Abstract
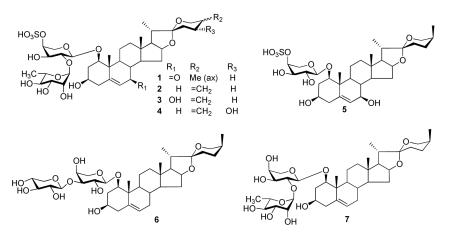
Six new steroidal saponins (1-6), angudracanosides A-F, were isolated from fresh stems of Dracaena angustifolia (Agavaceae), together with eight known compounds. The structures of compounds 1-6 were determined by detailed spectroscopic analyses and chemical methods. Antifungal testing of all compounds showed that 6 and 7 were active against Cryptococcus neoformans with IC50s of 9.5 and 20.0 μg/mL, respectively.
The genus Dracaena (Agavaceae) contains more than 60 species that are distributed from the Old World tropic region to the Canary Islands. Some species contain C27 steroidal saponins1, as evidenced in our previous phytochemical study of D. cochinchinensis.2 The C27 steroidal saponins have demonstrated antitumor1, anti-inflammatory3, and hypoglycemic4 activities as well as therapeutic potential for cardiovascular diseases.5 Some C27 steroidal saponins have antifungal activity, which varies with the aglycone structure and with the number and structure of monosaccharide units in their sugar chains.6
Dried roots and rhizomes of D. angustifolia Roxb (Agavaceae) collected in Vietnam have been studied, revealing the presence of several steroidal saponins with antiproliferative activity.7 Our interest in antifungal steroidal saponins prompted us to investigate fresh stems of this species that were collected in Southwest China. As a result, six new steroidal saponins (1-6), which were named angudracanosides A-F, and eight known compounds were isolated. The antifungal activity of these compounds against several human fungal pathogens was evaluated, and the results are presented in paper.
Results and Discussion
The fresh stems of D. angustifolia were extracted with methanol under reflux. A n-butanol-soluble portion of the methanol extract was chromatographed on the highly porous absorption resin Diaion HP-20, normal phase silica gel, Sephadex LH-20, and reversed-phase silica gel RP-18 to afford six new compounds (1-6). In addition, eight known compounds were identified as (25S)-spirost-5-en-1β,3β-diol-1-sulphate8, (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside9, 26-O-β-D-glucopyranosyl-furosta-5-ene-1β,3β,22ζ,26-tetrol-1-O-α-L-rhamnopyranosyl-(1-2)-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside10, (25S)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranosde10, terreside11,12, alliospiroside A (7)13, 3,4,5-trimethoxyphenyl-1-O-β-D-glucopyranoside14, and 3,4,5-trimethoxyphenyl-1-O-α-L-rhamnopyranosyl-(1-6)-β-D-gluco-pyranoside15, respectively, by comparison of the spectroscopic data with those reported in the literature. The new compounds were named angudracanoside A-F (1-6).
Angudracanoside A (1) was obtained as white amorphous powder. The negative ion HRFABMS and 13C NMR (DEPT) determined its molecular formula to be C38H58O16S. The IR (KBr) spectrum of 1 displayed the absorptions characteristic for hydroxy groups (3441 cm−1), a (25S)-spirostanol moiety [983, 918, 897, 838 cm−1 (intensity 918>897)], and a S-O stretching band at 1227 cm−1 indicative of a sulfate group.9 The 1H NMR spectrum of 1 (Table 2) displayed two methyl singlets [δ 0.80 (s, Me-18) and 1.51 (s, Me-19)], three methyl doublets [δ 2.11 (t-like, J = 6.8 Hz, Me-21), 1.03 (d, J = 6.3 Hz, Me-27), and 1.75 (d, J = 6.0 Hz, Me-Rha)], and two anomeric proton signals [δ 4.64 (d, J = 7.5 Hz) and 6.29 (br s)]. In the 13C NMR and DEPT spectra (Table 1), 38 carbon signals were observed, including 27 resonances assigned to a C27 steroidal skeleton and 11 carbon resonances from two sugars. Compared to the literature values of similar compounds16, the carbon resonances at δ 202.0, 162.7, 128.1, 109.7, 101.8, and 100.3 could be assigned to a ketone functionality, two olefinic carbons, one spirostanol carbon of the aglycone, and two anomeric carbons, respectively. These NMR data indicated that compound 1 was a spirostanol saponin with two sugar moieties. Acid hydrolysis of 1 with 1 M HCl in dioxane afforded L-arabinose and L-rhamnose. The absolute configurations of both sugars were determined GC analysis of their corresponding trimethylsilated L-cysteine adducts.17 The 1H and 13C NMR data of the sugar moiety of 1 were identical to those of (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside.9 The differences between 1 and the known compound were the molecular weight and the 13C NMR resonances of the B and F rings. Compound 1 was 14 mass units greater than (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside. The obvious spectroscopic differences at C-6, C-7 and C-8 on the B ring (δ 128.1, 202.0 and 42.0 for 1, respectively, versus δ 124.5, 33.5 and 33.0 for (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside, respectively) suggested that the ketone functionality was located at C-7 in 1. This was confirmed by the HMBC correlations of H-6, H-8, and H-9 with C-7 (δ 202.0). Other key HMBC correlations are shown in the Supporting Information. The IR absorption of 1 at 1655 cm−1 was consistent with that of 7-ketodiosgenin from Tamus edulis.18 Finally, the typical F ring carbon resonances of C-23, C-24, C-25 and C-27 at δ 26.5, 26.3, 27.7 and 16.5, respectively, strongly supported a (25S)-spirostanol skeleton for 1,9,16 in contrast to a (25R)-spirostanol skeleton for (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside with distinctly different chemical shifts of those carbons at δ 32.2, 28.2, 30.8, and 17.4, respectively. Thus, the aglycone of 1 was determined to be as shown. The sequence of the sugar chain in 1 was determined to be the same as that of (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside, by the HMBC and HSQC experiments. HMBC correlations from the anomeric signal at δ 6.29 (Rha H-1) to δ 76.0 (Ara C-2) and from δ 4.64 (Ara H-1) to δ 82.3 (Agly C-1) were observed. Moreover, the deshielded chemical shifts observed in the HSQC spectrum for the arabinosyl C-4 and H-4 at δ 76.1 and δ 5.32 indicated that the OH group at C-4 of the arabinosyl moiety in 1 was acylated, which was supported by the upfield shifts of the arabinosyl C-3 and C-5 at δ 75.2 and 66.2, respectively. This substituent was confirmed to be a SO3H group by the FABMS (fragment ion peaks at m/z 80 and 97, respectively). Therefore, compound 1 was characterized as (25S)-spirost-5-en-7-keto-1β,3β-diol-1-O-α-L-rhamnopyranosyl-(1-2)-(4-O-sulfo)-α-L-arabinopyranoside.
Table 2.
1H NMR Signals of Compounds 1-6 (in C5D5N, δ in ppm and J in Hz)a.
| C | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 3.71 dd (11.7, 3.8) | 3.76 dd (12.5, 3.0) | 3.76 dd (11.7, 3.8) | 3.78 dd (11.7, 2.6) | 3.77 dd (11.0, 3.5) | 3.87 dd (11.5, 3.5) |
| 2 | 2.78 br d (11.7) | 2.72 m | 2.64 br d (11.7) | 2.66 m | 2.66 m | 2.82 br d (11.5) |
| 2.37 br d (11.7) | 2.33 br d (12.5) | 2.28 br d (11.7) | 2.32 br d (11.7) | 2.02 q-like (11.9) | 2.16 br d (11.7) | |
| 3 | 3.86 m | 3.77 m | 3.84 m | 3.85 m | 3.82 m | 3.85 m |
| 4 | 2.21 br d (12.1) | 2.74 br d (9.0) | 2.51 dd (12.0, 4.8) | 2.56 br d (12.1) | 2.70 dd (11.5, 3.7) | 2.60 dd (11.8, 4.4) |
| 2.68 m | 2.67 m | 2.74 br d (12,0) | 2.66 m | 2.59 q-like (11.5) | 2.69 br d (11.8) | |
| 6 | 5.94 s | 5.59 br d (5.5) | 6.03 br d (5.2) | 5.55 br d (5.2) | 6.02 br d (6.5) | 5.57 br d (5.2) |
| 7 | - | 1.68 m, 1.52 m | 4.00 br s | 1.83 m,1.67 dd (11.9, 7.9) | 4.02 br s | 1.48 m, 1.88 dd (11.9, 4.7) |
| 8 | 2.49 dd (4.5, 5.0) | 1.75 m | 2.18 m | 1.49 m | 2.09 m | 1.72 m |
| 9 | 1.88 m | 1.46 m | 2.06 m | 1.47 m | 2.06 m | 1.46 m |
| 11 | 1.68 m, 2.97 brd (10.6) | 1.52 m, 2.87 br d (11.5) | 1.74 m, 2.95 br d (10.6) | 1.51 m, 2.86 br d (10.8) | 1.50 m, 2.84 br d (11.5) | 1.55 m, 2.88 br d (11.5) |
| 12 | 1.52 m, 1.06 m | 1.40 m, 1.21 ddd (11.5, 11.5, 3.6) | 1.40 m, 1.28 ddd (10.6, 10.6, 4.0) | 1.38 m, 1.20 ddd (11.8, 11.8, 3.6) | 1.43 m, 1.17 m | 1.69 m, 1.42 m |
| 14 | 1.60 m | 1.06 dd (7.0, 12.1) | 1.68 dd (6.8, 12.1) | 1.34 m | 1.68 m | 1.06 m |
| 15 | 3.24 m, 1.65 m | 1.46 m, 1.97 dd (8.0, 12.1) | 2.50 m, 1.51 ddd (12.5, 6.8, 8.6) | 1.98 m,1.38 m | 2.64 m, 1.54 m | 1.46 m, 1.99 ddd (6.7, 12.0, 7.3) |
| 16 | 4.50 q-like (8.6) | 4.47 q-like (9.0) | 4.59 q-like (8.6) | 4.46 q-like (8.5) | 4.54 q-like (7.0) | 4.49 q-like (7.3) |
| 17 | 1.67 m | 1.67 m | 1.64 m | 1.66 m | 1.79 dd (7.0, 6.5) | 1.70 m |
| 18 | 0.80 s | 0.81 s | 0.88 s | 0.80 s | 0.90 s | 0.81 s |
| 19 | 1.51 s | 1.46 s | 1.44 s | 1.40 s | 1.17 s | 1.40 s |
| 20 | 2.11 t-like (6.8) | 2.19 t-like (7.0) | 1.91 t-like (6.5) | 2.05 t-like (12.0) | 2.12 m | 1.91 t-like (6.8) |
| 21 | 1.06 d (7.0) | 1.02 d (7.0) | 1.00 d (7.0) | 1.03 d (6.8) | 1.07 d (7.0) | 1.05 d (7.0) |
| 23 | 1.85 m, 1.41 m | 2.67 m, 2.21 m | 1.68 m, 1.60 m | 1.98 dd (10.5, 12.3), 2.40 dd (6.3, 12.3) | 2.12 m, 1.87 m | 1.99 m, 1.85 m |
| 24 | 1.41 m, 1.31 m | 2.21 m, 1.88 m | 2.60 m, 2.18 m | 5.00 dd (6.3, 10.5) | 1.43 m, 1.24 m | 1.49 m, 1.33 m |
| 25 | 1.48 m | - | - | - | 1.30 m | 1.50 m |
| 26 | 4.00 br d (11.5) | 4.41 br d (12.0) | 4.37 br d (12.0) | 4.47 br d (11.5) | 3.98 br d (10.5) | 4.04 br d (11.5) |
| 3.32 d (11.5) | 3.98 d (12.0) | 3.94 d (12.0) | 4.24 dd (11.5, 3.3) | 3.28 d (10.5) | 3.66 d (11.5) | |
| 27 | 1.03 d (6.3) | 4.78 brs | 4.77 br s | 5.64 br s, 5.20 br s | 1.04 d (7.0) | 1.03 d (6.3) |
| Ara-1′ | 4.64 d (7.5) | 4.63 d (7.5) | 4.59 d, 7.5 | 4.72 d (7.5) | 4.57 d (7.0) | 4.77 d (7.5) |
| 2′ | 4.52 dd (9.2, 7.5) | 4.51 dd (9.0, 7.5) | 4.63 dd (8.2, 7.5) | 4.61 dd (9.3, 7.5) | 4.32 dd (9.0, 7.0) | 4.56 dd (8.2, 7.5) |
| 3′ | 4.23 dd (9.2, 3.6) | 4.24 dd (9.0 3.5) | 4.22 dd (8.2, 4.5) | 4.21 dd (9.3, 3.5) | 4.20 dd (9.0, 3.0) | 4.19 dd (8.2, 3.5) |
| 4′ | 5.32 m | 5.29 m | 5.27 m | 5.27 m | 5.48 br s | 4.46 br d (3.1) |
| 5′ | 4.65 dd (2.0, 12.0) | 4.72 dd (12.0, 2.5) | 4.63 m | 4.64 dd (11.5, 2.1) | 4.76 dd (12.0, 2.2) | 4.29 br d (12.3) |
| 3.71 brd (12.0) | 3.69 br d (12.0) | 3.58 br d (12.5) | 3.68 br d (11.5) | 3.63 d (12.0) | 3.71 dd (12.3, 3.1) | |
| Rha- 1′′ |
- Xyl-1′′ | |||||
| 6.29 brs | 6.22 brs | 6.27 br s | 6.30 br s | 5.30 d (7.5) | ||
| 2′′ | 4.65 m | 4.61 m | 4.71 br d (3.0) | 4.67 br d (3.5) | - 2′′ | 4.01dd (7.5, 8.2) |
| 3′′ | 4.61 m | 4.64 dd (3.5, 9.1) | 4.63 dd (3.0, 9.0) | 4.63 dd (3.5, 10.1) | - 3′′ | 4.22 dd (3.5, 10.1) |
| 4′′ | 4.54 dd (8.4, 8.4) | 4.31 dd (9.1, 9.1) | 4.35 dd (9.0, 9.0) | 4.21 m | - 4′′ | 4.15 m |
| 5′′ | 4.80 m | 4.77 m | 4.83 dq (9.0, 6.0) | 4.80 dq (10.1, 6.0) | - 5′′ | 4.35 dd (11.0, 4.6) |
| 6′′ | 1.75 d (6.0) | 1.71 d (6.0) | 1.75 d (6.0) | 1.72 d (6.4) | - | 3.75 dd (11.0, 5.6) |
Assignments were based on DEPT, HMQC, and HMBC experiments.
Table 1.
13C NMR Spectroscopic Data of Compounds 1-6 (C5D5N)a
| C | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 82.3 | 83.7 | 83.5 | 83.7 | 82.9 | 83.5 |
| 2 | 37.3 | 37.4 | 37.6 | 37.3 | 37.3 | 37.8 |
| 3 | 67.7 | 68.2 | 68.0 | 68.1 | 68.0 | 68.2 |
| 4 | 43.6 | 43.9 | 44.3 | 43.9 | 43.7 | 43.9 |
| 5 | 162.7 | 139.6 | 142.8 | 139.5 | 142.6 | 139.6 |
| 6 | 128.1 | 125.0 | 129.0 | 124.9 | 128.5 | 124.8 |
| 7 | 202.0 | 32.2 | 65.0 | 32.1 | 64.6 | 32.1 |
| 8 | 42.0 | 33.2 | 42.8 | 33.1 | 42.2 | 33.2 |
| 9 | 50.1 | 50.4 | 50.8 | 50.2 | 50.4 | 50.4 |
| 10 | 43.6 | 43.1 | 44.1 | 43.0 | 43.7 | 43.0 |
| 11 | 23.8 | 24.0 | 24.2 | 24.0 | 23.5 | 24.0 |
| 12 | 40.0 | 40.2 | 40.2 | 40.1 | 40.1 | 40.5 |
| 13 | 40.8 | 40.2 | 40.4 | 40.0 | 40.1 | 40.3 |
| 14 | 49.0 | 56.7 | 39.2 | 56.6 | 40.0 | 56.9 |
| 15 | 34.4 | 32.5 | 32.8 | 32.4 | 32.4 | 32.5 |
| 16 | 81.6 | 81.5 | 82.2 | 81.9 | 81.5 | 81.3 |
| 17 | 62.0 | 62.9 | 63.4 | 62.4 | 63.0 | 62.9 |
| 18 | 16.8 | 16.9 | 17.0 | 16.6 | 16.4 | 16.8 |
| 19 | 13.3 | 15.1 | 15.4 | 15.0 | 14.9 | 15.1 |
| 20 | 42.6 | 42.0 | 42.4 | 42.1 | 42.2 | 42.6 |
| 21 | 15.0 | 14.9 | 14.4 | 14.9 | 15.0 | 14.9 |
| 22 | 109.7 | 109.5 | 109.8 | 111.8 | 109.8 | 109.8 |
| 23 | 26.5 | 33.4 | 33.7 | 43.6 | 26.5 | 26.5 |
| 24 | 26.3 | 29.0 | 29.4 | 67.3 | 26.3 | 26.3 |
| 25 | 27.7 | 144.7 | 145.0 | 149.4 | 27.6 | 27.7 |
| 26 | 65.2 | 65.2 | 65.3 | 64.6 | 65.7 | 65.2 |
| 27 | 16.5 | 108.7 | 108.9 | 106.5 | 16.7 | 16.4 |
| Ara-1′ | 101.8 | 100.3 | 100.3 | 100.3 | 102.3 | 102.3 |
| 2′ | 76.0 | 75.9 | 76.3 | 76.2 | 72.9 | 71.7 |
| 3′ | 75.2 | 75.2 | 75.5 | 75.2 | 74.0 | 84.0 |
| 4′ | 76.1 | 76.4 | 76.3 | 76.2 | 76.4 | 69.3 |
| 5′ | 66.2 | 65.7 | 66.3 | 66.3 | 65.4 | 67.3 |
| Rha-1′′ | 100.3 | 101.3 | 101.3 | 101.7 | ||
| 2′′ | 72.4 | 72.6 | 72.9 | 72.6 | ||
| 3′′ | 72.4 | 72.5 | 72.8 | 72.6 | ||
| 4′′ | 74.3 | 74.7 | 74.7 | 74.5 | ||
| 5′′ | 69.6 | 69.5 | 69.4 | 69.8 | ||
| 6′′ | 19.0 | 19.0 | 19.0 | 19.0 | ||
| Xyl-1′′ | 106.7 | |||||
| 2′′ | 75.5 | |||||
| 3′′ | 78.3 | |||||
| 4′′ | 71.2 | |||||
| 5′′ | 67.3 |
Assignments were based on DEPT, 1H-1H COSY, HMQC, and HMBC experiments
Angudracanoside B (2) was obtained as white amorphous powder. The negative ion HRFABMS and 13C NMR (DEPT) analyses indicated an empirical molecular formula of C38H58O15S, which is two hydrogen atoms fewer than that of (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside. The overall 1H and 13C NMR data of 2 and (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside were very similar. The structural difference between them was in the F ring, i.e., an exo-double bond between C-25 and C-27 in 2 replaced the C-27 methyl group in (25R)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-4-O-sulfo-β-D-arabinopyranoside. The chemical shift values of the double bond carbons [δ 108.7 (CH2) and 144.7 (C)] were consistent with those of analogs reported in the literature.16 HMBC correlations of H-27 (δ 4.78) with C-24 (δ 29.0) and C-26 (δ 65.2) confirmed the presence of this double bond. Thus, compound 2 was formulated as spirost-5,25(27)-en-1β,3β-diol-1-O-α-L-rhamnopyranosyl-(1-2)-(4-O-sulfo)-α-L-arabinopyranoside.
The molecular formula of angudracanoside B (3) was determined as C38H58O16S from the negative ion HRFABMS and 13C NMR (DEPT) spectra, one oxygen atom more than that of 2. The NMR chemical shifts of 3 were closely related to those of 2, except for signals due to the B ring. Compared to 2, the olefinic proton and carbons in the B ring were shifted downfield to δ 6.03 and δ 129.0, 142.8 (versus δ 5.59 and δ 125.0, 139.6 in 2). In addition, the methylene carbon (δ 1.52/1.68, δ 32.2) linked to the olefinic carbon (C-6) disappeared and was replaced by an oxygenated methine carbon (δ 4.00 and δ 65.0). These data indicated the presence of an OH group at C-7, which was supported by HMBC correlations of H-7 (δ 4.00) with C-5 (δ 142.8), C-6 (δ 129.0) and C-8 (δ 42.8). The orientation of the 7-OH group was determined by the ROESY experiment. As H-7 showed strong NOE correlations with H-14 (δ 1.68), H-15α (δ 2.50), and H-6 (δ 6.03) it must be α-oriented; hence the 7-OH group must be β-oriented. Other key ROE correlations of 3 support the structure of compound 3 as being (25S)-spirost-5-en-1β,3β,7β-triol-1-O-α-L-rhamnopyranosyl-(1-2)-(4-O-sulfo)-α-L- arabinopyranoside (see Supporting Information).
Angudracanoside D (4) had molecular formula C38H58O16S, as deduced from the negative ion HRFABMS and 13C NMR (DEPT) spectra, the same as that of 3, and one more oxygen atom than that of 2. The NMR chemical shifts of 4 were similar to those of 2 with exceptions of signals due to the F ring. The difference between 4 and 2 was an additional OH at C-24 (δ 67.0), as indicated by a downfield shift of the C-27 olefinic carbon (Δδ, −2.2 ppm). HMBC correlations of H-27 (δ 5.64, 5.10) with C-24 (δ 67.0) and C-25 (δ 149.4) and H-23 (δ 1.98, 2.40) with C-24 (δ 67.0), C-25 (δ 149.4), and C-22 (δ 111.8) were also observed, which confirmed the OH at C-24. In addition, the 24-H proton [δ 5.00 (dd, J = 6.3, 10.5 Hz) was coupled to 23-H by 6.3 and 10.5 Hz. The J values implied that the 24-H was axial in orientation. Thus, compound 4 was deduced to be spirost-5,25(27)-en- 1β,3β,24α-triol-1-O-α-L-rhamnopyranosyl-(1-2)-(4-O-sulfo)-α-L-arabinopyranoside.
Angudracanoside D (5) had molecular formula C32H50O12S. In the 13C NMR and DEPT spectra of 5 (Table 1), 32 carbon signals were observed including 27 resonances assigned to a steroidal skeleton and five resonances assigned to a sugar. Acid hydrolysis of 5 confirmed the presence of L-arabinose in the sugar residue. The sulphate group attached to C-4 of the arabinopyranosyl unit, which in turn was attached to the OH at C-1 of the aglycone, was confirmed by comparison of the 1H and 13C NMR data with those of compounds 1-4 (Tables 1 and 2). The aglycone of 5 was identified as 7-hydroxy-(25S)-ruscogenin by comparison of its NMR data with those of compounds 3 and 1, as it shared the same A/B/C/D/E rings as those of 3 and the same F-ring as that of 1. 2D-NMR experiments including HMBC and ROESY spectra were also performed to facilitate the resonance assignments. Thus, the structure of compound 5 was determined to be (25S)-spirost-5-en-1β,3β, 7β-triol-1-O-(4-O-sulfo)-α-L-arabinopyranoside.
The molecular formula of angudracanoside E (6) was deduced as C37H58O12 from its quasi-molecular ion peak at m/z 693.3853 (calcd. for C37H58O12 693.3850) in the HRFABMS and the 13C NMR (DEPT) spectrum. Comparison of the 1H and 13C NMR spectra of 6 with those of (25S)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside 20 showed that both compounds possess the same aglycone with a sugar moiety at C-1. The only difference was that compound 6 lacked a set of 13C NMR resonances corresponding to a α-L-rhamnopyranosyl unit. Acid hydrolysis of 6 yielded L-arabinose and D-xylose as the sugar residues. HMBC correlation of the anomeric proton of xylose (δ 4.77) with C-3 of the arabinose (δ 84.0) confirmed the interglycosidic linkage. Therefore, compound 6 was determined to be (25S)-ruscogenin-1-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside.
The isolated steroidal saponins were tested for their activity against the fungal pathogens Candida albicans, C. glabrata, C. krusei, Cryptococcus neoformans, and Aspergillus fumigatus. Only compounds 6 and 7 exhibited antifungal activity selectively against C. neoformans, with minimum inhibitory concentrations (IC50s) of 9.5 and 20.0 μg/mL, respectively. Amphotericin B was used as a positive control and exhibited an IC50 of 2.5 μg/mL against C. neoformans. It was noted that both compounds possess (25S)-ruscogenin as their aglycone, and a different disaccharide sugar moiety at C-1. This study has further demonstrated that the antifungal activity of the C27 steroidal saponins is associated with individual aglycone and disaccharide sugar moieties at C-1.
It was interesting to note that the structures of steroidal saponins isolated in this study differed from those isolated from the same species that was collected in a different location.7 There was no single saponin that was isolated in both studies, although most saponins shared the basic 1,3-dihydroxylated steroidal skeleton with varying oxidation levels at different positions. The most distinguishing feature may be the presence of a sulphate group in the sugar moieties of some saponins isolated by us, while the saponins isolated by the Vietnam group had acetylated sugar moieties and a β-D-fucopyranosyl unit attached to the OH group at C-24 on the F-ring. It appears that geographic location, season of collection, and/or environmental conditions contribute to the chemical diversity of this plant species.
Experimental Section
General Experimental Procedures
Optical rotations were measured with a HORIBA SEPA-300 high-sensitive polarimeter. IR (KBr) spectra were measured on a Bio-Rad FTS-135 spectrophotometer. NMR spectra were measured in pyridine-d5 solution and recorded on a Bruker DRX-500 instrument (500 MHz for 1H NMR, and 125 MHz for 13C NMR) at 25 °C, using TMS as an internal standard. Mass spectra were recorded on a VG Auto Spec-3000 mass spectrometer using glycerol as matrix. Column chromatography (CC) was performed on Diaion HP20SS (Mitsubishi Chemical Co.), RP-18 (40-63 μm, Merck Chemical Co.), Sephadex LH-20 (Mitsubishi Chemical Co.), and Chromatorex ODS (100-200 mesh, Fuji Silysia Chemical Co. Ltd). Precoated silica gel plates (Qingdao Haiyang Chemical Co.) were used for TLC. Detection was done by spraying the plates with 10% sulfuric acid, followed by heating. GC analysis was run on an Agilent Technologies HP5890 gas chromatograph equipped with a H2 flame ionization detector. The column was a 30QC2/AC-5 quartz capillary column (30 m×0.32 mm) using the following conditions: column temperature: 180 °C/280 °C; programmed increase, 3 °C/min; carrier gas: N2 (1 mL/min); injection and detector temperature: 250 °C; injection volume: 4 μL, split ratio: 1/50. Silica gel (200-300 mesh and 10-40 μm) and reversed phase silica gel RP-8 (40-63 μm) were used for CC.
Plant Material
Fresh stems of D. angustifolia were collected in Xishuangbanna, Yunnan, China, during July 2005. The sample was identified by Prof. Chong-Ren Yang. A voucher specimen has been deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences.
Extraction and Isolation
The chipped fresh stems of D. angustifolia (5.0 kg) were extracted with MeOH under reflux. The MeOH extract was condensed under reduced pressure. The viscous concentrate was partitioned between H2O and n-BuOH. The n-BuOH fraction (27.938 g) was subjected to CC on highly porous absorption resin (Diaion HP-20) eluting with MeOH-H2O (0:1-1:0) to afford five fractions (A-E). Fraction A (1.201 g) was chromatographed on MCI gel eluting with MeOH-H2O (0:1-1:0), and then on silica gel with CHCl3-MeOH-H2O (9:1:0.1-7:3:0.5), to give compounds 6 (22 mg), (25S)-spirost-5-en-1β,3β-diol-1-sulphate (21 mg), (25S)-spirost-5-en-1β,3β-diol-1-sulphate (29 mg), 3,4,5-trimethoxyphenyl-1-O-β-D-glucopyranoside (21 mg) and 3,4,5-trimethoxy-phenyl-1-O-α-L-rhamnopyranosyl-(1-6)-β-D-glucopyranoside (29 mg). Fraction B (2.113g) was subjected to RP-18 CC eluting with MeOH-H2O (4:6-1:0) and silica gel CC eluting with CHCl3-MeOH-H2O (9:1:0.1-7:3:0.5) to give 4 (4 mg) and 5 (20 mg). Fraction C (5.30 g) was subjected to ODS CC eluting with MeOH-H2O (6:4-1:0), Sephadex LH-20 eluting with MeOH-H2O (0:1-6:4), and silica gel CC with CHCl3-MeOH-H2O (8:2:0.2-7:3:0.5) to give 1 (14 mg), 3 (274 mg), 2 (42 mg) and (25S)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside (14 mg). Fraction D (421 mg) was subjected to RP-18 CC eluting with MeOH-H2O (4:6-1:0), Sephadex LH-20 eluting with MeOH-H2O (0:1-7:3), and silica gel CC with CHCl3-MeOH-H2O (8:2:0.2-7:3:0.5) to give 26-O-β-D-glucopyranosyl-furosta-5-ene-1β,3β,22ζ,26-tetrol-1-O-α-L-rhamnopyranosyl-(1-2)-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside (29 mg) and terreside (95 mg); Fraction I (1.06 g) was subjected to RP-18 eluting with MeOH-H2O (3:7-1:0) and silica gel CC with CHCl3-MeOH-H2O (8:2:0.2-7:3:0.5) to give (25S)-ruscogenin-1-O-α-L-rhamnopyranosyl-(1-2)-O-β-D-xylopyranosyl-(1-3)-α-L-arabinopyranoside (4 mg) and alliospiroside A (7) (5 mg).
Angudracanoside A (1): white amorphous powder; (c 0.40, pyridine); IR (KBr) vmax: 3441 (OH), 2907 (CH), 1655 (C=O), 1227 (S-O), 983, 918, 897, 838 (intensity 918>897, (25S)-spiroketal) cm−1; 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 801 [M - 1]−. HRESIMS (negative ion mode): m/z 801.3379 [M - H]− (calcd. for C38H57O16, 801.3367).
Angudracanoside B (2): white amorphous powder; (c 0.14, pyridine); IR (KBr) vmax: 3439(OH), 2908 (CH), 1228 (S-O), 1092, 1050, 983, 917, 899, 837 cm−1; 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 785 [M - H]−; HRESIMS (negative ion mode): m/z 785.3430 [M - H]− (calcd. for C38H57O15S, 785.3418).
Angudracanoside C (3): white amorphous solid, (c 0.11, pyridine). IR (KBr) vmax: 3425 (OH), 2935 (CH), 1229 (S-O), 1049, 983, 918, 900, 850 cm−1; 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 801 [M - H]−; HRESIMS (negative ion mode): m/z 801.3357 [M - H]− (calcd. for C38H57O16S, 801.3367).
Angudracanoside D (4): white amorphous solid; (c 0.28, pyridine); IR (KBr) vmax: 3439(OH), 2908 (CH), 1224 (S-O), 1092, 1050, 983, 917, 899, 837 cm−1; 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 801 [M - H]−; HRESIMS (negative ion mode): m/z 801.3382 [M - H]− (calcd. for C38H57O16S, 801.3367).
Angudracanoside E (5): white amorphous powder; (c 0.05, pyridine); IR (KBr) vmax: 3439 (OH), 2927 (CH), 1217 (S-O), 1053, 986, 918, 899, 849 [intensity 912>899, 25(S)-spiroketal] cm−1; 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 657 [M - H]−. HRESIMS (negative ion mode): m/z 657.2947 [M - H]− (calcd. for C32H49O12S, 657.2944).
Angudracanoside F (6): white amorphous powder, (c 0.09, pyridine). IR (KBr) vmax: 3425(OH), 2920 (CH), 986, 919, 895, 873(intensity 919>895, (25S)-spiroketal) cm−1. 1H and 13C NMR (pyridine-d5): see Tables 1 and 2; FABMS (negative ion mode): m/z 694 [M - 1]−. HRESIMS (negative ion mode): m/z 693.3853 [M - H]− (calcd. for C37H57O12, 693.3850).
Acid Hydrolysis of 1-6: Compounds 1 (10 mg), 2-5 (5 mg), and 6 (8 mg) in 1M HCl-dioxane (1:1, v/v, 5 mL) were heated at 85° in a water bath for 6 h, respectively. The reaction mixtures were partitioned between CHCl3 and H2O four times. The aqueous layers were passed through Amberlite IRA-401 (OH− form), and the eluate was concentrated to dryness to give a saccharide mixture. Solutions of the sugar residues of these compounds in 1.5 mL of pyridine were added to L-cysteine methyl ester hydrochloride (1.0 mg) and kept at 60 °C for 1h, respectively. Trimethylsilylimidazole (1.5 mL) was added to the reaction mixtures and they were kept at 60 °C for 30 min. The supernatants (4 μL) were analyzed by GC, respectively.21 The monosaccharides of compounds 1-4 were determined by GC analysis of the derivatives to be L-arabinose and L-rhamnose, of which the retention times were 14.250 min and 15.823 min, respectively. The monosaccharide of compound 5 was determined to be L-arabinose, of which the retention times were 14.449 min. The monosaccharides of compound 6 were determined to be L-arabinose and L-xylose, of which the retention times were 14.250 min and 14.474 min, respectively.
Antifungal and Antibacterial Bioassay
Susceptibility testing was performed using a modified version of the NCCLS methods22,23 using organisms obtained from the American Type Culture Collection (Manassas, VA) including Candida albicans ATCC 90028, Candida glabrata ATCC 90030, Candida krusei ATCC 6258, Cryptococcus neoformansATCC 90113, and Aspergillus fumigatus ATCC 90906. The detailed procedures have been described in a previous paper.24
Supplementary Material
Acknowledgment
We are grateful to the staff of the analysis group of our institute for the measurement of spectroscopic data. This work was supported by the NSFC U0632010 and 2008ZX09401-004, and the State Key Laboratory of Phytochemistry and Plant Resources in West China, Chinese Academy of Sciences (P2008-ZZ08). Antifungal testing was supported by the grant NIH/NIAID AI 27094 and the USDA Agricultural Research Service specific cooperative agreement number 58-6408-2-009.
Footnotes
Supporting Information Available: The NMR spectra for angudracanoside A-F (1-6), structures of the known compounds (8-14) isolated from Dracaena angustifolia, important HMBC correlations for 1, and selected ROESY correlations of 3. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Sparg SG, Light ME, Staden JV. J. Ethnopharmacol. 2004;94:219–243. doi: 10.1016/j.jep.2004.05.016. [DOI] [PubMed] [Google Scholar]
- (2).Zheng QA, Zhang YJ, Li HZ, Yang CR. Steroids. 2006;71:160–164. doi: 10.1016/j.steroids.2005.09.007. [DOI] [PubMed] [Google Scholar]
- (3).da Silva BP, De Sousa AC, Silva GM, Mendes TP, Parente JP. Z. Naturforschung C. 2002;57:423–428. doi: 10.1515/znc-2002-5-603. [DOI] [PubMed] [Google Scholar]
- (4).Saito S, Nagamura Y. Therapeutic Agents for Hepatitis. 8059476 Japan patent. 1996 March;
- (5).Li BG, Zhou ZZ. New Drug Clin. Remed. 1994;13:75–76. [Google Scholar]
- (6).Yang CR, Zhang Y, Melissa RJ, Shabana IK, Zhang YJ, Li XC. Antimicrob Agents Chemother. 2006;50:1710–1714. doi: 10.1128/AAC.50.5.1710-1714.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tran QLT, Tezuka Y, Banskota AH, Tran QK, Saiki I, Kadota S. J. Nat Prod. 2001;64:1127–1132. doi: 10.1021/np0100385. [DOI] [PubMed] [Google Scholar]
- (8).Ali OA, Dominique G, Rene B, Bruno D, Robert A. Phytochemistry. 1996;42:895–897. [Google Scholar]
- (9).Yoshiaki W, Shuichi S, Yoshiteru I, Junzo S. Chem. Pharm. Bull. 1983;31:3486–3495. [Google Scholar]
- (10).Mimaki Y, Takaashi Y, Kuroda M, Sashida Y, Nikaido T. Phytochemistry. 1996;42:1609–1615. doi: 10.1016/0031-9422(96)00107-0. [DOI] [PubMed] [Google Scholar]
- (11).Xu YJ, Xie SX, Zhao HF, Dong H, Xu TH, Xu DM. Acta Pharmaceutica Sinica. 2001;36:750–753. [PubMed] [Google Scholar]
- (12).Ohtani K, Kasai R, Yamasaki K. Phytochemistry. 1996;42:1417–1422. doi: 10.1016/0031-9422(96)00131-8. [DOI] [PubMed] [Google Scholar]
- (13).Kravets SD, Vollerner YS, Gorovits MB, Shashkov AS, Abubakirov NK. Chem. Nat. Compd. 1986;22:174–181. [Google Scholar]
- (14).Shimonura H, Sashida Y, Oohara M, Tenma H. Phytochemistry. 1988;27:644–646. [Google Scholar]
- (15).Andrianaivoravelona JO, Terreaus C, Sahpaz S, Rasolondramanitra J, Hostettmann K. Phytochemistry. 1999;52:1145–1148. doi: 10.1016/s0031-9422(99)00158-2. [DOI] [PubMed] [Google Scholar]
- (16).Agrawal PK, Jain DC, Gupta PK, Thakur RS. Phytochemistry. 1985;24:2479–2496. [Google Scholar]
- (17).Hara S, Okabe H, Mihashi K. Chem Pharm Bull. 1987;35:501–506. [Google Scholar]
- (18).Gonzelez AG, Freire R, Salazar JA, Suarea E. Phtytochemistry. 2000;10:1339–1346. [Google Scholar]
- (19).Sashida Y. Steroidal Glycosides from Liliaceae Plants and their Biological Activities. In: Yang CR, Tanaka O, editors. Advances in Plant Glycosides, Chemistry and Biology. Elsevier; Amsterdam: 1999. pp. 201–211. [Google Scholar]
- (20).Mimaki Y, Kuroda M, Takaashi Y, Sashida Y. Phytochemistry. 1998;47:1351–1356. doi: 10.1016/s0031-9422(97)00545-1. [DOI] [PubMed] [Google Scholar]
- (21).Xu M, Zhang YJ, Yang CR. J. Nat. Prod. 2007;70:880–883. doi: 10.1021/np070012z. [DOI] [PubMed] [Google Scholar]
- (22).NCCLS . Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard M27-A2, 22(15) National Committee on Clinical Laboratory Standards; 2002. [Google Scholar]
- (23).NCCLS . Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard; M38-A, 22 (16) National Committee on Clinical Laboratory Standards; 2002. [Google Scholar]
- (24).Li XC, Hala NE, Alison CN, Alice MC. J. Nat. Prod. 1999;62:767–769. doi: 10.1021/np980469w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.