In an attempt to resolve the safety issues of viral vectors and the inefficiency issues of non-viral vectors, we are attempting to combine their advantages into an efficacious hybrid system through the use of vectorially-oriented, membrane-embedded, three component DNA packaging motors within a bioresponsive host vesicle. The ATP actuated DNA packaging motor from phi29 virus was chosen for testing this concept since one component, the 440 kD gp10 dodecamer, can be readily expressed and purified in histidine-tagged form, has a structure known to 2.1 Å resolution,1 and has been investigated for potential use in numerous applications.2,3 Although oligomeric protein assemblies have been incorporated into membranes for controlled transport applications,4 we are not aware of any previous efforts to transplant part of a functional viral motor into vesicle membranes with orientation control. The alternating hydrophilic-hydrophobic-hydrophilic trilayer structure of the gp10 connector array stem region (Figure 1) promotes aggregation in aqueous solutions via intermolecular association of the 2 nm band of hydrophobic residues.3 Incorporation of the phi29 connector array into a vesicle membrane, therefore, requires strategies that will accommodate both the hydrophobic domain in the dodecamer array and the required orientation for assembly of the other two motor components, pRNA and gp16 ATPase, to enable DNA packaging into the vesicle host. This work describes the development of a strategy to transplant the gp10 dodecamer arrays into vesicle membranes with orientation control using nitrilotriacetic acid-polyethylene glycol (NTA-PEG)-modified surfaces. The well-known property of PEG toward reducing nonspecific adsorption of proteins and vesicles onto solid substrates,5 as well as the deployment of a distal NTA unit for specific capture6 and orientation of N-his6-gp10, are key features of this approach.
Figure 1.
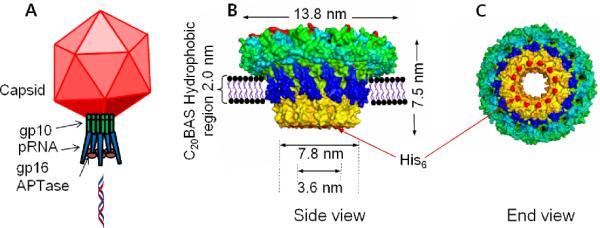
(A) Model of phi29 packaging motor showing the three motor components and their relationship with the capsid. (B) Conceptual diagram of phi29 N-his6-gp10 connector protein assembly in a C20BAS bolalipid membrane. (C) Exposed his6- sites at the narrow N-terminal end of the gp10 assembly that are available for Ni2+:NTA capture.
The method used for transplanting the gp10 array into C20BAS bolalipid vesicles is shown in Figure 2. The key to orientational control is fixation of the gp10 N-terminus to the glass surface via a short water-soluble polymer tether.7 A bias toward single insertions of N-his6-gp10 connector into C20BAS vesicles is achieved by producing low surface coverages of NTA-PEG so that surface-mediated reconstitution occurs with only a single immobilized gp10 connector array.
Figure 2.
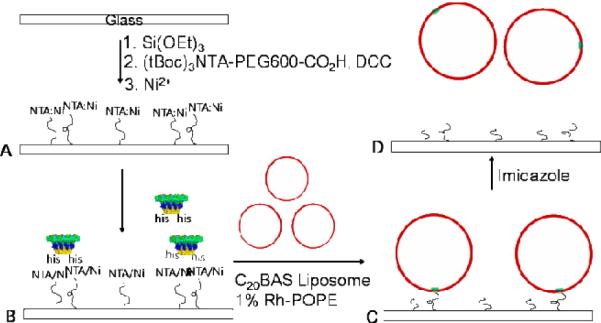
Transplantation strategy for incorporating oriented gp10 phi29 DNA motor into C20BAS bolalipid vesicles. A: Surface modification of glass with Ni2+:NTA-PEG600; B: Immobilization and orientation of N-his6-gp10 connector protein via Ni2+:NTA-PEG600 chelation; C: Transplantation of oriented N-his6-gp10 connector protein assembly into C20BAS vesicles via surface-mediated reconstitution; D: Release of immobilized C20BAS vesicles via Ni2+ stripping with imidazole. Final assembly of a functional phi29 motor would involve docking pRNA and gp16 ATPase with the vesicles obtained from D.
C20BAS bolalipid was chosen as the host membrane for the phi29 DNA motor since it has a similar hydrophobic thickness as the gp10 connector array stem domain (Figure 1) and is known to accommodate other hydrophobic peptides and integral membrane proteins.8 Since C20BAS prefers to adopt planar membranes in the absence of additional lipid components that can help stabilize a membrane structure with appreciable curvature,9 the inclusion of either cholesterol (Chol) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was attempted to enable the formation of stable C20BAS bolalipid-rich vesicles. TEM experiments (Figure S1, Supplementary Information) confirmed that 7:3 C20BAS:cholesterol and 9:1 C20BAS:POPC mixtures form stable vesicles upon probe sonication, in good agreement with DLS data showing that particles of 80–160 nm are formed by this procedure. These findings are also consistent with previous reports of homogeneous dispersions formed by binary C20BAS:POPC mixtures at low POPC molar ratios.10 We infer from these results that the monopolar lipids preferentially occupy the voids that would otherwise be present on the outer surface of the membranes formed by C20BAS in a transmembrane conformation,11 thus stabilizing curvature in the bolalipid vesicle membrane.
Confocal laser scanning microscopy (CLSM) images collected at NTA-PEG-grafted glass surfaces revealed little or no non-specific binding of the Ni2+:NTA-PEG surface (Figure 3A), however, extensive punctate surface staining was apparent on samples that had been pre-treated with N-his6-gp10 connector arrays (Figure 3B). Adjustment of the confocal plane to image the bulk phase revealed very little rhodamine (Rh) fluorescence in solution due to the absence of unbound vesicles. Addition of 2 M imidazole solution to these surfaces and incubation for 10 min prior to CLSM imaging revealed the opposite behavior, i.e., little or no surface fluorescence (Figure 3C) and extensive punctate Rh fluorescence in bulk solution (Figure 3D). TEM images of the imidazole-desorbed vesicles showed the presence of membrane-associated features whose size and shape are consistent with gp10 connector arrays (Figures 4 and S1). We infer from these observations that oriented insertion of N-his6-gp10 connector proteins occurred when they were exposed to small unilamellar C20BAS bolalipid vesicles. This conclusion is supported by the following observations: (1) absence of vesicle-associated fluorescence in samples that have not been exposed to connector protein (Figure 3A), (2) displacement of vesicle-associated Rh fluorescence upon imidazole stripping of Ni2+ from the his6:Ni2+:NTA-PEG interaction (Figure 3D), and (3) highly unfavorable interactions between the water-soluble PEG tether and the hydrophobic core of the C20BAS membrane that would be expected if the N-his6 terminus were oriented within the inner lumen of the vesicle. Similar results were obtained when 7:3 C20BAS:Chol vesicles were used in the surface-mediated reconstitution process. Taken together, these findings suggest that the sequence depicted in Figure 2 is a viable strategy for vectorial transplantation of N-his6-gp10 connector arrays into host vesicle membranes of appropriate hydrophobic thickness. The reversible adsorption of 9:1 C20BAS:POPC vesicles from Ni2+:NTA-PEG-grafted surfaces after transplantation of N-his6 gp10 connector protein was also confirmed by AFM. AFM images collected after rinsing with imidazole solution revealed a surface with few remaining vesicles (Figure S2, Supplementary Information). This result supports the conclusions drawn from CLSM experiments.
Figure 3.
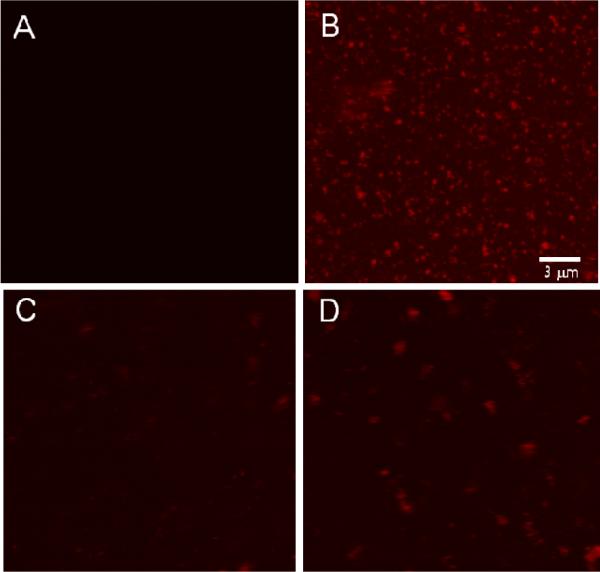
CLSM images of Ni2+:NTA-PEG-modified glass surfaces after exposure to 1% Rh-POPE labeled 9:1 C20BAS:POPC vesicles. A: Surface image taken after protein-free surfaces were exposed to bolalipid vesicles (control); B: Surface image taken after sequential exposure of the surface to N-his6 gp10 connector and bolalipid vesicles, respectively; C: Surface image of sample B taken after imidazole addition; D: Solution image of sample B after imidazole addition.
Figure 4.
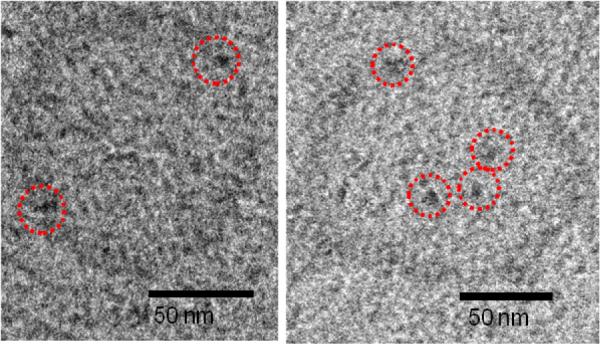
CryoEM of N-his6-gp10 connector array-containing 9:1 C20BAS:POPC vesicles produced as shown in Fig. 2. The membrane-embedded gp10 arrays are highlighted with dotted circles.
Since we seek the production of vesicles with a single gp10 array implanted in the membrane, low NTA-PEG600 surface coverages are required for enabling adequate spacing between individual surface-captured proteins to minimize vesicle-vesicle encounter at the interface during connector array insertion into the membrane. Increases in either the APTES concentration or treatment time produced surfaces that gave very large vesicles (> 10 μm diameter) that were difficult to displace from the surface, even with very high imidazole concentrations (CLSM data not shown). We infer from these findings that high surface coverages of gp10 connector protein lead to vesicle-vesicle fusion during the surface-mediated reconstitution step. Since these very large vesicles are presumed to possess numerous his6:Ni2+:NTA surface interactions, they would be expected to be difficult to competitively displace from the surface with imidazole.
In conclusion, Ni2+:NTA-PEG600-grafted glass surfaces are capable of immobilizing N-his6-gp10 from the phi29 DNA packaging motor. C20BAS bolalipid vesicles, stabilized by incorporation of 10 mol% POPC or 30 mol% cholesterol, can produce reconstituted vesicles with transplanted connector arrays embedded in the membrane upon exposure of the N-his6 gp10-primed surfaces to sonicated C20BAS dispersions. These oriented gp10 vesicles were detached by stripping the surface with excess imidazole, resulting in a construct that is primed for subsequent pRNA and gp16 ATPase binding to enable ATP-driven DNA packaging. Development of this simple DNA packaging motor implantation strategy is an important first step for the practical realization of nonviral vectors whose DNA cargo can be controllably encapsulated within a carrier system having properties that can be readily optimized for in vivo performance.
Supplementary Material
Acknowledgment
This work was supported by NIH PN2 EY 018230), CA112427 and KSF KRF-2007-357-D00084. The atomic coordinates for phi29 gp10 were obtained from the PDB under accession number 1FOU.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. J. Mol. Biol. 2002;315:663. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- (2).Liu H, Guo S, Roll R, Li J, Diao Z, Shao N, Riley MR, Cole AM, Robinson JP, Snead NM, Shen G, Guo P. Cancer Biol. Ther. 2007;6:697. doi: 10.4161/cbt.6.5.3962. [DOI] [PubMed] [Google Scholar]
- (3).Guo YY, Blocker F, Xiao F, Guo P. J. Nanosci. Nanotechnol. 2005;5:856. doi: 10.1166/jnn.2005.143. [DOI] [PubMed] [Google Scholar]
- (4).Graff A, Sauer M, Van GP, Meier W. Proc. Nat'l. Acad. Sci. USA. 2002;99:5064. doi: 10.1073/pnas.062654499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Otsuka H, Nagasaki Y, Kataoka K. Curr. Op. Coll. Interfac. Sci. 2001;6:3. [Google Scholar]
- (6).Kang E, Park J-W, McClellan SJ, Kim J-M, Holland DP, Lee GU, Franses EI, Park K, Thompson DH. Langmuir. 2007;23:6281. doi: 10.1021/la063719e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Christensen SM, Stamou D. Soft Matter. 2007;3:828. doi: 10.1039/b702849k. [DOI] [PubMed] [Google Scholar]
- (8).Febo-Ayala W, Morera-Felix SL, Hrycyna CA, Thompson DH. Biochemistry. 2006;45:14683. doi: 10.1021/bi061159c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Thompson DH, Svendsen CB, DiMeglio C, Anderson VC. J. Org. Chem. 1994;59:2945. [Google Scholar]
- (10).Brownholland DP, Longo GS, Struts AV, Justice MJ, Szleifer I, Petrache HI, Brown MF, Thompson DH. Biophys. J. 2009;97:2700. doi: 10.1016/j.bpj.2009.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Holland DP, Struts AV, Brown MF, Thompson DH. J. Am. Chem. Soc. 2008;130:4584. doi: 10.1021/ja710190p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


