Summary
Abscisic acid is a plant hormone with important functions in stress protection and physiology. Recently, the PYR/PYL/RCAR family of intracellular ABA receptors was identified. These receptors directly link ABA perception to a canonical ABA signaling pathway, in which ABA-bound receptors bind and inhibit type 2C phosphatases. High resolution crystal structures of members of this family have been solved in all relevant states: as apo receptors, bound to ABA, and as receptor-ABA-phosphatase complexes. Together, these structures provide a detailed gate-latch-lock mechanism of ABA recognition, receptor-PP2C interaction, and inhibition of the PP2C phosphatase activity and provide a basis for the design of synthetic ABA agonists for stress protection of crop plants.
Introduction
Plant growth and development is regulated by small molecule hormones and growth regulators that initiate signaling cascades upon binding to specific receptors. Abscisic acid (ABA) is an important phytohormone, which inhibits growth and development under unfavorable environmental conditions and protects plants against stresses, such as drought, salinity, cold, and pathogen exposure. Stresses induce strong increases in ABA levels by induction of ABA biosynthetic genes, enzymatic release from inactive glucose esters, and local increases by transport within plants. Elevated ABA levels in turn re-program plants to survive adverse conditions. ABA-mediated protective measures include closure of stomata, which are the pores in leaves for CO2/O2 exchange, to prevent water loss by transpiration, synthesis of osmo-protective substances, seed dormancy, inhibition of fruit ripening, shedding (“abscission”) of leaves, and down-regulation of enzymes needed for photosynthesis [for recent review see 1].
In addition to its fundamental roles in plant physiology, ABA also has the potential for important applications in agriculture. Globally, fresh water supplies are alarmingly decreasing and drought, salinity, cold, and other abiotic stresses are responsible for major crop harvest losses world-wide [2–4]. While survival under these conditions can be improved by timed exogenous application of ABA, ABA is costly, difficult to synthesize, and relatively unstable under field conditions. Therefore, there is interest in generating synthetic small molecule ABA receptor agonists for agricultural applications that are affordable, stable, and environmentally friendly.
Core ABA signaling pathway
Key components of the ABA signaling pathway that have been identified genetically in Arabidopsis are type 2C protein phosphatases (PP2Cs), subfamily 2 of Snf1-related protein kinases (SnRK2s), and phosphorylation-dependent basic leucine-zipper transcription factors called ABFs (ABA box-binding factors). However, genetic or biochemical identification of the receptors that bind ABA and initiate the ABA signal cascade had been hampered by high receptor redundancy and low ABA affinities. About a year ago, a subfamily of START domain-containing proteins were identified as best candidates for bona fide ABA receptors. These receptors bind to ABA and link their ABA-binding to the known components of the ABA signaling pathway. Members of this family were identified as PP2C interactors in yeast two-hybrid screens [5,6] and by resistance to a synthetic, ABA receptor-selective, agonist termed pyrabactin [7]. The remaining members of this family could readily be identified by sequence homology as PYR1-like (PYL) proteins or as regulatory component of ABA receptors (RCARs). ABA binds the PYR/PYL/RCAR receptors with micromolar affinities [5,6] and induces the receptors to bind to ABA-responsive PP2Cs, including ABI1, ABI2, and HAB1, and to inhibit their phosphatase activity. Notably, the affinity of ABA to receptor-PP2C complexes was at least an order of magnitude higher than to apo receptors [5,6], which raised the possibility that PP2Cs may function as ABA co-receptors.
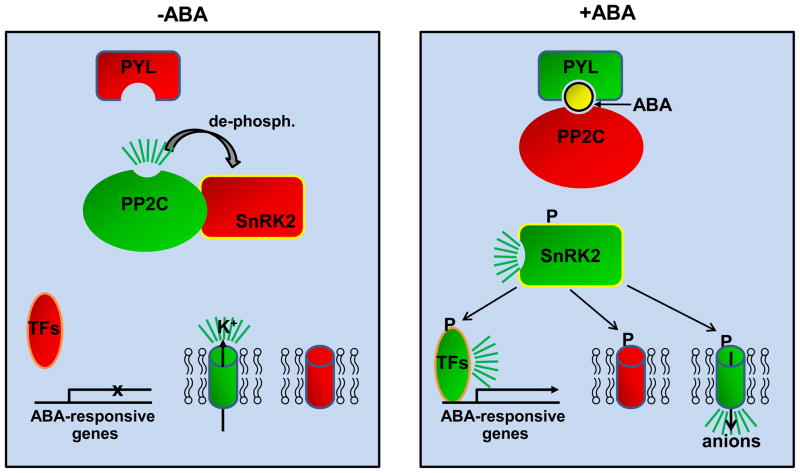
How do PYR/PYL/RCAR receptors affect the PP2C-SnRK2 signaling pathway? In the absence of ABA, the PP2Cs ABI1, ABI2, and HAB1 bind SnRK2s and dephosphorylate a phospho-serine residue in the SnRK2 activation loop, whose phosphorylation is required for SnRK2 kinase activity [7–12]. ABA/PYL-induced PP2C inhibition therefore restores phosphorylation of the SnRK2 activation loop, by autophosphorylation and/or phosphorylation by putative upstream kinases, and allows SnRK2s to phosphorylate and activate downstream targets, such as ABF transcription factors [11,13,14] and the ion channels responsible for turgor-mediated stomatal closure [15–17] (Fig. 1). Overexpression of PYL5 [6] or PYL9 [5] increase ABA sensitivity and triple pyr1/pyl1pyl4 and quadruple pyr1/pyl1/pyl2/pyl4 mutants display seed germination ABA insensitivity in vivo [7]. Similarly, a loss of function triple mutant defective in the three strongly ABA-activated kinases SnRK2.2, SnRK2.3, and SnRK2.6 is defective in all known ABA functions [18,19] and loss of function triple mutants defective in the PP2Cs ABI1, ABI2, and HAB1 are extremely ABA-hypersensitive [20]. While ABA signaling also involves subfamily 1 and 3 SnRK kinases, Ca-dependent kinases, MAP kinases and secondary messengers Ca2+, ROS, phosphatidic acid, nitric oxide, and others [21], the mutational data indicate that these additional signals at least largely feed into the core PYL/RCAR-PP2C-SnRK2 pathway. One example for signal integration involves the generation of ROS. SnRK2.6 phosphorylates and activates NADPH oxidase to generate H2O2 [22], which in turn inactivates the PP2C s ABI1 [23] and ABI2 [24] leading to increased ABA signaling and stomatal closure. In addition, an ABA signaling pathway can be reconstituted in vitro using only PYR1, ABI1, and SnRK2.6 for the ABA-dependent phosphorylation of the transcription factor ABF2 as well as in protoplast reporter gene assays using transient transfections of PYR1, ABI1, and SnRK2.6 expression plasmids, further demonstrating that PYR/PYL/RCAR receptors, PP2Cs, and SnRK2s are both necessary and sufficient to confer the ABA signal to an ABA-dependent transcription factor [8].
Figure 1.
Schematic presentation of the core ABA signaling pathway. Active components are shown in green, inactive ones in red, and ABA as yellow ball. Cylinders depict ion channels that regulate stomatal closure. TFs: transcription factors, P:phosphate.
PYR/PYL/RCAR crystal structures
Identification of the PYR/PYL/RCARs as ABA receptors led to a series of high profile publications of receptor structures that validated the PYR/PYL/RCARs as bona fide ABA receptors [25-29]. Solving the structures of apo and ABA-bound receptors as well as ABA/PP2C-bound complexes led to the identification of the detailed molecular mechanism of ABA recognition, receptor-PP2C interaction, and inhibition of the PP2C phosphatase activity. PYR/PYL/RCAR receptors belong to the large superfamily of START/Bet v I proteins, which share a common fold enclosing large hydrophobic ligand binding pockets that can bind a chemically diverse set of lipids, hormones, and antibiotics [30]. Crystal structures of PYR1, PYL1, and PYL2 receptors revealed a helix grip structure characteristic for START/Bet v I proteins, in which a large C-terminal α-helix (α3) is enfolded by a seven-strand anti-parallel β-sheet and two small α-helices [25,29] [26–28](Fig. 2a). Between the C-terminal helix and the β-sheet resides a large open ligand-binding pocket. The entry to the ligand binding pocket is flanked by two β-loops that we have named gate and latch [25] (alternatively named Pro-Cap and Leu-Lock [27], CL2 and CL3 [29], and β3-β4 and β5-β6 lid loops [26]) and whose amino acid sequences (SGLPA and HRL, resp.) are, with a single exception (PYL13), identical for all 14 members of the PYR/PYL/RCAR family. Unlike PYL1[25] and PYL2 [25,29], PYR1 has not been crystallized in the absence of ligand. However, PYR1 crystallized in the presence of equimolar [27] or slightly sub-equimolar [28] amounts of ABA revealed dimers in which only one of the two monomers was bound to ABA [27,28], therefore providing information on both ABA-bound and free PYR1 (see Fig. 2a for overlap of apo-PYL1, -PYL2, and -PYR1 structures). In the presence of excess ABA, PYR1, just as PYL1 [26] and PYL2 [25,29], bind ABA with 1:1 stochiometry as determined by ITC [28] and small-angle X-ray scattering [27].
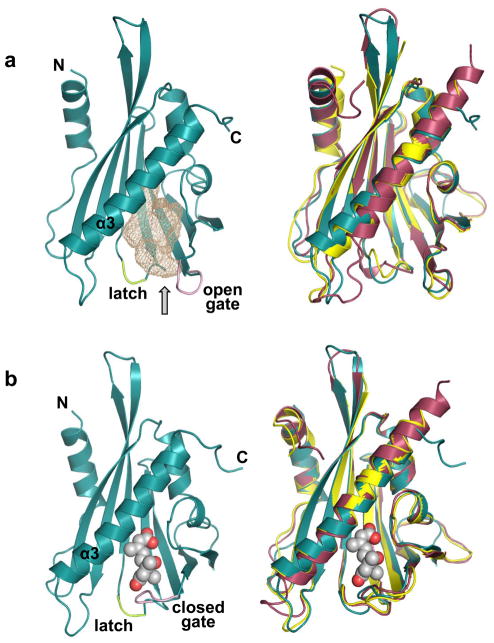
Figure 2.
Structures of apo and ABA-bound receptors. The gate loop is shown in pink and the latch loop in green. a. Structure of apo PYL2 (left) with the empty ligand binding pocket shown as pink mesh and overlay of the apo PYL2 (cyan), PYR1 (purple), and PYL1 (yellow) structures (right). N- and C-termini as well as the large helix (α3) are labeled. The arrow points to the entry to the ABA-binding pocket. b. Structure of ABA-bound PYL2 (left) and overlay of ABA-bound PYL2 (cyan), PYR1 (purple), and PYL1 (yellow) (right). ABA is shown as ball model.
The overall structures of ABA-bound PYL1, PYL2, and PYR1 are very similar to the structures of the apo receptors with the exception of the gate and latch loops. The gate loops of the PYL1, PYL2, and PYR1 receptors undergo a major conformational change shifting in its entirety towards the latch loop [25,27–29] (Fig. 2b). This shift has two important consequences. First, it closes the entrance to the ligand binding pocket, thereby shielding ABA inside of the receptors from solvent exposure. Second, closing onto the latch loop forms a new gate-latch interface, which is sterically facilitated by an outside flip of the glutamate that directly precedes the HRL latch [25,27–29]. Thus, ABA induces conformational changes of the gate and latch loop, which serves to transmit the hormone binding signaling to downstream effectors as detailed below.
ABA-receptor interactions
Fig. 2b gives an overview of ABA in the ligand binding pockets of PYR1, PYL1, and PYL2 and Fig. 3a shows a schematic presentation of all ABA-binding pocket interactions. ABA fits tightly into the shape of the pocket and makes a series of interactions with conserved receptor residues that involve all parts of ABA. The ABA carboxyl group is anchored opposite of the pocket entrance by two direct charge interactions with a conserved lysine (K64/K86/K59 in PYL2/PYL1/PYR1) as well as water-mediated interactions with, in the case of PYL2, N173, E98, and E147, residues that are identical in all receptors but PYL13 (which has a glutamine at the position of the lysine residue). In this position, the cyclohexene ring of ABA can pull the gate into the closed conformation by directly interacting with the three consecutive gate residues LPA, a conformation that is further stabilized by gate-latch interactions. The importance of gate-ligand interaction for gate closure is supported by structural and mutational data, demonstrating that the synthetic PYL ligand pyrabactin can adopt different orientations within PYL ligand binding pockets and only orientations that allow pyrabactin-gate interactions promote gate closure [31–34] (see below). Single amino acid mutations of the gate residues as well as the charged residues anchoring the ABA carboxyl group all result in compromised ABA-binding, PP2C interaction, and PP2C inhibition [25–27,29], demonstrating the importance of these residues for ABA binding and signaling.
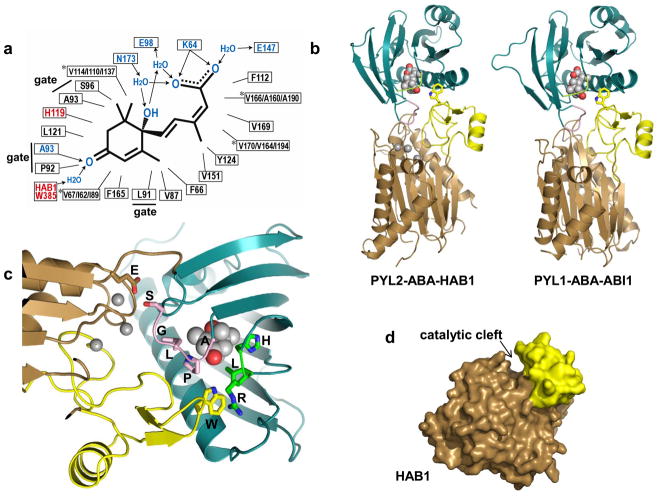
Figure 3.
Structure of ternary receptor-ABA-PP2C complexes. a. Schematic presentation of the interactions of ABA in receptor-ABA and receptor-ABA-PP2C complexes. Numbers refer to the amino acid positions within PYL2 (black for hydrophobic interactions and blue for polar interactions) and HAB1 (red). Note that the same PYL2 amino acids interact with ABA in the contexts of PYL-ABA and PYL2-ABA-HAB1. *: ABA-binding residues that differ between PYL2, PYL1, and PYR1, shown in the order PYL2/PYL1/PYR1. Arrows indicate the direction from electron donors to electron acceptors. b. Overview of the PYL2-ABA-HAB1 and PYL1-ABA-ABI1 structures. ABA is shown in ball presentation, PYL2 and PYL1 in blue with the gate and latch loop in pink and green. The catalytic PP2C domain is shown in brown with Mg2+ ions as balls and the protruding, receptor-interacting PP2C domain in yellow, with the key tryptophan residue in stick presentation. c. Close-up of the PYL2-HAB1 interface (same color code as above). Gate and latch residues as well as HAB1 E203 and W385 are shown in stick presentations and are labeled in single amino acid code. d. Surface presentation of HAB1 in the PYL2-ABA-HAB1 complex (PYL2-ABA removed). The protruding, receptor-interacting PP2C domain is shown in yellow
While the unnatural, biologically less active (−)-ABA isoform can be accommodated by the PYR1 binding pocket [27], the shape of the pocket sterically favors binding of the (+)-ABA isoform [25,27,28], providing a structural basis for the low efficiency of (−)-ABA binding [26]. In addition to understanding the atomic details of ABA perception, the detailed ABA-binding pocket interaction map may also provide a blueprint to generate synthetic ABA agonists to increase crop stress survival in agriculture.
PYL/ABA/PP2C ternary crystal structures reveal the mechanism of ABA-induced PP2C inhibition
The catalytic PP2C cores are sufficient for PYL/ABA-PP2C interaction [5,6] and crystal structures of ternary PYL2-ABA-HAB1 [25] and PYL1-ABA-ABI1 [26,29] PP2C core complexes showed overall catalytic PP2C folds consisting of a central β-sandwich flanked by two pairs of α-helices plus a small protruding domain of two anti-parallel β-strands and a short α-helix sticking out from the core (Fig. 3b), a fold very similar to that of human PP1A/PP2Cα [35]. Catalysis by PP2Cs is thought to require water-mediated binding of substrate phospho-serine/-threonine to Mg2+ or Mn2+ ions complexed at the bottom of an active site cleft [35] with the protruding domain forming the upper part of the cleft wall (Fig. 3b–d). ABA-bound PYL1 and PYL2 bind both the active site at the bottom of the cleft as well as to the inside of the protruding upper cleft wall, thereby inhibiting PP2C activity by blocking substrate entrance to the catalytic site [25,26,29]. These interactions are mediated by the gate-latch interface that is induced by ABA binding (Fig. 4) as well as the N-terminal part of the α3 helix. The SGLPA gate loop directly packs against the active PP2C site with the gate serine residue S89 [PYL2]/S112[PYL1]) contacting an Mg2+-coordinating glutamate residue in the PP2C active site loop. The protruding domain is the main interaction site with the PP2Cs. A tryptophan side chain (W385 in HAB1, W300 in ABI1) in the protruding domain sticks out from the surface and binds a hydrophobic cavity in ABA-bound PYL1 and PYL2 that is in part formed by and makes key contacts with the HRL latch loop. In this orientation, the tryptophan indole ring makes a water-mediated hydrogen bond with the ketone group of ABA and binds and causes conformational changes in both latch and gate residues (see Fig. 4). These interactions lock the gate and latch loops into the closed conformation, thus providing a structural rationale for the more than tenfold increase in ABA-binding affinity for PYL-PP2C complexes compared to PYL apo receptors [5,6,26,29,36]. PP2Cs can therefore function as ABA-co-receptors that interact with ABA in the receptor binding pocket and increase receptor ABA-binding affinity. Fig. 5 presents a summary cartoon of the structural states from apo receptor to ternary receptor-ABA-PP2C complex.
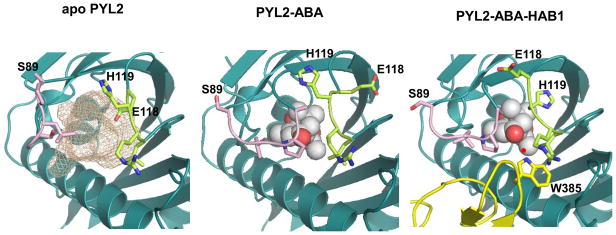
Figure 4.
Conformational changes in the gate (pink) and latch (green) loops. The PP2C protruding domain with W385 is shown in yellow, ABA as ball model. The small red ball represents the water molecule that mediates the interaction between HAB1 W385 and the ABA ketone group.
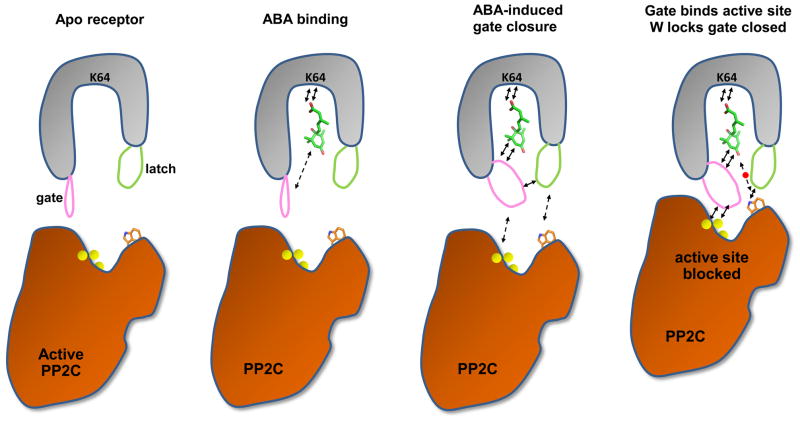
Figure 5.
Summary cartoon. ABA binding induces closure of the gate loop (pink) onto the latch (green). The new gate-latch interface binds in turn to the active site and the protruding domain of PP2Cs, which inhibits PP2C activity by blocking substrate access to the active site. Arrows indicate key interactions, yellow balls the catalytic Mg2+ ions, and the small red ball the water molecule that mediates the interaction between HAB1 W385 and the ABA ketone group.
The importance of the structural elements for receptor-PP2C interaction is supported by extensive mutagenesis data. Amino acid replacements in both the gate and latch loops of the receptors [25,26][29] as well as mutations of ABI1 W300 and the glutamate residue in the ABI1 active site [26] all compromise ABA-induced receptor-PP2C interactions. While most gate and latch residues stabilize PP2C interaction also indirectly by stabilizing ABA-binding, the histidine residue in the latch and the serine residue in the gate point away from the ABA binding pocket in apo and ABA-bound receptors, yet mutations of these residues significantly impair PP2C binding [25,29]. Finally, the ABA-insensitive abi1-1 and abi2-1 mutations [37] alter a glycine in the PP2C active site to an aspartic acid, which disrupts receptor-PP2C interactions [5–7,37]. In receptor-PP2C complexes, this glycine is in the immediate vicinity of the gate, and exchange of the glycine hydrogen with the bulky side chain of aspartate sterically interferes with gate binding [25,26,29].
Receptor dimerization may reduce receptor-PP2C interaction in the absence of ABA
Both apo and ABA-bound receptors are in dimeric configuration in crystal packing [25–29] and they form weak dimers in solution [27–29], yet they are monomeric in the PP2C complexes with 1:1:1 receptor:ABA:PP2C stochiometry[25–29]. The gate loops form part of the dimerization surface implying that the receptors need to dissociate in order to associate with PP2Cs. Receptor dimerization may therefore be a mechanism to reduce basal receptor-PP2C interaction [27–29]. While ABA-binding does not disrupt receptor dimerization, it slightly destabilizes receptor-receptor interaction [29], consistent with a possible role of receptor dimerization in blocking uninduced PP2C interaction [29]. Interestingly, some receptors have high basal activity [5–7,25] and PYL5 constitutively binds the PP2C HAB1, yet requires ABA to inhibit PP2C activity [6]. However, the basis for these receptor differences is unknown and currently functional data linking receptor dimerization to inhibition of PP2C interaction is lacking.
Pyrabactin is a receptor-selective ABA agonist and antagonist
Pyrabactin was identified as a synthetic, receptor-selective, ABA agonist that led to the identification of PYR1 [7,38; see above]. While pyrabactin activates PYR1, PYL1 and other receptors [5–7,25], it does not [32], or only to a limited fraction [33,34], activate PYL2. Unexpectedly, pyrabactin did not only fail to efficiently activate PYL2, but can also antagonize ABA-mediated activation of PYL2 [32]. Crystal structures of productive ABA receptor-pyrabactin complexes [PYL1-pyrabactin [31], PYR1-pyrabactin [33] and PYL1-pyrabactin-ABI1 [32]] as well as of the non-productive, ABA-antagonizing, PYL2-pyrabactin complex [32–34] revealed that pyrabactin can adopt different conformations within the highly conserved ligand binding pockets of these receptors. These different pyrabactin orientations either provide or lack the Van-der-Waals interactions required to induce gate closure [32–34] as demonstrated by introduction of a bulky phenylalanine residue into the PYL2 gate, which rescues formation of pyrabactin-gate interactions [32]. The importance of pyrabactin orientations is validated by mutational and structural data demonstrating that a single isoleucine (PYR1, PYL1) versus valine (PYL2) position in the ligand binding pockets is responsible for both pyrabactin orientation and agonism/antagonism properties [32–34]. In addition, the detailed structural information on pyrabactin-receptor interactions provided insights into “rules” required for the productive binding of small molecules into the ABA receptor ligand binding pockets [31]and allowed the structure-guided identification of novel pyrabactin-based synthetic ABA agonists [32]. The ability of pyrabactin to antagonize PYL2 raises the possibility that physiological ABA antagonist may exist to inhibit basal activities of ABA receptors [32].
Conclusions
The structural analyses have provided a detailed mechanistic understanding of ABA perception, ABA-induced PYL2/PYL1/PYR1-PP2C interaction, and the inhibition of PP2C phosphatase activity. However, important questions beyond ABA perception and the first step of ABA signaling remain to be addressed. The most important one of these will be to solve a PP2C-SnRK2 co-structure as well as PP2C and SnRK2 apo-structures. The Arabidopsis SnRK2 kinase domains are highly homologous to the kinase domain of human AMP-activated kinase (AMPK), an important drug target that is also regulated by PP2C-catalyzed dephosphorylation [39,40]. A SnRK2-PP2C co-structure may therefore also be relevant to the understanding of AMPK regulation. Second, it is currently poorly understood why the ABA signaling pathway in Arabidopsis involves so many different PYR/PYL/RCARs receptors. While individual receptors differ in their sensitivity, selectivity, and response towards ABA [5–7,25,36], the structural reasons for these differences are unknown. All structures solved today contain only members of the PYR1/PYL1/PYL2/PYL3 subclass of receptors. Structural and biochemical analysis of receptors of the two remaining PYL/RCAR subclasses are bound to provide important information to understand the differences between the three receptor subclasses. Third, the structures of ABA in the receptor binding pockets and in the context of receptor-PP2C complexes hold a promise for the development of ABA agonists and antagonists for use in agriculture that awaits tapping. Finally, we are just beginning to understand how other components of the ABA signaling pathway integrate into the PYL/RCAR-PP2C-SnRK2 pathway [41] and much work need to be done to gain a global mechanistic synthesis of the complete signaling network.
Acknowledgments
This work was supported by the Jay and Betty Van Andel Foundation and by National Institute of Health grants HL089301, GM087413, DK071662, and DK066202 to H.E.X.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Karsten Melcher, Email: karsten.melcher@vai.org.
X. Edward Zhou, Email: edward.zhou@vai.org.
H. Eric Xu, Email: eric.xu@vai.org.
References and recommended reading
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Delmer DP. Agriculture in the developing world: Connecting innovations in plant research to downstream applications. Proc Natl Acad Sci U S A. 2005;102:15739–15746. doi: 10.1073/pnas.0505895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, et al. Radically rethinking agriculture for the 21st century. Science. 2010;327:833–834. doi: 10.1126/science.1186834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. In this paper the family of intracellular ABA receptors was identified as ABI1 and ABI2 interactors and was named RCARs (regulatory components of ABA receptors). The authors demonstrated that RCARs directly bind (+)-ABA and that ABA mediates PP2C binding and inhibition. [DOI] [PubMed] [Google Scholar]
- 6•.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03981.x. The authors describe identification of PYL5, PYL6, and PYL8 as HAB1 interactors and characterize the ABA-dependent HAB1 interaction and inhibition by PYL5. [DOI] [PubMed] [Google Scholar]
- 7••.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. Park et al. exploited the ABA receptor subtype-selective synthetic ABA agonist pyrabactin to genetically identify PYR1 as founding member of the PYR1/PYL family. They demonstrated that PYR1 binds ABA and that PYR1 and PYR1-like PYL proteins bind and inhibit PP2Cs in the presence of ABA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. This paper biochemically demonstrates that PYL receptors, PP2C, SnRK2, and ABF2 are sufficient to initiate a cascade of ABA dependent activation of SnRK2 kinase and ABF2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 10.Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci U S A. 2009;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci U S A. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 18.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci U S A. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 20.Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Meinhard M, Grill E. Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 2001;508:443–446. doi: 10.1016/s0014-5793(01)03106-4. [DOI] [PubMed] [Google Scholar]
- 24.Meinhard M, Rodriguez PL, Grill E. The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta. 2002;214:775–782. doi: 10.1007/s00425-001-0675-3. [DOI] [PubMed] [Google Scholar]
- 25••.Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. This paper describes crystal structures of PYL1 and PYL2 in apo form as well as crystal structures of the binary PYL2-ABA and the ternary PYL2-ABA-HAB1 complexes. In combination with biochemical, mutational, NMR, and in planta data, it presents a comprehensive mechanism from ABA perception to PP2C inhibition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. Miyazono et al. report the structures of ABA-bound PYL1 and of the PYL1-ABA-ABI1 ternary complex. These structures, in combination with biochemical and mutational data, reveal the mechanism of ABA-induced PP2C inhibition. [DOI] [PubMed] [Google Scholar]
- 27••.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. The authors present the structure of dimeric PYR1 in the presence of ABA. The dimer contained one monomer in the apo form and one in the ABA-bound state, thereby providing details of ABA perception and ABA-induced conformational changes in PYR1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. Santiago et al. have also solved the structure of a PYR1/PY1-ABA heterodimers and provide biochemical data supporting a role of the gate loop in PP2C binding. [DOI] [PubMed] [Google Scholar]
- 29••.Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. This paper reports crystal structures of PYL2 in apo form, ABA-bound form, and as ternary complex with ABA and ABI1. Together with biochemical and mutational data, the authors present a comprehensive mechanism from ABA perception to PP2C inhibition. They also demonstrate that ABA binding weakens PYL2 dimerization, providing a model for a possible functional role in receptor dimerization. [DOI] [PubMed] [Google Scholar]
- 30.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Q, Yin P, Yan C, Yuan X, Li W, Zhang Z, Liu L, Wang J, Yan N. Functional mechanism of the ABA agonist pyrabactin. J Biol Chem. 2010 doi: 10.1074/jbc.M110.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, Suino-Powell KM, Kovach A, Tham FS, Cutler SR, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson FC, Burgie ES, Park SY, Jensen DR, Weiner JJ, Bingman CA, Chang CE, Cutler SR, Phillips GN, Jr, Volkman BF. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan X, Yin P, Hao Q, Yan C, Wang J, Yan N. Single amino acid alteration between Valine and Isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. J Biol Chem. 2010 doi: 10.1074/jbc.M110.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 36.Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 37.Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci U S A. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, Cutler SR. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol. 2007;3:716–721. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
- 39.Fogarty S, Hardie DG. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta. 1804:581–591. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 41.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010 doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]