Abstract
Chronic ethanol consumption in the Long-Evans (LE) rat has been associated with hepatic p53 activation, and inhibition of the insulin/PI3K/AKT signal transduction cascade due to increased expression of phosphatase and tensin homologue deleted on chromosome 10 (PTEN). We hypothesize that p53 activation and altered insulin signaling may influence the susceptibility of rats to ethanol-induced liver damage. Furthermore, p53 not only activates programmed cell death pathways and suppresses hepatocellular survival signals, but promotes gluconeogenesis to increase systemic insulin resistance due to a novel metabolic function. Here, we present evidence that chronic ethanol feeding in Fischer (F), Sprague-Dawley (SD) and LE rats promotes p53 activation, hepatic steatosis, oxidative stress, p53 up-regulated modulator of apoptosis (PUMA) and PTEN expression, which contribute to hepatocellular death and diminished insulin signaling in the liver. Such changes are pronounced in the LE, less prominent in SD, and virtually absent in the F rat strain. More importantly, there is activation of Tp53-induced glycolysis and apoptosis regulator (TIGAR) in the ethanol-fed LE rat. This event generates low hepatic fructose-2, 6-bisphosphate (Fru-2,6-P2) levels, reduced lactate/pyruvate ratio and may contribute to increased basal glucose turnover and high residual hepatic glucose production during euglycemic hyperinsulinemic clamp. Conclusions: p53 activation correlates with the susceptibility to ethanol-induced liver damage in different rat strains. p53 not only orchestrates apoptosis and suppresses cell survival, but by activating TIGAR and decreasing hepatic Fru-2,6-P2 levels it may also promote insulin resistance and therefore, contribute to the metabolic abnormalities associated with hepatic steatosis.
Keywords: Hepatocellular death; PTEN; PUMA; TIGAR; fructose-2,6-bisphosphate
INTRODUCTION
Alcoholic liver disease (ALD) is a disorder governed by gene-environment interactions [1]. A myriad of genetic factors have been linked to increased susceptibility towards ethanol-induced liver damage, many of which are involved in the regulation of insulin signaling, oxidative stress and apoptosis [2]. Hepatic insulin resistance, micro-and macrovesicular steatosis, increased oxidative stress, hepatocellular death, and suppressed cell survival signaling are the cardinal features of ALD produced by eight weeks of ethanol feeding in the LE rat [3, 4]. Chronic ethanol exposure impaired survival mechanisms in the liver by constitutive inhibition of insulin/PI3K signaling and downstream AKT activity, an effect that was mediated by increased PTEN expression and function [4]. Robust nuclear accumulation of p53 during chronic ethanol feeding was also observed [4]. It is established that PTEN is a downstream target of p53 [5] promoting hepatic insulin resistance [3]. However, the role of p53 in ALD has not been fully elucidated. Activation of p53 can be critical in ethanol-induced hepatocyte apoptosis, since genetic ablation of p53 abrogates ethanol-induced liver damage [6]. Furthermore, four weeks of ethanol feeding in rats was shown to increase mRNA abundance and acetylation of p53 [7]. It was recently demonstrated that the lifespan regulator p66shc that is indispensable for p53-dependent apoptosis [8] may have a pivotal role in ethanol-induced liver damage [9].
We compared rat strain susceptibility to ALD in the context of p53 activation [4]; this approach allowed us to correlate p53 alterations with liver damage. We demonstrated that p53 may have a broader role in the pathogenesis of ALD than previously determined. p53 not only orchestrated hepatocellular death and suppressed cell survival, but by upregulating TIGAR, caused perturbations in hepatic glucose metabolism that contributed to insulin resistance. Thus, p53 activation may be important in oxidative stress, apoptosis, impaired survival signaling, liver regeneration [10] and sustaining systemic and hepatic insulin resistance. Investigation of such pathways in different rat strains led to the hypothesis that ethanol-induced p53 expression may be crucial for the development of ALD.
MATERIALS AND METHODS
Animals and Treatment
The 150–200 gram, male Fischer, Sprague-Dawley and Long-Evans rats (Harlan Laboratories, Indianapolis, IN) were fed ad libitum with ethanol-containing (ethanol-derived calories were increased from 13% to 36% in the first two weeks) or isocaloric control liquid diet (Bioserv, Frenchtown, NJ) for eight weeks (following initial adaptation). Body weights and food intake were recorded weekly. After eight weeks, the animals were fasted for four hours and sacrificed. Blood and liver samples were collected. Five animals from each group were used for hepatocyte isolation. The Lifespan Animal Welfare Committee of Rhode Island Hospital, Providence, RI, approved all animal experiments.
Histological Studies and Image Analysis
Histological changes in the liver were assessed by routine hematoxylin-eosin, Oil Red O, Masson’s trichrome and Sirius red collagen staining. Slides were scanned with Aperio ScanScope CS (Aperio Technologies, Inc., Vista, CA). Area measurements were performed with iVision software (BioVision Technologies, Exton, PA). Positive Oil Red O staining was defined through intensity thresholding. Apoptosis was assessed by Apoptag® ISOL Dual Fluroescence Apoptosis Detection Kit (Millipore, Billerica, MA). Cells stained for apoptosis were visually counted and recorded. Results were expressed as number of apoptotic cells per square millimeter.
Chemicals
All chemicals used in these studies were purchased from Sigma (Sigma-Aldrich, St. Louis, MO), unless otherwise specified.
Biochemical Assays
Serum alanine aminotransferase (ALT) levels were measured by using a commercially available kit (Thermo Fisher Scientific, Inc., Waltham, MA). Liver lysates were used to measure triglyceride, lactate and pyruvate with corresponding kits (BioVision, Inc., Mountain View, CA). Serum alcohol was measured by Analox GM7 analyzer (Analox Instruments, Lunenburg, MA). Alcohol dehydrogenase (ADH) activity in liver lysates was measured with an assay kit (Biomedical Research Center, SUNY Buffalo, Buffalo, NY). Fru-2,6-P2 content was measured by the previously described endpoint enzymatic assay [11]. ADH, triglyceride, lactate, pyruvate and hepatic Fru-2,6-P2 assays were normalized to the protein content of the liver lysates. Protein concentration was determined by the bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc./Pierce, Waltham, MA).
Assessment of Basal Glucose Kinetics and Euglycemic Hyperinsulinemic Clamp in Rats
The euglycemic hyperinsulinemic clamp in rats has been carried out as previously described [12]. The detailed description of these methods can be found in the Supplementary Material.
Caspase-3 Assay
Caspase-3 activity was measured by the CPP32/Caspase-3 Fluorometric Assay Kit (BioVision, Inc., Mountain View, CA) following the manufacturer’s instructions. The optical density values were normalized to the protein content of the lysates.
Hepatocyte Isolation and Cell Fractionation
Hepatocyte isolation was carried out as previously described [13]. Nuclear and cytosolic proteins were separated by using NE-PER Kit (Thermo Fisher Scientific, Inc./Pierce, Waltham, MA) from fresh liver tissue. The purity of these fractions was assessed by performing Western-blots with nuclear lamin A (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and cytosolic β-actin (Sigma-Aldrich, St. Louis, MO) or GAPDH (Cell Signaling Technology, Inc., Danvers, MA) antibodies.
Measurement of Intracellular ROS
Intracellular ROS formation was assessed by using 2′,7′-dichlorodihydrofluorescin (DCF) diacetate (Invitrogen Corporation, Carlsbad, CA), as previously described [14].
Long-Extension Polymerase Chain Reaction (LX-PCR) to Detect Mitochondrial DNA (mtDNA) Damage
DNA was extracted from flash-frozen liver tissue samples by using the EZ1® DNA Tissue Kit with BioRobot EZ1 (Qiagen, Inc., Valencia, CA). The LX-PCR and rat mtDNA specific primer sequences have been previously described [15]. PCR products less than 16-kb were considered damaged/rearranged mtDNA species.
p53 and Sterol Regulatory Element-Binding Protein 1c Electrophoretic Mobility Shift Assays
The LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Inc./Pierce, Waltham, MA) was used to assess the in vitro binding activity of p53 and Sterol Regulatory Element-Binding Protein 1c (SREBP1c). 5′-end biotin labeled oligonucleotides with the corresponding unlabeled controls were obtained from Sigma (Sigma-Aldrich, St. Louis, MO). The sequences of the p53 and SREBP1c consensus oligonucleotides are available upon request. The binding reaction and subsequent steps were carried out following the manufacturer’s instructions.
Western-blot Analysis
Whole tissue lysate was prepared from livers, as previously described [14]. Protein lysates were separated on 6 % to 15 % SDS-PAGE and transferred onto PVDF membrane (Thermo Fisher Scientific, Inc./Pierce, Waltham, MA). The following primary antibodies were used: CYP2E1 (Abcam, Inc., Cambridge, MA), p53, PUMA-α, PTEN, p110 subunit of PI3K, AKT1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), IRS-2, p85 subunit of PI3K (Millipore, Billerica, MA), p-AKTS473 (R&D Systems, Inc., Minneapolis, MN), p-AKTT308, cleaved-PARP (Cell Signaling Technology, Inc., Danvers, MA), TIGAR (Lifespan Biosciences, Inc., Seattle, WA).
Statistical analysis
Data are presented as mean ± SEM and analyzed by using GraphPad Instat® software (GraphPad Software, Inc., La Jolla, CA); unpaired Student’s t or Tukey-Kramer tests were executed unless otherwise stated. Data pertaining to basal glucose metabolism and euglycemic hyperinsulinemic clamp were analyzed by ANOVA followed post hoc by Student-Neuman-Keuls test for multiple group comparisons. Statistical consideration related to weight gain in the various experimental groups can be found in the Supplementary Material. Differences among the groups were considered significant when p < 0.05.
RESULTS
Hepatic Steatosis and ALT Elevation
The level of steatosis and hepatocellular damage, as measured by ALT levels, induced by chronic ethanol feeding was compared in three rat strains. We compared the ethanol-sensitive LE rat [4] to two other frequently used laboratory rat strains (F and SD) to assess the differences in ethanol consumption, blood ethanol levels, ethanol metabolism and weight gain during feeding. Experiments were repeated three times, using 5–10 animals per group each time. There was no difference in ADH activity (Supplementary Figure 1A) and CYP2E1 abundance in the liver of various rat strains after eight weeks of ethanol feeding (Supplementary Figure 1B). There were statistically significant interactions between strain and week (p<.0001) and between diet and week (p<.0001), indicating that, overall, strains grew at different rates and that rats fed with ethanol grew at a lower rate than those on control diet. Additionally, the non-significant three-way interaction between strain, diet, and week (p=0.3065) indicated that ethanol influenced weight gain over time similarly in all strains (Supplementary figure 1C). Comparable food intake in the various strains was coupled with no statistically significant differences in the mean blood ethanol levels (LE; 130 ± 9, F; 107 ± 18, SD; 115 ± 15 mg/dl, p=NS) during the experiment. Despite these similarities, LE rats developed significantly more micro-and macrovesicular steatosis, cell dropout and disorganized liver structure than F or SD (Figure 1A). Oil Red O staining (Supplementary Figure 2A and 2B) and triglyceride quantification (Figure 1B) confirmed pronounced steatosis. No evidence of hepatic fibrosis was found by Masson’s trichrome or Sirius red staining (data not shown). Hepatocellular damage was observed in LE rats by statistically significant elevation in ALT levels as compared to pair-fed controls and the other ethanol-fed strains (SD and F) (Figure 1C).
Figure 1. Chronic alcohol-fed LE rats exhibit steatosis and ALT elevations.
Hematoxylineosin staining revealed micro- and macrovesicular steatosis, cell dropout and disorganized liver architecture in the LE rat. These changes were less pronounced in the other two (F, SD) ethanol-fed strains. (panels F&I vs A-H in Fig. 1A). Enhanced steatosis was confirmed by a liver triglyceride assay (Fig. 1B). Histopathological changes in the LE model were associated with ALT elevation (Fig. 1C). (*): LE control vs LE ethanol, (#): LE ethanol vs SD ethanol, p<0.05
Steatosis is Associated with Enhanced SREBP1c Activity
SREBP1c activation plays a pivotal role in ethanol-induced fatty liver disease [16, 17]. The effect of ethanol on SREBP1c is mediated by both AMPK-dependent [17] and –independent mechanisms [16]. The SREBP-1c isoform activates a battery of genes involved in fat synthesis. More importantly, SREBP1c also acts as a repressor for the transcription of insulin receptor substrate-2 (IRS-2) in the liver; thus, its abnormal expression may suppress hepatic insulin signaling [18]. Chronic activation of hepatic SREBP1c has been observed in insulin resistant murine models [19]. In this regard, the nuclear abundance of transcriptionally active SREBP1c was significantly increased in the LE ethanol-fed rats (Supplementary Figure 3A), indicating that de novo fat synthesis may contribute to hepatic triglyceride accumulation. Significantly reduced IRS-2 levels were also found (Supplementary Figure 3B).
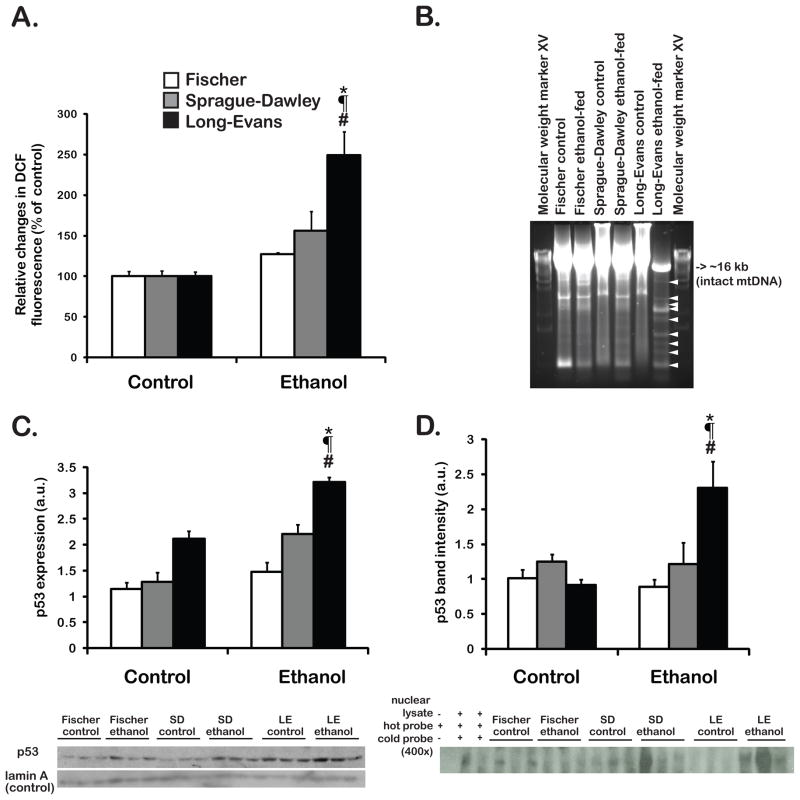
Steatosis is Associated with ROS, MtDNA Damage and p53 Activation
The impact of increased fat accumulation on ROS and subsequent mtDNA damage in the liver were explored. Fatty acid deposition in hepatocytes is known to elicit a “lipotoxic” pathologic response [20]. Oxidative stress may be a central component of lipotoxicity in fatty liver disease [21]. The levels of H2O2 formation were assessed in isolated hepatocytes. As shown in Figure 2A, chronic ethanol feeding significantly increased H2O2 levels (~ 2.5 fold) in the LE strain (p<0.05), and caused lesser ROS elevations in the F and SD (1.3 and 1.5 fold increase, respectively, p=NS). An adverse cellular consequence of chronic oxidative stress is deletion/rearrangement of mtDNA. These changes in the mitochondrial genome can subsequently lead to disturbed oxidative phosphorylation, further increasing the likelihood of aberrant ROS formation [22, 23]. The previously described LX-PCR method [15] was employed to amplify the entire rat mitochondrial genome; PCR products less than 16 kb represented deleted/rearranged mtDNA species as indicators of DNA damage. These were most abundant in the liver of the ethanol-fed LE rat (Figure 2B, white arrowheads). In addition, ROS generation may lead to the stabilization, activation and accumulation of p53 in the cytoplasm and nucleus [4, 14, 24]. Thus, the nuclear abundance and DNA binding activity of p53 were evaluated by Western-blot analysis (Figure 2C) and EMSA (Figure 2D), respectively. p53 accumulation was significantly increased (~2 fold) in LE, but only marginally upregulated in SD (p=NS). Chronic ethanol feeding did not induce p53 in the F strain. The p53 was biologically active as shown by EMSA, depicted in Figure 2D.
Figure 2. Oxidative stress, mtDNA damage and p53 activation.
Oxidative stress was measured in isolated hepatocytes (Fig. 2A). Mitochondrial DNA damage was detected in the liver of LE rats (Fig. 2B). Nuclear abundance of p53 was assessed by Western blot analysis and quantified by densitometry (Fig. 2C). In vitro binding activity of p53 to DNA target sites was assessed by EMSA. The results of densitometry are shown in Fig. 2D. (*): LE control vs LE ethanol, (#): LE ethanol vs SD ethanol, (¶): LE ethanol vs F ethanol, p<0.05.
p53 Promotes PUMA, Caspase-3 Activation, Poly (ADP-ribose) Polymerase (PARP)-Cleavage and Apoptosis
The expression and function of downstream p53 target genes that influence apoptosis, cell survival and glucose metabolism were evaluated. PUMA is a BH3-only pro-apoptotic member of the bcl-2 family that is involved in hepatocyte lipoapoptosis [25]. Chronic ethanol consumption led to p53 activation and accumulation of PUMA-α in the cytoplasm; the highest levels were observed in LE rats (Figure 3A). Activation of the apoptotic cascade was demonstrated by enhanced caspase-3 activity and PARP cleavage. Finally, double-stranded DNA breaks were observed with in situ oligo ligation (ISOL) dual fluorescent labeling (Figure 3B–D). Taken together, we believe that the p53-dependent apoptosis was the most active in the chronic ethanol-fed LE rat, moderate in the SD and inactive in the F strain.
Figure 3. p53 activation by chronic ethanol promotes PUMA-mediated apoptosis, caspase-3 and PARP-cleavage.
Cytosolic PUMA-α accumulation was assessed by Western-blot analysis (Fig. 3A). Caspase-3 activity was measured using liver lysates (Fig. 3B). PARP cleavage was assessed by Western-blot analysis (Fig. 3C). Results for apoptosis were expressed as number of apoptotic cells per square millimeter (Fig. 3D). (*): LE control vs LE ethanol (#): LE vs SD ethanol, (¶): LE ethanol vs F ethanol, p<0.05.
Hepatic p53 Activation Induces Downstream PTEN Expression and Reduced AKT Phosphorylation
Evidence suggests that p53 transcriptionally regulates PTEN [5]. Upregulation of PTEN by chronic ethanol feeding leads to suppression of the insulin stimulated PI3K/AKT prosurvival pathway [4]. In addition, acute ethanol exposure increases the association of PTEN with PI3K p85α subunit, which curbs survival signaling in the liver [26]. We confirmed increased PTEN expression (Figure 4A and 4B) associated with reduced AKT phosphorylation (Figure 4A) only in the ethanol-fed LE rat where p53 has been previously shown to be highly expressed [4].
Figure 4. Chronic ethanol consumption activates p53, induces PTEN and alters AKT phosphorylation.
Elements of AKT signaling were assessed by Western-blot analysis using whole liver lysates (Fig. 4A). The result of PTEN densitometry is shown (Fig. 4B) (*): LE control vs LE ethanol, p<0.05.
The p53 Downstream Target TIGAR Impairs Fru-2,6-P2 Activity and Promotes Hepatic Glucose Production
It is noteworthy that p53 has important metabolic functions in hepatocytes [27]. TIGAR has been shown to hydrolyze Fru-2,6-P2 [28], a critical activator of glycolysis and inhibitor of gluconeogenesis that couples glucose homeostasis to AKT signaling [29]. TIGAR expression was most evident in the ethanol-fed LE rat. In contrast, ethanol feeding failed to induce TIGAR expression in the F strain (Figure 5A). TIGAR upregulation was followed by hepatic Fru-2,6-P2 depletion (Figure 5B) and impaired glycolysis, as evidenced by marked suppression of the hepatic lactate/pyruvate ratio in the LE rat (Figure 5C). Since it has been shown that decreasing hepatic Fru-2,6-P2 content in vivo alters various aspects of glucose metabolism including hepatic glucose production (HGP) [29], we were prompted to assess basic parameters of glucose metabolism in our experimental groups under basal and hyperinsulinemic conditions. The basal plasma glucose concentration in control and ethanol-fed rats did not differ regardless of the strain (Table 1). In contrast, while the ethanol-fed F and SD rats demonstrated 30–35% decrease, the LE rats exhibited a 15% increase in basal plasma insulin compared to the respective controls (Table 1). Our data indicate that all three strains of control rats had comparable basal rates of glucose Ra and Rd (rate of glucose appearance and rate of glucose disappearance) and that ethanol feeding did not significantly alter these rates (Table 1). However, the LE rats did show a trend (+10–15%) for an increase in basal glucose turnover upon ethanol feeding, which may suggest augmented gluconeogenesis. After determining basal glucose kinetics, euglycemic hyperinsulinemic clamp was initiated. The primed continuous infusion of insulin uniformly increased plasma insulin concentration regardless of strain or diet (Table 1). This dose of insulin was selected to produce a new steady-state insulin concentration, which would be expected to reduce endogenous HGP by 70–90% [12, 30]. During the last hour of clamp the plasma glucose was maintained at ~5.5 mmol/l in all groups (Table 1). While the glucose infusion rate necessary to maintain euglycemia during the clamp did not differ between control rats of various strains, it was 35%, 46%, and 63% lower in the alcohol-fed F, SD and LE rats, respectively, compared to their time-matched pair-fed controls (Figure 5D). During the hyperinsulinemic clamp, HGP was comparable in all control animals, but was significantly greater in all ethanol-fed rats. More importantly, the ethanol-fed LE rats had the highest residual HGP during the clamp (Figure 5E). As a result of these changes, the percent suppression of HGP by insulin was blunted in all ethanol-fed animals with the LE rats demonstrating the least insulin-induced suppression of HGP (Figure 5F).
Figure 5. The p53 downstream target gene TIGAR reduces hepatic Fru-2,6-P2 content and alters glucose metabolism. Chronic ethanol consumption produces hepatic insulin resistance in alcohol-fed rats.
TIGAR expression was assessed by Western-blot analysis in whole liver lysates (Fig. 5A). Hepatic fructose-2,6-bisphosphate (Fig. 5B), lactate and pyruvate levels were determined (Fig. 5C). A euglycemic hyperinsulinemic clamp was performed on catheterized conscious unrestrained control and alcohol-fed rats (n=8–9). The rate of 25% D-glucose infused exogenously to maintain euglycemia during the hyperinsulinemic clamp is illustrated (Fig. 5D). The absolute rate of HGP was quantitated as the difference between the 3H-glucose determined rate of total glucose Rd and the exogenous glucose infusion rate (Fig. 5E). The percent suppression of HGP during the clamp was calculated as described (Fig. 5F). (*): control vs ethanol (#): LE ethanol vs SD ethanol, (¶): LE ethanol vs F ethanol (&) SD ethanol vs F ethanol, p<0.05
Table 1.
Plasma glucose and insulin concentrations under basal conditions and during final hour of euglycemic hyperinsulinemic clamp in control and ethanol-fed rats
| Strain | Diet | Basal Glucose(mmol/l) | Basal Insulin(pmol/l) | Basal Glucose Ra/Rd(μmol/min/kg) | Clamp Glucose(mmol/l) | Clamp Insulin(pmol/l) |
|---|---|---|---|---|---|---|
| Fischer | Control | 5.1±0.2 | 281±14 | 48.1±2.3 | 5.5±0.1 | 780±71 |
| Ethanol | 5.3±0.2 | 177±12* | 47.6±2.8 | 5.5±0.2 | 882±69 | |
| Sprague-Dawley | Control | 5.4±0.3 | 308±18 | 45.6±3.2 | 5.5±0.2 | 750±59 |
| Ethanol | 5.6±0.2 | 205±25* | 43.7±3.0 | 5.6±0.2 | 845±74 | |
| Long-Evans | Control | 5.5±0.2 | 323±20 | 42.6±2.3 | 5.6±0.2 | 751±85 |
| Ethanol | 5.9±0.2 | 366±18# | 48.3±3.1 | 5.5±0.2 | 865±84 |
Values are means ± SEM (n=8–9/group).
control vs ethanol,
LE ethanol vs F/SD ethanol, p< 0.05
Role of p53 in ALD
Our findings are consistent with the hypothesis that chronic ethanol consumption activates the p53/TIGAR axis, which then contributes to hepatic and systemic insulin resistance. Another consequence of reduced glycolysis and promotion of oxidative phosphorylation may be the amplification of ROS formation leading to mtDNA damage. A model based on these studies is presented in Figure 6.
Figure 6. Diagram illustrating a role for p53 in ALD.
Activation of p53 is linked to hepatic steatosis and oxidative stress. Enhanced p53 expression promotes apoptosis, suppresses insulin signaling and cell survival, as well as shifts energy metabolism towards ROS-prone oxidative phosphorylation, decreases glycolysis and favors gluconeogenesis. In this context, increased HGP enhances systemic insulin resistance and further promotes steatosis by activating SREBP1c. Grey boxes represent the results of signaling abnormalities during chronic ethanol consumption.
DISCUSSION
Although, it is increasingly recognized that p53 has broad-spectrum effects on ROS, apoptosis, and cellular metabolism, there is limited information on its exact role in ALD. We propose a novel hypothesis that p53 activation may be important in the pathogenesis of ALD and facilitate disease progression by multiple mechanisms, as outlined in Figure 6. Indeed, p53 appeared to modulate ethanol-induced hepatocyte apoptosis, since it was completely abrogated in mice with a p53 null background [6]. It is known that p53 is directly involved in the regulation of oxidative stress. In addition, p53-mediated impairment of insulin/IGF-1 signaling not only curbs regenerative capacity and survival signaling in the liver, but also increases oxidative stress by facilitating hepatic steatosis [3, 10, 31]. In this regard, the nuclear accumulation of p53 correlated with the levels of ROS and the severity of ethanol-induced liver damage: p53 activation was the most prominent in the LE rat; the F strain showed minimal expression and had little to no evidence of liver damage, steatosis, oxidative stress and apoptosis.
The generation of ROS provides a major stimulus for p53 stabilization and subsequent induction of apoptosis by a feed-forward regulatory loop [24]. Recent observations revealed that p53 not only orchestrates various forms of cell death, but also regulates cellular energy metabolism and suppresses the PI3K/AKT pathway. Suppression of the hepatic insulin signaling by PTEN inhibited the insulin/PI3K/AKT axis [3, 4]. Here, we report for the first time that chronic ethanol consumption activated SREBP1c, which led to steatosis and suppressed IRS-2, further compromising insulin signaling in the LE rat. Activation of metabolic targets (e.g. TIGAR) of p53 and interference with the PI3K/AKT pathway may reduce glycolysis and promote gluconeogenesis. TIGAR functions as a fructose-bisphosphatase [32] and depletes Fru-2,6-P2, a change that has been linked to elevated hepatic gluconeogenesis, impaired AKT phosphorylation and insulin resistance [29]. In our study, we demonstrate that unlike other rat strains, in which ethanol induces mild reduction in basal insulin levels [30], the LE rat exhibits a detectable increase in basal plasma insulin in conjunction with normal glucose levels, indicating systemic insulin resistance. Furthermore, our data suggest that ethanol feeding elicits a slight increase in basal glucose Ra (e.g.: primarily hepatic gluconeogenesis) in the presence of elevated plasma insulin that is suggestive of hepatic insulin resistance. More importantly, the euglycemic hyperinsulinemic clamp revealed that all investigated ethanol-fed rat strains develop some degree of hepatic insulin resistance, but that the impairment in insulin action is most marked in the LE strain.
This study provides evidence that p53 may be an important metabolic regulator that affects insulin resistance in the context of ALD (Figure 5). Increased hepatic glucose production favors SREBP1c activation [33] contributing to suppression of IRS-2 (Supplementary Figure 3B). Nonetheless, hepatic upregulation of p53 and the subsequent propensity for insulin resistance is not specific to ALD; it has also been demonstrated in non-alcoholic steatohepatitis [33]. Thus, p53 may similarly contribute to hepatic and systemic insulin resistance in that disease, as well. Taken together, these findings suggest that p53 may be a central element in the pathogenesis of ALD.
Our study also underlines the importance of rat strain variability in ALD studies. For example, many investigated biochemical and pathologic changes related to p53 function were largely absent in F, prominent in LE and intermediate in SD strain. These inter-strain differences were observable even with similar blood ethanol levels during eight weeks of consumption and equal expression of two ethanol metabolizing (ADH and CYP2E1) enzymes in the three rat strains. Although changes in the p53 system are certainly important and associated with various degree of vulnerability, we cannot exclude the possibility that p53-independent mechanisms may also influence rat strain susceptibility to ALD and therefore further studies are necessary. However, it is important to emphasize that different conclusions may be reached regarding the pathogenesis of ALD when the effects of ethanol on the liver varies in rat strains with respect to the generation of steatosis, oxidative stress, insulin resistance, p53 activation, cellular injury and apoptosis. Based on this study, researchers may find the Long-Evans model the most attractive one to study changes in p53 activation, subsequently followed by apoptosis, oxidative stress and insulin resistance even after only eight weeks of ethanol feeding.
Supplementary Material
Acknowledgments
We thank Dr. Murray B. Resnick, Dr. Patricia Meitner, Dr. Jason T. Machan, Virginia Hovanesian and Paul Monfils for their technical assistance. The excellent technical assistance of Susan Lang in performing the euglycemic hyperinsulinemic clamps is gratefully acknowledged.
Financial support
Supported in part by NIH Grants AA002666, AA008169 (J.R.W.), AA011431, AA012908 (S.M.D.) and AA11290 (C.H.L.)
List of abbreviations
- LE
Long-Evans
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- F
Fischer
- SD
Sprague-Dawley
- PUMA
p53 up-regulated modulator of apoptosis
- TIGAR
Tp53-induced glycolysis and apoptosis regulator
- Fru-2
6-P2, fructose-2,6-bisphosphate
- ROS
reactive oxygen species
- ALD
alcoholic liver disease
- ALT
alanine aminotransferase
- ADH
ethanol dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- DCF
2′,7′-dichlorodihydrofluorescin diacetate
- LX-PCR
long-extension polymerase chain reaction
- mtDNA
mitochondrial DNA
- EMSA
electrophoretic mobility shift assay
- SREBP1c
sterol regulatory element-binding protein 1c
- CYP2E1
cytochrome P450 2E1
- IRS-2
insulin receptor substrate 2
- PARP
poly (ADP-ribose) polymerase
- ISOL
in situ oligo ligation
- SCO2
synthesis of cytochrome c oxidase 2
- HGP
hepatic glucose production
- Ra
rate of glucose appearance
- Rd
rate of glucose disappearance
Footnotes
The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsukamoto H. Conceptual importance of identifying alcoholic liver disease as a lifestyle disease. J Gastroenterol. 2007;42:603–609. doi: 10.1007/s00535-007-2075-3. [DOI] [PubMed] [Google Scholar]
- 2.Wilfred de Alwis NM, Day CP. Genetics of alcoholic liver disease and nonalcoholic fatty liver disease. Semin Liver Dis. 2007;27:44–54. doi: 10.1055/s-2006-960170. [DOI] [PubMed] [Google Scholar]
- 3.de la Monte SM, Yeon JE, Tong M, Longato L, Chaudhry R, Pang MY, et al. Insulin resistance in experimental alcohol-induced liver disease. J Gastroenterol Hepatol. 2008;23:e477–486. doi: 10.1111/j.1440-1746.2008.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeon JE, Califano S, Xu J, Wands JR, De La Monte SM. Potential role of PTEN phosphatase in ethanol-impaired survival signaling in the liver. Hepatology. 2003;38:703–714. doi: 10.1053/jhep.2003.50368. [DOI] [PubMed] [Google Scholar]
- 5.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 6.Pani G, Fusco S, Colavitti R, Borrello S, Maggiano N, Cravero AA, et al. Abrogation of hepatocyte apoptosis and early appearance of liver dysplasia in ethanol-fed p53-deficient mice. Biochem Biophys Res Commun. 2004;325:97–100. doi: 10.1016/j.bbrc.2004.09.213. [DOI] [PubMed] [Google Scholar]
- 7.Lieber CS, Leo MA, Wang X, Decarli LM. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373:246–252. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, Migliaccio E, et al. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 9.Pani G, Koch OR, Galeotti T. The p53-p66shc-Manganese Superoxide Dismutase (MnSOD) network: a mitochondrial intrigue to generate reactive oxygen species. Int J Biochem Cell Biol. 2009;41:1002–1005. doi: 10.1016/j.biocel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Pang M, de la Monte SM, Longato L, Tong M, He J, Chaudhry R, et al. PPARdelta agonist attenuates alcohol-induced hepatic insulin resistance and improves liver injury and repair. J Hepatol. 2009;50:1192–1201. doi: 10.1016/j.jhep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruni P, Vasta V, Farnararo M. An endpoint enzymatic assay for fructose 2,6-bisphosphate performed in 96-well plates. Anal Biochem. 1989;178:324–326. doi: 10.1016/0003-2697(89)90646-5. [DOI] [PubMed] [Google Scholar]
- 12.Lang CH, Dobrescu C, Bagby GJ. Tumor necrosis factor impairs insulin action on peripheral glucose disposal and hepatic glucose output. Endocrinology. 1992;130:43–52. doi: 10.1210/endo.130.1.1727716. [DOI] [PubMed] [Google Scholar]
- 13.Fulop P, Derdak Z, Sheets A, Sabo E, Berthiaume EP, Resnick MB, et al. Lack of UCP2 reduces Fas-mediated liver injury in ob/ob mice and reveals importance of cell-specific UCP2 expression. Hepatology. 2006;44:592–601. doi: 10.1002/hep.21310. [DOI] [PubMed] [Google Scholar]
- 14.Derdak Z, Mark NM, Beldi G, Robson SC, Wands JR, Baffy G. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 2008;68:2813–2819. doi: 10.1158/0008-5472.CAN-08-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Z, Wanagat J, McKiernan SH, Aiken JM. Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection. Nucleic Acids Res. 2001;29:4502–4508. doi: 10.1093/nar/29.21.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP) J Biol Chem. 2002;277:29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- 17.You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798–1808. doi: 10.1053/j.gastro.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 18.Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol. 2004;6:351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- 19.Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 20.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 21.Younossi ZM. Review article: current management of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2008;28:2–12. doi: 10.1111/j.1365-2036.2008.03710.x. [DOI] [PubMed] [Google Scholar]
- 22.Brown MD, Wallace DC. Molecular basis of mitochondrial DNA disease. J Bioenerg Biomembr. 1994;26:273–289. doi: 10.1007/BF00763099. [DOI] [PubMed] [Google Scholar]
- 23.Johns DR. Seminars in medicine of the Beth Israel Hospital, Boston. Mitochondrial DNA and disease. N Engl J Med. 1995;333:638–644. doi: 10.1056/NEJM199509073331007. [DOI] [PubMed] [Google Scholar]
- 24.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Werneburg NW, Akazawa Y, et al. JNK1-dependent PUMA expression contributes to hepatocyte lipoapoptosis. J Biol Chem. 2009;284:26591–26602. doi: 10.1074/jbc.M109.022491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007;46:1791–1800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- 27.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Jogl G. Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator) J Biol Chem. 2009;284:1748–1754. doi: 10.1074/jbc.M807821200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Khan SA, Peng LJ, Li H, Carmella SG, Lange AJ. Perturbation of glucose flux in the liver by decreasing F26P2 levels causes hepatic insulin resistance and hyperglycemia. Am J Physiol Endocrinol Metab. 2006;291:E536–543. doi: 10.1152/ajpendo.00126.2006. [DOI] [PubMed] [Google Scholar]
- 30.Kang L, Sebastian BM, Pritchard MT, Pratt BT, Previs SF, Nagy LE. Chronic ethanol-induced insulin resistance is associated with macrophage infiltration into adipose tissue and altered expression of adipocytokines. Alcohol Clin Exp Res. 2007;31:1581–1588. doi: 10.1111/j.1530-0277.2007.00452.x. [DOI] [PubMed] [Google Scholar]
- 31.Pessayre D, Berson A, Fromenty B, Mansouri A. Mitochondria in steatohepatitis. Semin Liver Dis. 2001;21:57–69. doi: 10.1055/s-2001-12929. [DOI] [PubMed] [Google Scholar]
- 32.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 33.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.