via remodeling of a helical intermediate and that ZapA promotes the helix to ring transition. [DOI] [PubMed] [Google Scholar]
- 19.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 21**.Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. Mapping of the Noc binding sites on the B.subtilis chromosome shows they are absent from the terminus region and that placing them there causes a delay in cell division. The results suggest that nucleoid occlusion not only spatially regulates cell division, but also serves to temporally coordinate division with chromosome replication/segregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 23.Michie KA, Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- 24**.Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. The authors use rheometry and other methods to show that MinC antagonizes FtsZ assembly in vitro in at least two ways; by straining longitudinal bonds within FtsZ polymers, as well as by reducing lateral associations between polymers. [DOI] [PubMed] [Google Scholar]
- 25*.Shen B, Lutkenhaus J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinCC/MinD. Mol Microbiol. 2009;72:410–424. doi: 10.1111/j.1365-2958.2009.06651.x. It is shown that MinC/MinD engages FtsZ via its C-core tail. Competition of MinC/MinD with FtsA and ZipA represents a third manner by which MinC/MinD can antagonize Z-ring assembly/stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Dajkovic A, Mukherjee A, Lutkenhaus J. Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J Bacteriol. 2008;190:2513–2526. doi: 10.1128/JB.01612-07. Kinetic and theoretical considerations indicate that the SOS-induced division inhibitor SulA sequesters monomeric FtsZ that is in a polymerization-competent form, and lead to a kinetic model of FtsZ polymerization that includes a monomer isomerization step to generate the polymerization-competent form and that can account for cooperativity of FtsZ assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Shen B, Lutkenhaus J. Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol Microbiol. 2010;75:1285–1298. doi: 10.1111/j.1365-2958.2010.07055.x. The authors identify residues at the subunit interface of FtsZ polymers as critical for the division inhibitory activity of the N-terminal domain of MinC. It is proposed that MinC specifically weakens those subunit interfaces where GTP hydrolysis has already taken place. [DOI] [PubMed] [Google Scholar]
- 28.Kruse K, Howard M, Margolin W. An experimentalist's guide to computational modelling of the Min system. Mol Microbiol. 2007;63:1279–1284. doi: 10.1111/j.1365-2958.2007.05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. Purified MinD and MinE are shown to spontaneously form dynamic patterns, such as traveling waves, on planar phospholipid layers. It is shown that patterning involves cycles of protein-membrane attachment/detachment, diffusion, and protein-membrane reattachment, as assumed in many theoretical models. [DOI] [PubMed] [Google Scholar]
- 30**.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci U S A. 2010;107:8071–8078. doi: 10.1073/pnas.0911036107. Besides traveling waves, the authors describe a number of other dynamic patterns formed by purified MinD and MinE on planar phospholipid layers. The experimental set-up argues against bulk diffusion coefficients of MinD and MinE to be critical determinants in generating surface patterns, indicating the need for more refined models of Min protein oscillation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh CW, Lin TY, Lai HM, Lin CC, Hsieh TS, Shih YL. Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli. Mol Microbiol. 2010;75:499–512. doi: 10.1111/j.1365-2958.2009.07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma LY, King G, Rothfield L. Mapping the MinE site involved in interaction with the MinD division site selection protein of Escherichia coli. J Bacteriol. 2003;185:4948–4955. doi: 10.1128/JB.185.16.4948-4955.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. The authors find that membrane affinity of proteins that peripherally bind membrane via an amphiphatic helix, such as MinD and FtsA, is significantly affected by membrane potential. Besides more general implications, this could have important implications for the design of in vitro systems that include these division proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci U S A. 2002;99 :6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: I. Changes in the membrane properties induced by MinD binding. Biochim Biophys Acta. 2008;1778:2496–2504. doi: 10.1016/j.bbamem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: II. Domain structure of the membrane enhances MinD binding. Biochim Biophys Acta. 2008;1778:2505–2511. doi: 10.1016/j.bbamem.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Moriya S, Rashid RA, Rodrigues CD, Harry EJ. Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Mol Microbiol. 2010;76:634–647. doi: 10.1111/j.1365-2958.2010.07102.x. Work with germinating spores of B.subtilis suggests that in addition to Min and NO, the placement of Z-rings is biased by something that is correlated with the initiation of chromosome replication. [DOI] [PubMed] [Google Scholar]
- 38.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 39.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two–step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Pedro MA, Quintela JC, Höltje J-V, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 42*.van den Ent F, Vinkenvleugel TM, Ind A, West P, Veprintsev D, Nanninga N, den Blaauwen T, Lowe J. Structural and mutational analysis of the cell division protein FtsQ. Mol Microbiol. 2008;68:110–123. doi: 10.1111/j.1365-2958.2008.06141.x. Presents the structure and mutational analyses of the periplasmic domain of E.coli FtsQ. [DOI] [PubMed] [Google Scholar]
- 43*.Masson S, Kern T, Le Gouellec A, Giustini C, Simorre JP, Callow P, Vernet T, Gabel F, Zapun A. Central domain of DivIB caps the C-terminal regions of the FtsL/DivIC coiled-coil rod. J Biol Chem. 2009;284:27687–27700. doi: 10.1074/jbc.M109.019471. Structural and interaction studies on S.pneumoniae DivIB (FtsQ) FtsL and DivIC (FtsB) yield a compelling picture of how the proteins are arranged in the highly conserved FtsQ/L/B subcomplex of the septal ring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, Wong CH, Ma C. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. Provides the complete structure of PBP1B, one of the two major bifunctional murein synthases in E.coli that is active in generating both cylindrical and septal murein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Tarry M, Arends SJ, Roversi P, Piette E, Sargent F, Berks BC, Weiss DS, Lea SM. The Escherichia coli cell division protein and model Tat substrate SufI (FtsP) localizes to the septal ring and has a multicopper oxidase-like structure. J Mol Biol. 2009;386:504–519. doi: 10.1016/j.jmb.2008.12.043. Shows that FtsP accumulates at constriction sites, as well as provides its structure. Together with [54] below, establishes FtsP as a genuine division protein in E.coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 48.Maggi S, Massidda O, Luzi G, Fadda D, Paolozzi L, Ghelardini P. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology. 2008;154:3042–3052. doi: 10.1099/mic.0.2008/018697-0. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez MD, Beckwith J. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J Bacteriol. 2009;191:2815–2825. doi: 10.1128/JB.01597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J Bacteriol. 2010;192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Alexeeva S, Gadella TW, Jr, Verheul J, Verhoeven GS, den Blaauwen T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol. 2010;77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. The authors use FRET on whole fixed cells to assay proximity of various components in the septal ring and detect new interactions involving FtsN. The approach is a valuable addition to other methods aimed at elucidating the architecture of the division apparatus. [DOI] [PubMed] [Google Scholar]
- 52.Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65 :1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derouaux A, Wolf B, Fraipont C, Breukink E, Nguyen-Disteche M, Terrak M. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J Bacteriol. 2008;190:1831–1834. doi: 10.1128/JB.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samaluru H, SaiSree L, Reddy M. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol. 2007;189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karimova G, Robichon C, Ladant D. Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J Bacteriol. 2009;191:333–346. doi: 10.1128/JB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. Shows that in addition to EnvC, the other LytM-domain proteins in E.coli are also involved in septal murein splitting. Establishes NlpD as an important player and as a new outer membrane septal ring component. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potluri L, Karczmarek A, Verheul J, Piette A, Wilkin JM, Werth N, Banzhaf M, Vollmer W, Young KD, Nguyen-Disteche M, et al. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol Microbiol. 2010;77:300–323. doi: 10.1111/j.1365-2958.2010.07205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at division sites, and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. Supports a role for FtsN in triggering cell constriction, identifies the SPOR domain as an autonomous septal localization domain that likely recognizes a transient form of septal murein, and identifies three new components of the division apparatus. See also [59] and [63] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arends SJ, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61*.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2010;192:4847–4858. doi: 10.1128/JB.00607-10. Establishes that the Tol-Pal complex is essential in Caulobacter. As in E.coli (see[60]), the complex is found to be required for efficient OM invagination during cell fission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa T, Priyadarshini R, Jacobs–Wagner C. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol. 2008;70:634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Moll A, Thanbichler M. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol. 2009;72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. The authors find that FtsN is much better conserved than previously thought and identify and characterize FtsN of Caulobacter crescentus. SPOR domains are identified as autonomous septal localization determinants in several species. See also [58] and [59] [DOI] [PubMed] [Google Scholar]
- 64.Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 65.Ursinus A, van den Ent F, Brechtel S, de Pedro M, Holtje JV, Lowe J, Vollmer W. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol. 2004;186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67**.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. Polymers of artificially membrane-tethered FtsZ are shown to distort membrane surfaces in a direction that depends on the placement of the membrane-tether on the FtsZ molecule. The results imply that FtsZ polymers bend in a fixed direction relative to the long axis of the polymer, and support the idea that polymer bending might provide the force driving membrane constriction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erickson HP. Modeling the physics of FtsZ assembly and force generation. Proc Natl Acad Sci U S A. 2009;106:9238–9243. doi: 10.1073/pnas.0902258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Disteche M, den Blaauwen T, Holtje JV, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 71.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 72.Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Priyadarshini R, Popham DL, Young KD. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol. 2006;188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74**.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. It is shown that murein amidase activities in the septal ring that are required for septal murein splitting, and for efficient killing of cells with β-lactam antibiotics, are directly controlled by the LytM-domain proteins EnvC and NlpD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76**.Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. Identifies unique, and mutually enhanced, murein hydrolase activities of the SpoIID and SpoIIP proteins, indicating they form a murein-degrading engine that helps pull the septal membranes around the forespore during endospore formation in B.subtilis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78*.Fleming TC, Shin JY, Lee SH, Becker E, Huang KC, Bustamante C, Pogliano K. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 2010;24:1160–1172. doi: 10.1101/gad.1925210. Provides sophisticated evidence that SpoIIIE is required for septal membrane fission during endospore formation in B.subtilis, but only when entrapped DNA blocks progress of the constricting septal ring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79*.Dubarry N, Barre FX. Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 2010;29:597–605. doi: 10.1038/emboj.2009.381. Provides evidence that FtsK transports DNA from the septal pore before pore closure. It is proposed that FtsK might actively delay pore closure when DNA blocks progression of membrane fission. Together with [80], it is clear that FtsK-independent mechanism(s) of inner-membrane scission must exist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]