Abstract
The U.S. Tox21 collaborative program represents a paradigm shift in toxicity testing of chemical compounds from traditional in vivo tests to less expensive and higher throughput in vitro methods to prioritize compounds for further study, identify mechanisms of action, and ultimately develop predictive models for adverse health effects in humans. The NIH Chemical Genomics Center (NCGC) is an integral component of the Tox21 collaboration due to its quantitative high throughput screening (qHTS) paradigm, in which titration-based screening is used to profile hundreds of thousands of compounds per week. Here, we describe the Tox21 collaboration, qHTS-based compound testing, and the various Tox21 screening assays that have been validated and tested at the NCGC to date.
Keywords: Tox21, National Research Council, National Toxicology Program, toxicity testing, in vitro assays, NIH Roadmap, NIH Chemical Genomics Center, quantitative high-throughput screening
Introduction
Traditionally, the toxicological evaluation of environmental chemicals has largely relied on animal models that have been used to extrapolate to potentially harmful events in humans. These models have been developed to evaluate specific toxicological endpoints such as oral, dermal and ocular toxicity; immunotoxicity; genotoxicity; reproductive and developmental toxicity; and carcinogenicity. While these animal models have provided useful information on the safety of chemicals, they are relatively expensive, low-throughput, and sometimes inconsistently predictive of human biology and pathophysiology. Recently, several major new initiatives have begun to utilize in vitro methods and a variety of new technologies to develop in vitro signatures and computational models predictive of in vivo response. These initiatives should enable researchers to identify a battery of in vitro assays that will detect perturbations in cellular pathways that are expected to contribute to or result in adverse health effects [1]. Furthermore, these initiatives represent a welcome movement away from traditional in vivo high dose hazard studies [1]. To appreciate the scientific and technological advancements that are shaping toxicity testing today, it is important to appreciate where this new paradigm fits in the context of historical testing.
Traditional Toxicity Testing Methods
Since its inception, toxicity testing has relied on animal models treated at maximum tolerated dose levels, with the results extrapolated to human health outcomes at lower doses. This approach dates back to the 1950s, when the utilization of more specific or mechanistic animal models, and knowledge of the underlying mechanisms for any particular toxicological response, were relatively unknown [2]. Such in vivo testing is costly, time consuming and low throughput [3]. The complete toxicological profiling of one chemical in standard in vivo assays consisted of the following toxicity tests: acute, sub-chronic, and chronic toxicity; reproductive toxicity; developmental toxicity, ocular and skin irritation, hypersensitivity; phototoxicity; and toxicokinetic studies [4]. Despite the disadvantages associated with testing in animals, the majority of the understanding regarding chemical toxicity has come from data obtained in such systems [5]. However, even extensive animal testing does not provide a mechanistic understanding of toxicity, and knowledge concerning adverse risks to humans is still inadequate [6]. Hence, a need for more mechanistic data and “theoretical framework for rational decision making” was noted in the early 1980s [6].
More recently, there have been numerous studies highlighting intra- and inter-species differences in mammals, including humans. Williams and Weisburger [7] pointed out that intra-species differences among different mouse strains affect the severity and incidence of neoplasms, making extrapolation of various cancers from mice to humans difficult. Inherent resistance to spontaneous and malignant tumors in nonhuman primate models has also led to the variation in the manifestation of disease across these species [8]. In addition to inter- and intra-species differences in disease models, other species-specific differences that affect disease outcome and extrapolations include differences in basal metabolic rate, metabolic pathways, cancer type (sarcomas in mice versus carcinomas in humans), genetic aberrations associated with tumors, and telomere biology, especially with regard to humans and mice [9]. In addition to physiologic differences, the difference in observed high dose toxicity in rodents and low dose risks in humans will require knowledge of physiological differences with regard to mode, tissue of exposure, mechanism of action, and knowledge of previous in vitro data regarding the agent in question.
Toxicity Testing in the 21st Century and the U.S. Tox21 Partnership
The advent of technological innovations in molecular and cellular biology prompted the National Toxicology Program (NTP) to propose a new Roadmap in 2004, “A National Toxicology Program for the 21st Century,” [10] focusing on three main areas: refining traditional toxicology assays, developing rapid mechanism-based predictive screens, and improving the overall utility of data for making public health decisions. This Roadmap placed an increased emphasis on the use of alternative assays for identifying key pathways and molecular mechanisms linked to disease [10] The U.S. Environmental Protection Agency (EPA) started its ToxCast program in 2006 to address many of the same issues [11]. While these programs were in their early stages, a 2007 report from the National Research Council (NRC) entitled Toxicity Testing in the 21st Century (Tox21) enunciated what has become a widely accepted vision for future toxicology testing, calling for the development and utilization of in vitro models in human cells of toxicological response based on automated high throughput screening (HTS) of pathway-based cellular assays related to toxicity and computational modeling [12]. The report envisioned that initially, such less expensive and higher throughput assays could be used to evaluate the modes of action of chemicals for more comprehensive testing programs and that eventually these data would allow for the rapid and mechanism-based prediction of in vivo biological responses [2,13,14].
In order to move this research agenda forward, the NTP partnered with the NIH Chemical Genomics Center (NCGC) in 2005 to pilot the chemical, biological, and informatics processes required for the transition from predominantly in vivo to in vitro toxicology. In 2006, this partnership was expanded to include the EPA. In 2008, in recognition of successful proof-of-principle studies [3,15] and galvanized by the NRC report, the “Tox21” collaboration was formally established via a Memorandum of Understanding among the agencies (see: http://ntp.niehs.nih.gov/28213) and publication of a policy paper from the senior leadership of the three organizations [16].
The Tox21 collaboration takes advantage of the complementary strengths of the three partners (Figure 1). The NTP, a trans-Department of Health and Human Services (DHHS) program headquartered at the NIH National Institute of Environmental Health Sciences (NIEHS), has enormous experience in experimental toxicology. The NCGC, a trans-NIH program administered by the National Human Genome Research Institute (NHGRI), has unparalleled capacity and expertise in vitro assays, titration-based screening, and informatics. The EPA National Center for Computational Toxicology (NCCT), part of the EPA’s Office of Research and Development, has deep computational toxicology expertise [14]. This combined expertise has allowed for rapid implementation of the NRC vision, applying of novel methodologies to evaluate a large number of chemicals in a range of in vitro assays in a short period of time [17]. While realization of the NRC vision may ultimately require a research effort on the scale of the Human Genome Project [18], success of this effort would be transformational for toxicology testing for environmental and pharmaceutical chemicals, providing cheaper, faster, and more accurate assessment of the toxicological potential of new chemicals.
Figure 1.
The Tox21 collaboration. The Tox21 collaboration brings together the experimental toxicology expertise of the NTP, high throughput screening technology at the NCGC and computational toxicology expertise at the EPA.
Role of the NCGC in the Tox21 Collaboration
The NCGC was established in 2004 as the first assay development, screening, informatics, and chemistry center of what was to become the NIH Roadmap Molecular Libraries Probe Production Center Network. The Molecular Libraries Initiative (MLI), a component of the NIH Roadmap for Medical Research, was born from the need for new approaches to determine function and therapeutic potential of human genes on the heels of the Human Genome Project, and to accelerate the pace that basic research is translated into small molecule therapeutics [17] (see: http://nihroadmap.nih.gov/molecularlibraries/). As part of the NCGC’s technology development program, a platform for automated testing of hundreds of thousands of compounds in titration-based format over a short period of time was developed [13,19], and this quantitative high-throughput screening (qHTS) platform has become a central aspect of the Tox21 program.
Traditional biological assays have been low-throughput, employing animal models and labor-intensive testing of samples. Furthermore, the growth of small molecule collections required the development of HTS technologies to test a large number of compounds in a timely manner [20]. While HTS has successfully allowed the screening of large chemical libraries to generate hits for medicinal chemistry optimization in the setting of drug discovery, HTS as traditionally practiced is not suitable for toxicity testing since it assays each compound at only single concentration [19], and thus generates large numbers of false positives (FP) and false negatives (FN) [21]. In contrast, the qHTS paradigm tests each compound at multiple (7-15) concentrations across a ~4-log concentration range, thus producing concentration-response based activity profiles of all compounds from the primary screen with greatly reduced FN and FP rates. Miniaturized assay volumes (<10 μL/well) in a 1,536 well-plate format provides the throughput to generate concentration response curves (CRCs) for every compound library member tested [22]. Curve fitting and CRC classification characterizes each curve based on parameters, such as curve fit and efficacy after the primary screening in a qHTS format, allowing for the identification of structure-activity relationships (SAR). The NCGC makes its screening data publicly available through PubChem (see: http://pubchem.ncbi.nlm.nih.gov), and its software available on its website (see: http://www.ncgc.nih.gov/pub/openhts/) to enable the scientific community to utilize the data in their own research [23].
The ability of qHTS to produce reliable activity profiles of chemicals has also allowed the NCGC to profile large libraries of chemicals for their propensity to produce assay artifacts which would otherwise be interpreted as true biological effects [24]. NCGC has taken advantage of titration-based screening to identify compounds that produce a wide variety of different artifactual activities, including apparent enzyme inhibition through compound aggregation [25], compound autofluorescence [26] and firefly luciferase inhibition [27]. These profiling examples demonstrate the utility of qHTS in distinguishing true effects from artifacts for more reliable toxicity screening and efficient chemical probe development.
NCGC Chemical Library Collection used for Tox21 Assays
An essential component of NCGC’s qHTS paradigm is the availability of large chemical libraries in a titration-based format. The availability of several concentrations across different plates gives the user flexibility to utilize concentrations relevant to the assays. In total, the NCGC has well over 400,000 compounds from the NIH Molecular Libraries Small Molecule Repository (MLSMR) and NCGC-specific compound collections. The latter currently includes approximately 1,400 compounds each from the NTP and EPA compound libraries, and 2,816 clinically used drugs in the NCGC Pharmaceutical Collection (NPC). Overall, compound selection is based on having a defined chemical structure and known purity, in addition to the extent of each compound’s solubility in dimethylsulfoxide (DMSO) [13]. One limitation of compound storage in DMSO is precipitation (augmented by DMSO water content and number of freeze-thaw cycles) [28]. Compound integrity studies using P450 assays and qHTS at NCGC revealed decreased compound potency over time and lower efficacy of older samples stored in DMSO [29]. For this reason, compounds are used in screening collections at the NCGC for no longer than 4-6 months.
The current NTP compound collection consists of 1,408 compounds, with more than 50 of the compounds represented twice to assess assay reproducibility. The NTP collection includes solvents, fire retardants, dyes, preservatives, plasticizers, therapeutic agents, inorganic and organic pollutants, drinking water disinfection by-products, pesticides, and natural products [3]. Selection of the 1,408 compounds was partly based on the availability of toxicological data from standard tests of carcinogenicity, genotoxicity, immunotoxicity, and/or reproductive and developmental toxicity [3]. Compounds were prepared as 10 mM stock solutions in DMSO and 14 plates representing 2.23-fold dilutions in 1536-well compound plates from 384-well plates [30]. The current EPA collection consists of 1,462 compounds prepared similarly as the NTP compound collection. Compounds were primarily selected based on the need to screen and prioritize environmental chemicals to which humans are exposed through the environment or food. These chemicals include those known to be bioactive, those manufactured or used in large quantities and those to which humans are exposed on a routine basis [31]. In the near future, approximately 1,400 additional compounds will be added to each of the NTP and EPA libraries for testing as an integrated Tox21 library [14].
The NPC collection (R. Huang et al., unpublished) was prepared at the NCGC and currently contains 2,816 small molecules, 52% which are approved by the Food and Drug Administration (FDA) for human or animal use in the United States. The remaining drugs are either approved for use in other countries, such as Europe, Canada, or Japan, or are compounds that have been tested in clinical trials. The majority of the NPC collection is prepared as a 10 mM stock in DMSO and prepared as fifteen 2.23-fold dilution plates in 1,536-well format [30]. Currently, an additional 1,400 compounds are being added to the NPC collection. The next phase of Tox21 testing will begin later this year utilizing the combined NTP, EPA, and NPC compound sets, totaling over 10,000 chemicals.
NCGC Technology
The NCGC characterizes toxicity endpoints in cell-based assays utilizing integrated robotic systems combined with batteries of in vitro assays and computational analysis [32]. The NCGC robotic platform stores large compound collections, performs distinct assay steps and measures user defined assay outputs in an integrated fashion [23]. The compound storage unit is capable of storing approximately 300,000 compounds in 7-point titrations, which correlates to over 2.2 million compound samples [23]. A Pin Transfer Station performs the transfer of 23 nL of compound from a 1,536-well compound plate to a 1,536-well assay plate, with each plate holding up to 1,408 compounds (located in columns 5-48). Assay specific controls (located in columns 1-4) are located on an additional 1,536-well compound plate and transferred simultaneously with the test compounds to the assay plate [23]. Solenoid dispensers, having the capability of dispensing volumes ranging from 200 nL to 20 μL, are used for reagent and cell dispensing. Furthermore, up to 8 tips can be used for dispensing in either 90° direct dispense or 45° angled head dispense with regard to the well. This allows for the modification of straight head and angled dispense, depending on the reagent type and condition of cells (i.e., if they are grown in a delicate monolayer, then it may be suitable to use the angled head dispense). For the aspiration of liquid, each dispenser comes equipped with an aspiration head made out of 32 stainless-steel tubes for column-wise removal of reagents or media from the plate. The aspirator head enables cell washing and fixing in 1,536-well format [33] for cell cycle protocols or protocols involving antibody steps. There are several factors that can be optimized with the dispenser and aspirators, such as dispense volumes, aspiration depth, and aspiration speed [23].
There are four different types of detectors that are currently integrated into the robotic system. These detectors essentially enable a wide variety of assays to be performed at NCGC and accommodate various assay technologies. Furthermore, these readers cover the entire spectrum of speed and information content. While charged-coupled device (CCD)-based camera imagers are capable of very fast read times per 1,536-well plate (<1 minute/plate), they provide the least information regarding characteristics of individual cells. The converse is true for confocal-based imaging readers, which can provide detailed information on sub-cellular structures with longer read times (~ 1 hour/plate). Specifically, the EnVision and ViewLux (PerkinElmer) are photomultiplier tube (PMT) and CCD-based instruments, respectively, which cover a wide range of fluorescence, absorbance, and luminescence [3,15,34,35] bulk well readouts commonly used in high throughput assays [23]. The ViewLux can be utilized for luminescence, fluorescence, absorbance, time-resolved fluorescence resonance energy transfer (TR-FRET) [36], and fluorescence polarization assays [23]. The Envision is best suited for AlphaScreen assays, ß-lactamase reporter assays [37,38], and can be customized for the detection of multiple wavelength regions [23] and TR-FRET assays [36]. Each assay format/readout has its own set of advantages. For example, the use of ß-lactamase as a reporter has several advantages, such as ratiometric readouts from dual emissions (460 and 530 nm), which minimizes well-to-well and plate-to-plate variation caused by differences in plating density. Additionally, the 530 nm fluorescent signal can be used as in indication of cell viability (and a proxy for compound cytotoxicity) and auto fluorescence [37].
For imaging assays, the user must sacrifice speed to obtain more information about individual cells and characterized cell populations. Two imaging platforms that are utilized at the NCGC are the Acumen Explorer (TTP Labtech) and the IN Cell Analyzer 1000 (GE Healthcare). The Acumen Explorer is a PMT-based laser scanning microplate cytometer that is equipped with three excitation and four emission lasers for enumeration and characterization of fluorescent objects [39]. The Acumen is able to provide total well and individual cell fluorescence readings. Compared to the ViewLux, where the entire plate is read in less than one minute, Acumen read times can vary between 10-20 minutes per plate, depending on the number of wavelengths required by the assay of interest. Thus far, the Acumen has been used in several assays performed at NCGC, such as GFP-based assays [33] and multiplexing of dual fluorescent drug sensitive and drug resistant cell lines [40]. For higher resolution automated fluorescent imaging, the IN Cell Analyzer is designed to collect data either at the single cell or sub-cellular level. The data collected is often complementary to that collected from the Acumen, since additional orthogonal phenotypes can be identified. Furthermore, the instrument comes with own algorithm to analyze the data acquired [41].
Once data is obtained for each assay, the CRCs for each compound are analyzed and classified as previously described [3,19,36]. Briefly, raw plate reads for each titration point are normalized relative to an assay-specific positive control (100% or -100%) and DMSO-only wells (0%), and then corrected by applying a pattern correction algorithm using DMSO-only plates at the beginning and end of each stack [3]. Half-maximal inhibition/activation concentration (AC50) and efficacy values are obtained from fitting the concentration-response titration points to the Hill equation [42]. Compounds are classified as curve classes 1-4 according to the characteristics of the CRC, such as efficacy and quality of curve fit (R2). Class 1 curves display two asymptotes, while class 2 curves display one asymptote. Class 1 and 2 curves are further subdivided into subclasses a (efficacy ≥ 6SD) and b (efficacy < 6SD). These curves have statistically significant curve fits and are usually selected for follow-up analyses. Compounds in curve class 3 only display activity at the highest concentration tested and compounds with class 4 curves show no concentration response and are deemed as inactive. The ability to decipher curves using qHTS for every compound tested is important since many responses, such as toxicity, are measured over broad concentration ranges (typically between 0.5 nM to 92 μM), which may greatly decrease the FP and FN rates.
Assay Implementation for Tox21
Various assays (Table 1) in different cell types or lines (Table 2) have been successfully developed, miniaturized, and validated for 1,536-well plate format at NCGC and screened against the initial Tox21 compound collection. For the majority of assays, compound incubation durations were limited to 48 hours due to evaporation-induced edge effects observed in 1,536-well plates [3]. Stainless steel assay lids with rubber gaskets were also used to allow air exchange and minimize edge effects [23]. The assays described below demonstrate the adaptation of existing low-throughput assays to higher throughput, reliable 1,536-well format assays. These studies show the ability to modify existing assays to profile larger numbers of compounds for toxicity-based safety studies.
Table 1.
Tox21 Assays Currently Available at NCGC
| Assay | Assay endpointa | Cell type | Assay readout | References |
|---|---|---|---|---|
| Cell viability | Intracellular ATP content |
Hek293; Jurkat; HepG2; SH-SY5Y; SK-N-SH; BJ; HUV-EC-C; MRC-5; Mesangial; Kidney proximal tubules; N2a; H-4-II-E; NIH3T3 |
Luminescence | [3,15] |
|
| ||||
| Apoptosis | Caspase 3/7 | |||
|
| ||||
| Membrane integrity |
LDH release |
Hek293; Mesangial | Fluorescence |
[34] |
| Protease release | Luminescence | |||
|
| ||||
| Mitochondrial toxicity |
Membrane potential | HepG2 | Fluorescence | |
|
| ||||
| DNA damage | Micronucleus | CHO | Fluorescence | |
|
| ||||
| Cytokine | IL-8; TNF-α | THP-1 | Homogeneous time resolved fluorescence |
|
|
| ||||
| Nuclear receptor |
AR; ERα; FXR; PPARδ; PPARγ; RXR; TRβ; VDR; |
Hek293 | β-lactamase reporter | |
|
|
|
|||
| GR | HeLa | |||
|
| ||||
| hPXR; AhR; rPXR | HepG2 | Luciferase reporter | ||
|
| ||||
| Toxicity pathway |
AP-1; HIF-1α; SIE; NFκB |
ME180 | β-lactamase reporter | [37,38,59,60] |
|
|
|
|||
| HSR; ESRE | HeLa | [61] | ||
|
|
|
|||
| ARE/Nrf2; | HepG2 | |||
|
|
|
|||
| CREB | Hek293, CHO | [53] | ||
|
|
|
|||
| p53 | HCT-116 | |||
|
| ||||
| ARE/Nrf2; HSR; ESRE |
HepG2 | Luciferase reporter | [58] | |
|
| ||||
| hERG channel | Thallium influx | U-2OS | Fluorescence | [45] |
LDH, lactate dehydrogenase ; IL-8, interleukin-8 ; TNFα, tumor necrosis factor α; AR, androgen receptor; ERα, estrogen receptor α; FXR, farnesoid X receptor; PPARδ, peroxisome proliferator-activated receptor δ; PPARγ, peroxisome proliferator-activated receptor γ; RXR, retinoid X receptor; TRβ, thyroid hormone receptor β; VDR, vitamin D receptor; GR, glucocorticoid receptor; hPXR, human pregnane X receptor; AhR, aryl hydrocarbon receptor; rPXR, rat pregnane X receptor; AP-1, activator protein-1; HIF-1α, hypoxia inducible factor -1α; SIE, sis-inducible element; NFκB, nuclear factor kappa B ; HSE, heat shock response element; ESRE, endoplasmic reticulum stress response element ; ARE/Nrf2, antioxidant response element/NF-E2 related factor 2; CREB, cAMP response element binding.
Table 2.
Cell Lines Tested
| Cell type | Origin | Species |
|---|---|---|
| BJ | Foreskin fibroblasts | Human |
| CHO | Chinese hamster ovary | Hamster |
| HCT-116 | Colorectal carcinoma | Human |
| Hek293 | Embryonic kidney cells | Human |
| HeLa | Cervical carcinoma | Human |
| HepG2 | Hepatocellular carcinoma | Human |
| HUV-EC-C | Vascular endothelial cells | Human |
| H-4-II-E | Hepatoma | Rat |
| Jurkat | T-cell leukemia | Human |
| Kidney proximal tubules | Freshly isolated from kidney | Rat |
| MRC-5 | Lung fibroblasts | Human |
| Mesangial | Renal glomeruli | Human |
| ME180 | Cervical carcinoma | Human |
| N2a | Neuroblastoma | Mouse |
| NIH3T3 | Embryonic fibroblasts | Mouse |
| SH-SY5Y | Neuroblastoma | Human |
| SK-N-SH | Neuroblastoma | Human |
| THP-1 | Monocytic leukemia | Human |
| U-2OS | Osteosarcoma | Human |
To assess chemical effect on cell membrane integrity, a newly developed cytotoxicity assay that measures released intracellular proteases upon membrane damage with a bioluminescent assay readout was evaluated [34]. Although there have been similar assays developed for lower density formats, few have been miniaturized and validated in a high-throughput format with a robust assay signal [34]. This protease release assay for membrane damage detection was miniaturized in 1,536-well format and was screened against the initial NTP compound collection in HEK 293 and human renal mesangial cells (Tables 1 and 2) [34]. All compounds were tested in 14-point titration series to identify compounds that disrupt membrane integrity. The assay performed well in miniaturized format, with high reproducibility of the control compound across every plate in both cell lines, high signal-to-background ratios and Z’ values (a statistical measure of assay performance) [43].Additionally, replicate compounds within the NTP compound collection demonstrated high intra-experimental reproducibility, further indicating the reliability of the qHTS assay. The compounds identified from this assay were known membrane disrupters, including α-solanine and zinc pyrithione. The majority of compounds active in both cell lines were detergents such as digitonin, tetra-N-octylammonium bromide, and p-n-nonylphenol, which are known to disrupt membrane integrity, while additional non-detergent compounds known to be membrane disrupters were also identified in the assay. Furthermore, some compounds were shown to be uniquely active in one cell line or the other, thus revealing cell line-specific membrane disruption potential. Overall, this study demonstrated the successful miniaturization of an existing cytotoxicity assay using a luminescent readout. Furthermore, the application of qHTS to this assay format validated the need to characterize the biological activities of compounds over a broad concentration range [34].
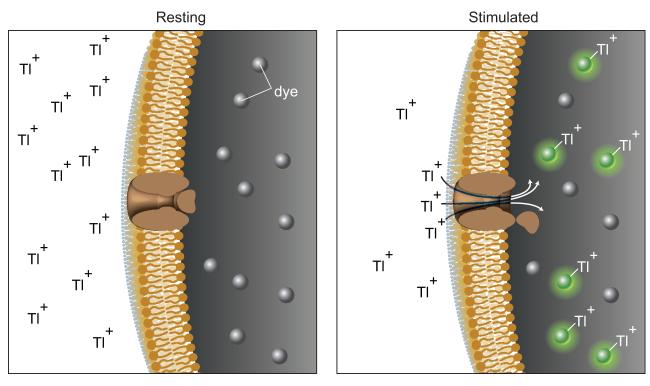
Cardiotoxicity has been commonly examined in the human ether-a-go-go-related gene (hERG) potassium channel. The hERG channel is responsible for the repolarization of cardiac action potential, which is associated with certain forms of inherited and acquired long QT syndrome (LQTS) [44,45]. LQTS may lead to sudden death through a rare ventricular arrhythmia [45]. Some drugs have been removed from the market due to their potential to induce LQTS by inhibiting the hERG channel, which warrants the need for pre-marketing screening of drugs to minimize the risk of sudden death in the treatment of non-life threatening diseases [45,46]. Patch clamp electrophysiology technique is still the gold standard for hERG activity in drug development [47,48], but this assay is low-throughput and costly, and requires specialized training for personnel [45]. The radioligand binding assay is also commonly used to test compound binding to the hERG channel, but this assay gives little or no information on the functional effect of the ligand on the channel (blocker, activator, or no effect) and the allosteric effect of the ligands [49]. To overcome these limitations, a functional assay was developed for the hERG channel by measuring thallium influx into the cells and validated this assay in a 1,536-well plate format. The assay principle is shown in Figure 2, where thallium ions enter the cells through open hERG channels after stimulation and bind to the dye, yielding an increase in fluorescence. This fluorescence signal is inhibited in the presence of hERG channel blockers [45]. The qHTS screen identified a group of known hERG inhibitors, such as pimozide, amiodarone, and verapamil, from a library of 1,280 pharmacologically active compounds (LOPAC1280). Furthermore, the activities of the hERG channel inhibitors in the thallium influx assay are well correlated with those obtained from automated patch clamp experiments [45].
Figure 2.
Principle of the thallium flux assay. At resting state, cells expressing hERG channels are loaded with dye from the assay kit. Upon stimulation, thallium ions enter the cells through open hERG channels and bind to the dye, yielding green fluorescence upon excitation, proportional to the bound dye.
In Vitro Assays for Cytotoxicity
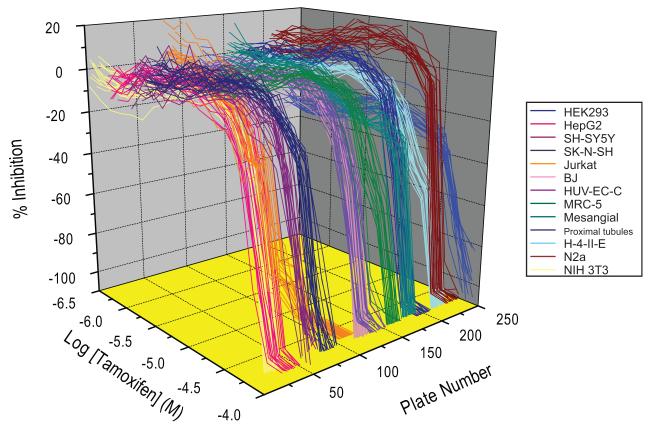
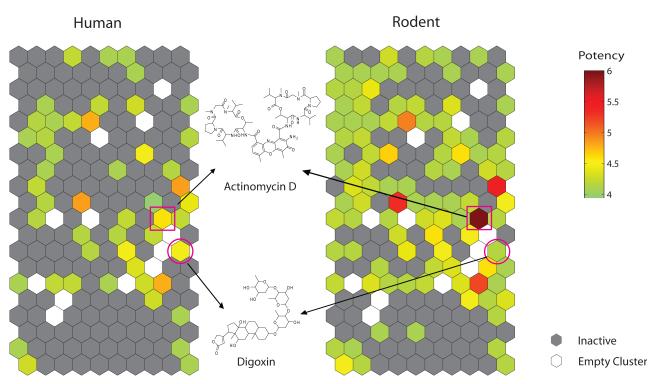
One of the goals of the Tox21 collaboration is to establish in vitro signatures of in vivo human and rodent toxicity. In order to create in vitro signatures of compound cytotoxicity across species, 1,408 compounds from the initial NTP collection were profiled for cytotoxicity across 13 different human and rodent cell types [3]. These human and rodent cell types were derived from 6 tissue types that are common targets of xenobiotic toxicity (Tables 1 and 2); thus, this study aimed to develop species and cell-specific cytotoxicity profiles for each compound [3]. Each compound was tested at 14 concentrations (0.5 nM to 92 μM) in a luminescent cell viability assay that measures adenosine triphosphate (ATP) levels of metabolically active cells. The luminescent ATP quantitation assay worked well in 1,536-well format with robust Z’, signal-to-background ratio, and coefficient of variation (CV) values for the positive control compound (tamoxifen). Tamoxifen CRCs for each cell line were consistent across the plates, however, the response pattern was cell-line specific, with Jurkat cells being most sensitive and mesangial cells being least sensitive (Figure 3) [3]. Furthermore, the correlation of IC50 values for the 55 duplicate compounds present in the NTP compound collection across all 13 cell lines was significant (0.71, p < 0.001). There were 428 compounds that displayed cytotoxicity in at least one cell type and clustering analysis was performed in order to decipher cytotoxicity profiles across cell types. Within the subgroup of active compounds, multiple effects from the compounds were identified within and across compound types, cell types, and species. For example, human and rodent-derived cells including SH-SY5Y, Jurkat, H-4-II-E, NIH 3T3, N2a, HEK293 and rat renal proximal tubule cells were most sensitive to compound-induced cytotoxicity, whereas human fibroblast and skin cells were the least sensitive. Overall, the rodent cells used in this study demonstrated more sensitivity to the compounds tested than the human cells. Compounds that showed activity in at least one cell type were clustered according to their IC50 values, revealing clusters and specific compounds that were selectively cytotoxic in a particular cell type and species. For example, digoxin was more cytotoxic in human HEK293 cells than rat renal proximal tubule cells. However, actinomycin D was much more cytotoxic in rat renal proximal tubules than human HEK293 cells (Figure 4). Overall, a striking finding was the lack of concordance in the patterns of compound activity in cells derived from the same tissue but from different species (there were also instances where cells with similar tissue origin in the same species showed discordance in compound activity profiles), highlighting inter-species differences in response. Thus, an important finding from the study is that in vitro cytotoxicity in a particular cell type, even if from the same tissue/species, does not necessarily predict cytotoxicity in another cell type [3]. Thus, the combination of in vitro profiling with qHTS allows for the generation of hypotheses related to mechanisms of toxicity and prioritization for more intense toxicological investigation related to in vivo toxicity.
Figure 3.
Positive control titration across human and rodent cell lines. The reproducibility of tamoxifen, used as a positive control in ATP-mediated cytotoxicity testing, is shown for each cell line for 234 plates tested. Jurkat cells were most sensitive to tamoxifen while NIH3T3, HEK293, BJ and mesangial cells were least sensitive.
Figure 4.
Species-selective compounds. Compound activity patterns were obtained through hierarchical clustering of compound IC50 values. Shown are compound activity patterns for human HEK 293 cells and rat renal proximal tubule cells. Two compounds, actinomycin D and digoxin, illustrate species-specific cytotoxicity for those particular compound clusters.
The application of clustering to data with multiple endpoints can help uncover underlying mechanisms involved in broad phenotypes such as cytotoxicity. To examine the mechanism of compound-induced cytotoxicity in various cell types, two different endpoints (cytotoxicity and caspase-3/7 activation) were assessed by testing the 1,408 NTP compounds for both endpoints across 13 different cell types [15]. The cytotoxicity and caspase-3/7 assays performed well in a 1,536-well format and the quality of the data was suitable for use in computational efforts. The overall active rate for the 13 caspase assays (0.4-3.5%) was lower than the rate for the 13 cytotoxicity assays (4-11%). Hierarchical clustering based on compound cytotoxicity and caspase EC50/IC50 patterns revealed similar clustering based on endpoints rather than cell type, indicating that cytotoxicity and caspase activation assays provide distinct sets of information, and that most compounds induce cytotoxicity through mechanisms other than caspase-3/7 activation. The only exception was the Jurkat cell line, where the caspase and cytotoxicity assays clustered together. One explanation may be that the cell death induced by most compounds in Jurkat cells is dependent on caspase-3/7 activation [15]. The N2a cell line appeared to have the least number of active compound overlap between the cytotoxicity and caspase assays, indicating the contribution of mechanisms outside of caspase activation for cytotoxicity. This approach will be useful for generating hypotheses for compound mechanism of action. However, hypothesis generation will be strengthened by the inclusion of more compounds and endpoints to build stronger models predictive of in vivo toxicity.
Computational Modeling
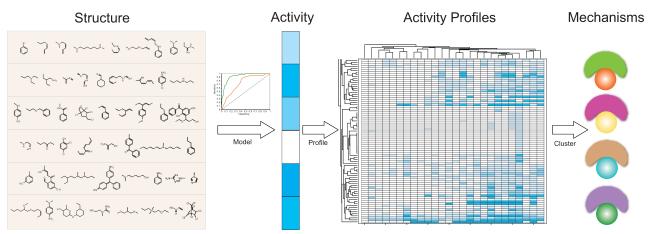
Due to the rapidly increasing number of environmental chemicals that need to be tested and the need for prioritization of those compounds for in vivo studies, more computational modeling is needed to complement experimental approaches in order to decrease the time associated with testing and accelerate prioritization of the data [50,51]. Some challenges associated with computational modeling of toxicity data include the diversity of compounds and structures that can produce the same outcome [50]. However, the production of CRCs for every compound tested provides a data-rich resource for SAR analysis, computational modeling, and chemical prioritization for more extensive toxicological evaluation. The NCGC recently developed a weighted feature significance (WFS) algorithm, a fragment-based approach that does not rely on whole molecule similarity to model toxicity (Figure 5), which is designed to achieve good prediction with structurally diverse sets of compounds [50]. Such approaches can be applied to generate testable hypotheses on mechanisms of compound toxicity. Starting with the structure of a compound, one could model and predict its toxicity in one assay or cell type and in multiple cell types, which essentially forms the activity pattern or signature that indicates the compound’s mechanism of toxicity.
Figure 5.
Weighted feature significance (WFS) algorithm. The algorithm has been developed to build fragment based models to predict various toxicity endpoints, such as cell viability, caspase-3/7 activation, mutagenicity and hepatotoxicity, based on compound structure. The sensitivity and specificity of these models have been rigorously tested using ROC curves. Activity or toxicity profiles can be generated by testing compounds against a battery of cell lines or assays measuring the same toxicity endpoint. The activity pattern of a compound across such a battery of assays can be viewed as the compound signature, which can be used subsequently to group compounds into different activity clusters, each representing a distinct toxicity mechanism or mode of action. The underlying assumption is that compounds exhibiting similar activity patterns or signatures are likely to share the same biological target or mode of action. Such approaches can be applied to generate testable hypotheses on mechanisms of compound toxicity. Starting with the structure of a compound, one could then model and predict its toxicity in one assay or cell type and in multiple cell types, which essentially forms the activity pattern or signature that indicates the compound’s mechanism of toxicity.
Models were developed for two aforementioned [3,15] assays, with rigorous performance evaluation of all models using receiver operating characteristic (ROC) curves [52]. One advantage of the WFS approach is its ability to identify structural features responsible for toxicity [50]. For example, structural features significantly enriched in the pan-cytotoxic compounds include substituted/activated benzenes, imines, 1,3-dienes, heavy metals and imines [50]. Toxic features were also identified for caspase-3/7-activating compounds [15], such as cyclic alkyl ketones and alkyl halides. Thus, the significant toxic features present in compounds could be used to predict a particular mechanism of toxicity, such as caspase-3/7 activation [50]. Overall, the WFS approach can be applied to model other toxicity endpoints such as mutagenicity and hepatotoxicity and may be applicable to a larger array of compounds. Unlike other modeling methods, WFS can also be utilized even when compound structures are highly diverse. Additionally, WFS was shown to have comparable or better predictive power when compared to Native Bayesian clustering or a support vector machine approach in most test cases [50]. An analysis of the initial Tox21 collection of 2,800 compounds revealed that additional chemicals are required for enhancement of compound diversity in these collections to increase the number of robust structural predictors of the WFS, which validates the previously described initiative to expand the compound collection to more than 10,000 chemicals.
Cellular Pathway Assays
Although attractive, target-based screens may lead to the identification of active compounds that do not retain their activity in a physiological environment [53]. Thus, cell-based assays offer an alternative assay format in which the readout is dependent on specific components acting on a single signaling pathway. Furthermore, the combination of toxicity pathways associated with adverse health events with engineered cellular assays designed to measure the perturbation of these pathways in response to a chemical is a critical implementation of the National Academy of Sciences Toxicity Testing in the 21st Century report [54]. As an example of such an approach, a ß-lactamase reporter gene assay was employed to identify compounds that inhibit [37] or induce [38] hypoxia-inducible factor 1α (HIF-1α) activity (Figure 6). Hypoxia, the reduction in the normal level of tissue oxygen tension within a tissue, is associated with a number of pathologies including cancer and inflammation [55]. Under hypoxic conditions, HIF-1 subunits heterodimerize and translocate into the nucleus prior to binding to a hypoxia-response element (HRE) upstream of target genes that activate angiogenesis and vascular endothelial growth factor (VEGF) [56]. Hypoxic conditions attenuate the degradation of HIF-1α, leading to the transcription of survival genes in many solid tumors and poor cancer prognosis [57]. Thus, compounds that inhibit HIF-1α responsive tumor hypoxia may be a valuable chemotherapeutic approach [37]. Furthermore, a separate screen for inducers of hypoxia is also valuable to identify those compounds that may serve as hypoxia mimetics [38].
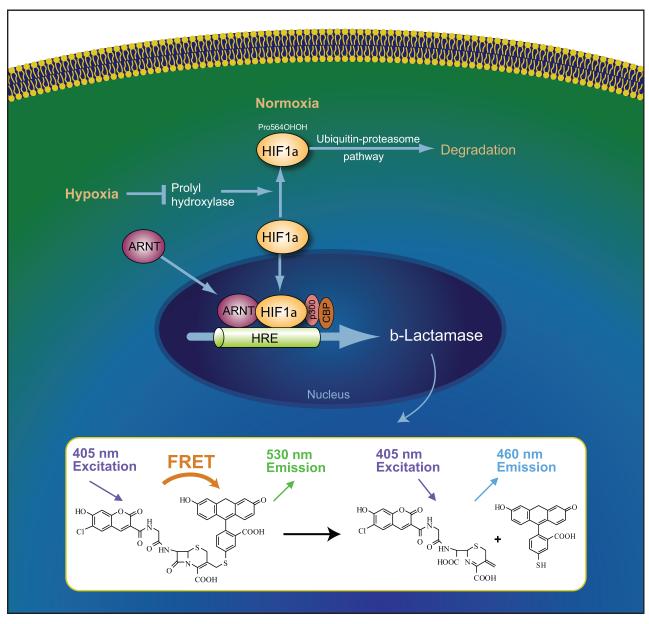
Figure 6.
Reporter gene assay for HIF-1 activity. During normoxia, the HIF-1α subunit is degraded by the ubiquitin-proteosome pathway. In this case, the absence of ß-lactamase expression leaves the fluorescent substrate molecule, which contains coumarin and fluorescein. Excitation of the coumarin results in fluorescence resonant energy transfer to the fluorescein moiety, resulting in the emission of green fluorescent signal (530 nm). However, under hypoxic conditions, HIF-1α heterodimerizes with the HIF-1ß subunit and translocates to the nucleus. Next, the HIF-1 complex binds to HRE regulatory sequences upstream of target genes. In this assay, stimulation under hypoxic conditions results in the transcription of ß-lactamase, which cleaves the fluorescent substrate molecule, disrupting energy transfer. Excitation of the coumarin molecule in the presence of ß-lactamase enzyme activity results in a blue fluorescence signal (460 nm). The ratio of the blue:green signals provides a normalized reporter response.
To identify inhibitors of hypoxia, 73,000 compounds from the MLMSR compound collection [37] were screened between 7-15 concentrations in an HRE-bla assay performed in ME-180 cells (Tables 1 and 2). Additionally, to identify HIF-1α inducers, 1,408 compounds from the NTP collection were screened at 14 concentrations also utilizing the HRE-bla assay. The screening for both assays performed well and indicated the suitability for qHTS to identify inhibitors and activators of HIF-1α. Three hundred and fifty inhibitors with reliable curve classes were identified and SAR analysis of these compounds yielded 18 structural series sharing a common scaffold. Several follow-up studies, such as the evaluation of compound effects on low oxygen induced HIF-1 signaling and VEGF secretion, were employed to ensure the specificity of compound activity in the HIF-1α assay [37]. Overall, the primary qHTS and follow-up compounds identified from SAR analysis demonstrated specificity for inhibition of HIF-1α activity and little to no cytotoxicity, and thus appear to be good candidates for further testing in other cancer cell lines or animal models [37]. Conversely, 10 compounds were identified and confirmed as inducers of HIF-1α activity from the primary screening using the NTP compound collection [38]. In the follow-up studies, 5 of 10 compounds significantly induced VEGF secretion in human ME180 cells. These 5 compounds were further tested for their dependence on HIF-1 with regard to VEGF secretion by testing them in HIF-1α wild type and knockout mouse embryonic fibroblast (MEF) cell lines. Compounds involved in VEGF secretion dependently (such as phenathroline) or independently (such as 7,12-DMBA) of HIF-1α were identified. Finally, these 5 compounds were tested against a battery of reporter genes driven by hypoxia-responsive gene promoters [38,58] to establish the promoter activity profiles. For example, three compounds (including phenathroline) produced promoter activity profiles very similar to those produced by standard hypoxic conditions (1% O2) used in cell-based studies [38,58]. Furthermore, this study highlights the use biological profiling data with hierarchical clustering to group compounds that operate under a similar mode of action [58].
Conclusions and Future Directions
The Tox21 collaboration is combining technology, biology, and computational methods in order to advance in vitro testing for toxicology [12]. The NCGC is working with its Tox21 partners to develop next-generation testing methods, alternative approaches to existing methods, and modeling in vitro and in vivo responses.
The examples given here are only a few of the assays that have been utilized to date to study specific endpoints or pathways in human and rodent in vitro assays; in total, the NCGC has generated over 6 million data points and over 400,000 concentration-response curves for Tox21 chemicals in specific assays. The qHTS-driven production of CRCs for every compound tested provides a data-rich resource for SAR analysis, computational modeling and chemical prioritization for more extensive toxicological evaluation. This directly points to the advantages of using qHTS with regard to compound hazard identification, since qHTS will allow for a more accurate assessment of compound-induced toxicity using cell-based studies and an idea of starting doses to use in in vivo studies. SAR analysis will also enable the identification of toxic compounds with similar structures for follow-up testing. Thus, a basis of hazard characterization with regard to toxicity will emerge prior to in vivo studies. Furthermore, in vitro toxicity tests performed in human-derived cell lines may provide important biomarkers of exposure which can be directly tested in human populations [2]. Human risk assessment from some in vitro studies may prove difficult, thus a step-wise approach starting with qHTS, computational modeling, and carefully designed tissue and species-specific cell-based assays will provide a stronger and mechanistically predictive approach for in vivo testing and human risk assessment [2].
Building on the solid foundation described here, future Tox21 goals include the inclusion of new platforms for qHTS (such as high content screening and nanotechnology), assessment of genetic variation involved in human and rodent toxicity, incorporation of metabolism/biotransformation capability into the current and future assays, identification and prioritization of critical cellular pathways and key targets for screening, expansion of the compound libraries including compounds that are DMSO or water insoluble, discerning the link between observed perturbations in vitro and pathologies in exposed humans, and creation of relational public databases and tools to interrogate the screening data. Table 1 comprises some of the current and future Tox21 assays at the NCGC, such as those for oxidative stress response [54]. Data production in Tox21 is now moving into its exponential growth phase, and over the next several years the interdisciplinary Tox21 collaboration will continue to innovate in assay biology, screening, and computation, to usher in a new era of efficient, mechanistic, and predictive chemical toxicology.
Teaser: A synopsis of the Tox21 initiative and a focus on the NIH Chemical Genomics Center’s efforts within this program using in vitro methods and quantitative high throughput screening
Acknowledgements
We gratefully acknowledge Paul Shinn for compound management. We also thank Dr. Raymond Tice for critical reading of the manuscript and Darryl Leja for illustrations.
Abbreviations
- NTP
National Toxicology Program
- EPA
U.S. Environmental Protection Agency
- NRC
National Research Council
- Tox21
Toxicity Testing in the 21st Century
- HTS
high-throughput screening
- NCGC
NIH Chemical Genomics Center
- DHHS
Department of Health and Human Services
- NIEHS
National Institute of Environmental Health Sciences
- NHGRI
National Human Genome Research Institute
- NCCT
National Center for Computational Toxicology
- MLI
Molecular Libraries Initiative
- qHTS
quantitative high-throughput screening
- FP
false positive
- FN
false negative
- CRCs
concentration response curves
- SAR
structure activity relationships
- MLSMR
Molecular Libraries Small Molecule Repository
- NPC
NIH Pharmaceutical Collection
- DMSO
dimethylsulfoxide
- FDA
Food and Drug Administration
- CCD
charged-coupled device
- PMT
photomultiplier tube
- TR-FRET
time-resolved fluorescence resonance energy transfer
- AC50
half-maximal inhibition
- hERG
human ether-a-go-go
- LQTS
long QT syndrome
- LOPAC1280
library of pharmacologically active compounds
- ATP
adenosine triphosphate
- CV
coefficient of variation
- WFS
weighted feature significance
- ROC
receiver operating characteristic
- HIF-1α
hypoxia-inducible factor 1 alpha
- HRE
hypoxia-response element
- VEGF
vascular endothelial growth factor
- MEF
mouse embryonic fibroblasts
Biography
 Dr. Sunita J. Shukla is currently a postdoctoral fellow at the NIH Chemical Genomics Center (NCGC). She is currently working under the guidance of Dr. Menghang Xia in developing in vitro toxicity based assay screening using a quantitative high throughput screening platform. Furthermore, she is also working under the guidance of Dr. Doug Auld in the area of assay development and high content screening. Dr. Shukla is the first recipient of the Humane Society/Procter and Gamble Fellowship honoring the advancement of alternatives to animal testing. Prior to joining NCGC, she received her Ph.D in Human Genetics, with a focus on pharmacogenetics of anticancer agents, from the University of Chicago in the lab of Dr. M. Eileen Dolan in 2007. Additionally, she received a Master of Public Health degree at Saint Louis University with a focus on Epidemiology and Environmental/Occupational Health in 2001. She has authored or co-authored 13 peer-reviewed publications.
Dr. Sunita J. Shukla is currently a postdoctoral fellow at the NIH Chemical Genomics Center (NCGC). She is currently working under the guidance of Dr. Menghang Xia in developing in vitro toxicity based assay screening using a quantitative high throughput screening platform. Furthermore, she is also working under the guidance of Dr. Doug Auld in the area of assay development and high content screening. Dr. Shukla is the first recipient of the Humane Society/Procter and Gamble Fellowship honoring the advancement of alternatives to animal testing. Prior to joining NCGC, she received her Ph.D in Human Genetics, with a focus on pharmacogenetics of anticancer agents, from the University of Chicago in the lab of Dr. M. Eileen Dolan in 2007. Additionally, she received a Master of Public Health degree at Saint Louis University with a focus on Epidemiology and Environmental/Occupational Health in 2001. She has authored or co-authored 13 peer-reviewed publications.
 Dr. Christopher Austin is Director of the NIH Chemical Genomics Center (NCGC) and Senior Advisor to the Director for Translational Research at NHGRI. The NCGC is an ultra-high-throughput screening, informatics, and chemistry center that develops novel compounds as probes of biology and starting points for development of new drugs for rare and neglected diseases, profiles small molecule libraries for biological and toxicological activities, and develops new paradigms to increase the efficiency and genome-wide reach of assay, screening, chemistry, and informatics technologies. Dr. Austin received his A.B. from Princeton and M.D. from Harvard, trained in neuroscience and genetics at Harvard, and came to NIH in 2002 from Merck.
Dr. Christopher Austin is Director of the NIH Chemical Genomics Center (NCGC) and Senior Advisor to the Director for Translational Research at NHGRI. The NCGC is an ultra-high-throughput screening, informatics, and chemistry center that develops novel compounds as probes of biology and starting points for development of new drugs for rare and neglected diseases, profiles small molecule libraries for biological and toxicological activities, and develops new paradigms to increase the efficiency and genome-wide reach of assay, screening, chemistry, and informatics technologies. Dr. Austin received his A.B. from Princeton and M.D. from Harvard, trained in neuroscience and genetics at Harvard, and came to NIH in 2002 from Merck.
 Dr. Menghang Xia is group leader of cellular toxicity and signaling at the NIH Chemical Genomics Center (NCGC). Dr. Xia and her research group are currently focused on the target-specific and mechanism-based toxicological studies, in collaboration with the Biomolecular Screening Branch at the National Toxicology Program (NTP) and National Center for Computational Toxicology at the U.S. Environmental Protection Agency (EPA). Her group has developed and validated a battery of in vitro toxicological assays using a quantitative high through screening platform, and investigated the mechanism of action of chemicals in multiple cellular signaling pathways. Dr. Xia received her Ph.D from State University of New York at Buffalo, did postdoctoral training at University of California at San Francisco, and joined NCGC in 2005 from Merck.
Dr. Menghang Xia is group leader of cellular toxicity and signaling at the NIH Chemical Genomics Center (NCGC). Dr. Xia and her research group are currently focused on the target-specific and mechanism-based toxicological studies, in collaboration with the Biomolecular Screening Branch at the National Toxicology Program (NTP) and National Center for Computational Toxicology at the U.S. Environmental Protection Agency (EPA). Her group has developed and validated a battery of in vitro toxicological assays using a quantitative high through screening platform, and investigated the mechanism of action of chemicals in multiple cellular signaling pathways. Dr. Xia received her Ph.D from State University of New York at Buffalo, did postdoctoral training at University of California at San Francisco, and joined NCGC in 2005 from Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andersen ME, Krewski D. Toxicity testing in the 21st century: bringing the vision to life. Toxicol Sci. 2009;107(2):324–330. doi: 10.1093/toxsci/kfn255. [DOI] [PubMed] [Google Scholar]
- 2.Krewski D, et al. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009;29(4):474–479. doi: 10.1111/j.1539-6924.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- 3.Xia M, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116(3):284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg AM, Frazier JM. Alternatives to animals in toxicity testing. Sci Am. 1989;261(2):24–30. doi: 10.1038/scientificamerican0889-24. [DOI] [PubMed] [Google Scholar]
- 5.Zurlo J, et al. Animals and Alternatives in Testing: History, Science, And Ethics. Mary Ann Liebert, Inc.; 1993. [Google Scholar]
- 6.Rowan AN. Alternatives: interaction between science and animal welfare. In: Goldberg AM, editor. Product Safety Evaluation. Mary Ann Liebert, Inc.; 1983. pp. 113–133. [Google Scholar]
- 7.Williams G, Weisburger G. Toxicology, The Basic Science of Poisons. McGraw-Hill; 1993. Chemical carcinogenesis. [Google Scholar]
- 8.Beniashvili D. Experimental Tumors in Monkeys. CRC Press; 1994. [Google Scholar]
- 9.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3(12):952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 10.National Toxicology Program A National Toxicology Program for the 21st Century: A Roadmap for the Future. 2004.
- 11.Dix DJ, et al. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci. 2007;95(1):5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 12.National Research Council (NRC) Toxicity Testing in the 21st Century: A Vision and a Strategy. National Academy Press; 2007. [Google Scholar]
- 13.Austin CP, et al. Tox21: Putting a Lens on the Vision of Toxicity Testing in the 21st Century. 2008. [Google Scholar]
- 14.Kavlock RJ, et al. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009;29(4):485–487. doi: 10.1111/j.1539-6924.2008.01168.x. discussion 492-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R, et al. Characterization of diversity in toxicity mechanism using in vitro cytotoxicity assays in quantitative high throughput screening. Chem Res Toxicol. 2008;21(3):659–667. doi: 10.1021/tx700365e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins FS, et al. Toxicology. Transforming environmental health protection. Science. 2008;319(5865):906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austin CP, et al. NIH Molecular Libraries Initiative. Science. 2004;306(5699):1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 18.Seidle T, Stephens ML. Bringing toxicology into the 21st century: a global call to action. Toxicol In Vitro. 2009;23(8):1576–1579. doi: 10.1016/j.tiv.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103(31):11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnecke V, Bostrom J. Computational chemistry-driven decision making in lead generation. Drug Discov Today. 2006;11(1-2):43–50. doi: 10.1016/S1359-6446(05)03703-7. [DOI] [PubMed] [Google Scholar]
- 21.Malo N, et al. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24(2):167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 22.Thomas CJ, et al. The pilot phase of the NIH Chemical Genomics Center. Curr Top Med Chem. 2009;9(13):1181–1193. doi: 10.2174/156802609789753644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael S, et al. A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol. 2008;6(5):637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorne N, et al. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 14(3):315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng BY, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50(10):2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 26.Simeonov A, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51(8):2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 27.Auld DS, et al. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51(8):2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 28.Oldenburg K, et al. High throughput sonication: evaluation for compound solubilization. Comb Chem High Throughput Screen. 2005;8(6):499–512. doi: 10.2174/1386207054867364. [DOI] [PubMed] [Google Scholar]
- 29.MacArthur R, et al. Monitoring compound integrity with cytochrome P450 assays and qHTS. J Biomol Screen. 2009;14(5):538–546. doi: 10.1177/1087057109336954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasgar A, et al. Compound Management for Quantitative High-Throughput Screening. JALA Charlottesv Va. 2008;13(2):79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Judson R, et al. The toxicity data landscape for environmental chemicals. Environ Health Perspect. 2009;117(5):685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt CW. TOX 21: new dimensions of toxicity testing. Environ Health Perspect. 2009;117(8):A348–353. doi: 10.1289/ehp.117-a348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auld DS, et al. Fluorescent protein-based cellular assays analyzed by laser-scanning microplate cytometry in 1536-well plate format. Methods Enzymol. 2006;414:566–589. doi: 10.1016/S0076-6879(06)14029-X. [DOI] [PubMed] [Google Scholar]
- 34.Cho MH, et al. A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol In Vitro. 2008;22(4):1099–1106. doi: 10.1016/j.tiv.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanega C, et al. Comparison of bioluminescent kinase assays using substrate depletion and product formation. Assay Drug Dev Technol. 2009;7(6):606–614. doi: 10.1089/adt.2009.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla SJ, et al. Identification of pregnane X receptor ligands using time-resolved fluorescence resonance energy transfer and quantitative high-throughput screening. Assay Drug Dev Technol. 2009;7(2):143–169. doi: 10.1089/adt.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia M, et al. Identification of small molecule compounds that inhibit the HIF-1 signaling pathway. Mol Cancer. 2009;8:117. doi: 10.1186/1476-4598-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia M, et al. Identification of chemical compounds that induce HIF-1alpha activity. Toxicol Sci. 2009;112(1):153–163. doi: 10.1093/toxsci/kfp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowen WP, Wylie PG. Application of laser-scanning fluorescence microplate cytometry in high content screening. Assay Drug Dev Technol. 2006;4(2):209–221. doi: 10.1089/adt.2006.4.209. [DOI] [PubMed] [Google Scholar]
- 40.Brimacombe KR, et al. A dual-fluorescence high-throughput cell line system for probing multidrug resistance. Assay Drug Dev Technol. 2009;7(3):233–249. doi: 10.1089/adt.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norton JT, et al. Automated high-content screening for compounds that disassemble the perinucleolar compartment. J Biomol Screen. 2009;14(9):1045–1053. doi: 10.1177/1087057109343120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol (London) 1910;40:4–7. [Google Scholar]
- 43.Zhang JH, et al. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 44.Sanguinetti MC, et al. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81(2):299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 45.Titus SA, et al. A new homogeneous high-throughput screening assay for profiling compound activity on the human ether-a-go-go-related gene channel. Anal Biochem. 2009;394(1):30–38. doi: 10.1016/j.ab.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Ponti F, et al. Safety of non-antiarrhythmic drugs that prolong the QT interval or induce torsade de pointes: an overview. Drug Saf. 2002;25(4):263–286. doi: 10.2165/00002018-200225040-00004. [DOI] [PubMed] [Google Scholar]
- 47.Diaz GJ, et al. The [3H]dofetilide binding assay is a predictive screening tool for hERG blockade and proarrhythmia: Comparison of intact cell and membrane preparations and effects of altering [K+]o. J Pharmacol Toxicol Methods. 2004;50(3):187–199. doi: 10.1016/j.vascn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Tao H, et al. Automated tight seal electrophysiology for assessing the potential hERG liability of pharmaceutical compounds. Assay Drug Dev Technol. 2004;2(5):497–506. doi: 10.1089/adt.2004.2.497. [DOI] [PubMed] [Google Scholar]
- 49.Leishman D, Waldron G. Assay Technologies: Techniques Available for Quantifying Drug-Channel Interactions. In: Triggle DJ, et al., editors. Voltage-Gated Ion Channels as Drug Targets. Wiley-VCH; 2006. pp. 37–63. [Google Scholar]
- 50.Huang R, et al. Weighted feature significance: a simple, interpretable model of compound toxicity based on the statistical enrichment of structural features. Toxicol Sci. 2009;112(2):385–393. doi: 10.1093/toxsci/kfp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pritchard JF, et al. Making better drugs: Decision gates in non-clinical drug development. Nat Rev Drug Discov. 2003;2(7):542–553. doi: 10.1038/nrd1131. [DOI] [PubMed] [Google Scholar]
- 52.Schoonjans F, et al. Presentation of receiver-operating characteristics (ROC) plots. Clin Chem. 1996;42(6 Pt 1):986–987. [PubMed] [Google Scholar]
- 53.Xia M, et al. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci U S A. 2009;106(7):2412–2417. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons SO, et al. Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol Sci. 2009;111(2):202–225. doi: 10.1093/toxsci/kfp140. [DOI] [PubMed] [Google Scholar]
- 55.Mole DR, Ratcliffe PJ. Cellular oxygen sensing in health and disease. Pediatr Nephrol. 2008;23(5):681–694. doi: 10.1007/s00467-007-0632-x. [DOI] [PubMed] [Google Scholar]
- 56.Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16(9):4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redell MS, Tweardy DJ. Targeting transcription factors for cancer therapy. Curr Pharm Des. 2005;11(22):2873–2887. doi: 10.2174/1381612054546699. [DOI] [PubMed] [Google Scholar]
- 58.Simmons SO. Hypoxia response: a model toxicity pathway for high-throughput screening. Toxicol Sci. 2009;112(1):1–3. doi: 10.1093/toxsci/kfp193. [DOI] [PubMed] [Google Scholar]
- 59.Johnson RL, et al. A quantitative high-throughput screen for modulators of IL-6 signaling: a model for interrogating biological networks using chemical libraries. Mol Biosyst. 2009;5(9):1039–1050. doi: 10.1039/b902021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller SC, et al. Identification of known drugs that act as inhibitors of NF-kappaB signaling and their mechanism of action. Biochem Pharmacol. 79(9):1272–1280. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hancock MK, et al. HTS-Compatible beta-Lactamase Transcriptional Reporter Gene Assay for Interrogating the Heat Shock Response Pathway. Curr Chem Genomics. 2009;3:1–6. doi: 10.2174/1875397300903010001. [DOI] [PMC free article] [PubMed] [Google Scholar]