Abstract
Metazoan replication-dependent histone pre-mRNAs undergo a unique 3′-cleavage reaction which does not result in mRNA polyadenylation. Although the cleavage site is defined by histone-specific factors (hairpin binding protein, a 100-kDa zinc-finger protein and the U7 snRNP), a large complex consisting of cleavage/polyadenylation specificity factor, two subunits of cleavage stimulation factor and symplekin acts as the effector of RNA cleavage. Here, we report that yet another protein involved in cleavage/polyadenylation, mammalian cleavage factor I 68-kDa subunit (CF Im68), participates in histone RNA 3′-end processing. CF Im68 was found in a highly purified U7 snRNP preparation. Its interaction with the U7 snRNP depends on the N-terminus of the U7 snRNP protein Lsm11, known to be important for histone RNA processing. In vivo, both depletion and overexpression of CF Im68 cause significant decreases in processing efficiency. In vitro 3′-end processing is slightly stimulated by the addition of low amounts of CF Im68, but inhibited by high amounts or by anti-CF Im68 antibody. Finally, immunoprecipitation of CF Im68 results in a strong enrichment of histone pre-mRNAs. In contrast, the small CF Im subunit, CF Im25, does not appear to be involved in histone RNA processing.
INTRODUCTION
Pre-messenger RNAs undergo a series of processing reactions, such as capping, splicing and 3′-cleavage before they are exported from the nucleus and serve as templates for translation. During 3′-end processing, the majority of pre-mRNAs acquire a poly(A) tail in a concerted cleavage/polyadenylation reaction (1). Several cis-acting elements direct the required factors to the cleavage site. In vertebrates, cleavage and polyadenylation specificity factor (CPSF) recognizes the highly conserved AAUAAA hexamer located 10–30-nt upstream of the cleavage site (2), and cleavage stimulation factor (CStF) binds to a G/U rich downstream sequence element (3). In addition, symplekin, cleavage factors I and II, poly(A) polymerase and nuclear poly(A) binding protein (PABN1) are required during the reaction (1).
In contrast, the transcripts of replication-dependent histone genes in metazoans lack introns and poly(A) tails (4). Instead of the poly(A) tail, their mRNAs end in a conserved 26-nt long sequence, 16 nt of which form a hairpin structure. The processing at the 3′-end requires only one endonucleolytic cut that usually occurs 5 nt after the hairpin. This cleavage reaction is mediated by several trans-acting factors, among them the U7 small nuclear ribonucleoprotein (snRNP) which binds by RNA:RNA base pairing to a conserved histone downstream element (HDE) located a few nucleotides after the processing site. The U7 snRNP exhibits a unique ring-shaped protein complex composed of five Sm proteins and two U7-specific Sm-like proteins, termed Lsm10 and Lsm11 (5,6). Other important processing factors are the hairpin binding protein (HBP; also termed stem-loop binding protein, SLBP) (7,8), a 100-kDa zinc-finger protein (ZFP100) (9) that connects HBP bound to the upstream hairpin with the U7 snRNP bound to the HDE, as well as an essential heat-labile protein complex (10,11).
Thus, in the past, the two types of processing reactions seemed to be very different, except, perhaps, for the chemistry of the ends produced and the preference for certain nucleotides at the cleavage sites (12–14). However, in 2005, the endonuclease executing the cleavage after the hairpin was identified as the 73 kDa subunit of CPSF (CPSF-73) (15) and the heat-labile factor (HLF) was characterized as a complex composed of all five subunits of CPSF, two subunits of CstF as well as symplekin (16). In addition, the U2 snRNP was found to bind to an RNA element in the coding region of replication-dependent histone mRNAs and thereby to stimulate the U7-snRNP-dependent 3′-end formation (17), similarly to interactions that occur between the last 3′-splice site and CPSF bound to the polyadenylation site during cleavage and polyadenylation (18).
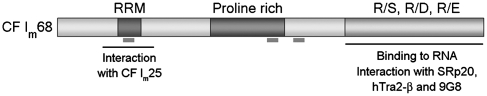
However, certain components of the cleavage/polyadenylation machinery, including mammalian cleavage factor I (CF Im), were not detected in the heat-labile complex. CF Im is a heterodimer consisting of a small 25-kDa subunit and one of three different larger subunits of 59, 68 and 72 kDa, respectively (19). All three large polypeptides can be UV crosslinked to a cleavage and polyadenylation substrate. In addition, analysis of the kinetics of the cleavage reaction indicated that interaction of CF Im with the RNA is one of the earliest steps in the assembly of the cleavage/polyadenylation complex. It facilitates the recruitment of other processing factors and stabilizes the binding of CPSF (20). The 68 kDa subunit has a domain organization that is reminiscent of spliceosomal SR proteins with an N-terminal RNA recognition motif (RRM) and a C-terminal RS-like domain (Figure 1). The RRM does not bind strongly to RNA, but seems to function in the interaction with the small subunit CF Im25 (21). The RS-like domain of CF Im68 is sufficient for the localization in nuclear speckles (and in the nucleoplasm) and mediates the interaction in vitro with a subset of shuttling SR proteins (21,22).
Figure 1.
Domain organization of CF Im68. CF Im68 has a structure reminiscent of spliceosomal SR proteins. It contains an N-terminal RNA recognition motif (RRM) and a C-terminal charged domain. Over its RRM, it interacts with the 25 kDa subunit of CF Im. Its C-terminal charged domain can bind to RNA and interacts with a subset of spliceosomal SR proteins. The positions of the peptides identified by microsequencing are indicated by thick bars.
Surprisingly, we have identified CF Im68 in our most highly purified U7 snRNP preparations (5,6). We could validate this association which appears to depend on the N-terminus of Lsm11 in vitro and in vivo. In addition CF Im68 co-precipitates histone mRNA precursors (as well as some mature histone mRNA), and the efficiency of histone RNA 3′-processing seems to depend on the presence of CF Im68 both in vivo and in vitro.
MATERIALS AND METHODS
Plasmids
Complementary DNA clones of mouse Lsm11 or its MPL>AAA mutant in the pcDNA3-HA vector as well as pGex-4 T-derived plasmids containing the first 156 amino-acid codons of Lsm11 or the MPL mutant or the first 202 amino acids of Nxf1 have been described (6,23,24). pcDNA3-Flag-Lsm11 was constructed by replacing the HA-tag of the pcDNA3-HA-Lsm11 vector (6) with a linker encoding the Flag epitope. The cDNA for CF Im68, kindly provided by Walter Keller (Biozentrum, University of Basel), was subcloned into pcDNA3-HA and pGex-4T1 (Amersham Biosciences). pMal-Tev-HA-CF Im25 was constructed by inserting the CF Im25 open reading frame together with a linker encoding the TEV protease cleavage site and the HA epitope into pMal-c (New England Bioloabs). pCMV-Flag-CF Im25 has been described elsewhere (24).
To produce plasmids encoding HA-CF Im68-VC155, HA-CF Im25-VC155, Flag-Lsm11-VN173 and Flag-Lsm11/MPL-VN173, the open reading frames of CF Im68, CF Im25, Lsm11 and Lsm11/MPL were amplified by PCR from plasmids described above and subcloned in the Venus vectors pHA-VC155 and pFlag-VN173 (25). In this way, all the open reading frames are inserted between an N-terminal tag (HA or Flag) and a C-terminal Venus (VC or VN) fragment. The empty pHA-VC155 and pFlag-VN173 were used as mock controls. As colocalization controls in the absence of bimolecular fluorescence complementation (BiFC) signals, we used the following plasmids lacking Venus fragments: pcDNA3-HA-Lsm11, pcDNA3-HA-MPL, pCMV-Flag-CF Im25. To test for an interaction with HA-CF Im68-VC155, the CF Im25 open reading frame of pGEM-CF Im25 was cleaved by EcoRI/XhoI and inserted into pHA-VC155.
Sequences coding for short hairpin RNAs (shRNAs) were inserted as double-stranded oligonucleotides into pSUPERpuro between the BglII and HindIII sites as described (26,27). Two different target sequences were used for Lsm11 (5′-GGTATTCACTCGACACATA-3′ and 5′-GTATTCACTCGACACATAA-3′) and for Tcrβ (5′-TAACCATGACTATATGTAC-3′ and 5′-GGCTGATCCATTACTCATA-3′). In each construct, the sense and antisense sequences are separated by a 9-nt spacer (TTCAAGAGA) that allows the formation of a hairpin loop. These vectors are referred to as pSUPuro-Lsm11-1 and -2 as well as pSUPuro-Tcrβ-1 and -2. To obtain a puromycin-selectable HA-CF Im68 expression plasmid (HA-CFIm68-SUPuro), a fragment of pcDNA3-HA-CFIm68 containing CMV promoter, HA-CF Im68 cDNA and bovine growth hormone polyadenylation signal was inserted into pSUPERpuro (thereby replacing the shRNA expression cassette). All clones were verified by DNA sequencing. Details of the constructs are available on request.
Antibodies
An antibody to GST-CF Im68 was raised in rabbits and affinity purified by binding to the recombinant protein immobilized on nitrocellulose filters. On western blots, this antibody was detected with anti-rabbit antibody coupled to horse radish peroxydase (HRP; Promega). It revealed a single ∼68-kDa protein in whole-cell extract from human 293-T cells (data not shown). Other previously described antibodies were directed against Lsm10 and Lsm11 (5,6), CF Im25 (19) and SmB/B′ (46). Flag-tagged proteins were detected by anti-Flag M2 monoclonal antibody (Sigma) coupled to HRP. HA-tagged proteins were detected either directly with anti-HA antibody (12CA5, Roche) coupled to HRP or indirectly with a species-specific antibody coupled to HRP (Promega). Hexahistidine-tagged proteins were detected by using an anti-His antibody (HIS1, Sigma). Endogeneous β-actin was detected with polyclonal anti-actin antibody (20–33, Sigma).
For immunofluorescence, we used anti HA-tag (6E2, mouse monoclonal antibody, Cell Signaling) or anti Flag M2 (mouse monoclonal antibody, SIGMA) as primary antibodies and goat-anti-mouse Alexa fluor 488 and 633 (Molecular Probes) as secondary antibodies.
U7 snRNP purification and peptide sequencing
The purification of U7 snRNPs and peptide microsequencing (performed at the Harvard Microchemistry Facility) have been described (5).
Cell culture and transfection
Human HeLa and 293-T cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, AG, Basel, Switzerland) supplemented with 10% fetal calf serum (BioConcept, Allschwil, Switzerland), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, AG, Basel, Switzerland) at 37°C in a wet atmosphere containing 5% CO2. For plasmid transfection, they were grown to 60–70% confluency in 10 cm dishes and transfected with 10 µg of the appropriate plasmids complexed with Dreamfect (OZ Biosciences). Cells were usually harvested 48 h post-transfection.
For the fluorescence experiments of Figure 4 and Supplementary Figure S3, HeLa cells were grown on glass coverslips in Dulbecco’s modified Eagle’s Medium High Glucose supplemented with 10% foetal bovine serum, 2 mM l-glutamine and 100 U/ml penicillin/streptomycin (all components from Euroclone) at 37°C, 5% CO2. Plasmid transfections were performed after cells had reached 80% confluency by using Escort V transfection reagent (Sigma) as recommended by the manufacturer.
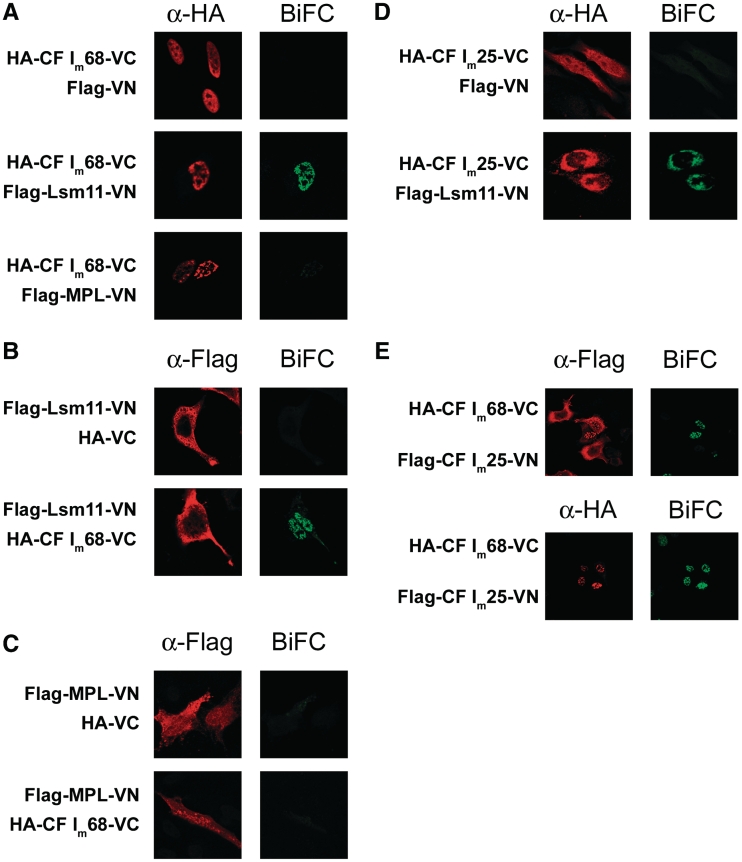
Figure 4.
Interaction of CF Im68 with Lsm11 in live, unperturbed cells. (A), (B) and (C) Analysis of Lsm11/CF Im68 heterodimerization. The left column shows immunostaining images (red) of HA-CF Im68-VC with anti-HA antibody (A) and Flag-Lsm11-VN (B) or Flag-MPL-VN (C) with anti-Flag antibody. The right column shows BiFC (green) that occurs when the two proteins fused to the N- and C-terminal halves of Venus interact with each other. (D) Analysis of Lsm11/CF Im25 interaction. The left column shows HA-CF Im25-VC detected with anti-HA antibody, whereas the right column shows BiFC. (E) Analysis of CF Im68/CF Im25 heterodimerization. The left column shows Flag-CF Im25-VC detected with anti-HA antibody (upper row) or HA-CF Im68-VC detected with anti-HA antibody (lower row), whereas the right column shows BiFC.
Depletion and overexpression experiments
Hela cells were transfected with either two siRNAs against CF Im68 (Ambion, IDs: s223186 and s21775) at 25 nM concentration each, with a single siRNA against CF Im25 (Ambion, ID: s21770) or with a non-targeting siRNA control (Ambion), each at 50 nM concentration, by using Lullaby reagent (OZ Biosciences) according to the manufacturer’s instructions. The cells were split 1:1 at 24 h post-transfection. After another 24 h, a second round of transfection with half the initial amount of siRNAs was performed. Cells were harvested 4 days after the primary transfection for RNA isolation and western blot analysis of CF Im68.
For depletion of Lsm11, the cells were transfected in six-well plates with a mixture of 0.5 µg each of pSUPuro-Lsm11-1 and -2 or, as control, a similar mixture of pSUPuro-Tcrβ-1 and -2. For the overexpression of CF Im68, the cells were transfected with HA-CFIm68-SUPuro in 10 cm dishes by using the standard Dreamfect protocol described above. Control cells were either transfected with an empty pSUPERpuro or with a pSUPERpuro derivate expressing an HA-tagged EGFP. Both controls yielded similar results, indicating that the strong expression from the CMV promoter has no effect on its own. Culturing the cells in the presence of 1.5 µg/ml puromycin eliminated untransfected cells. Three to 5 days after transfection, cells were harvested for RNA isolation and western blot analysis of CF Im68.
To assess the depletion of Lsm11, cDNA corresponding to 100 ng reverse transcribed RNA was subjected to a Lsm11-specific PCR. The PCR was performed in a volume of 25 μl containing 12.5 μl 2× FastStart PCR Mastermix (Roche) supplemented with a primer pair specific for Lsm11 at a final concentration of 200 nM each, and 35 cycles were performed (for primer sequences see Supplementary Table S1).
Interaction studies by co-immunoprecipitation
The preparation of whole cell or small-scale nuclear extracts and immunoprecipitations were performed as described (5). The extract from one 10 cm dish (100 µl) was incubated with 100 µg of RNase A (Sigma) for 20 min at 30°C prior to immunoprecipitation. Proteins were resolved on 12% high-TEMED SDS–polyacrylamide gels (28), analysed by western blots with appropriate antibodies (see below) and developed by the enhanced chemiluminescence method (Amersham).
Interaction studies by GST pull-down assays
To study protein–protein interactions in vitro, recombinant GST (negative control) or GST fusion proteins were expressed in Escherichia coli BL21 Gold or BL21(DE3)LysS transformed with pGex-4 T-derived plasmids encoding GST-fusions with the N-terminus of Lsm11, the N-terminus of the Lsm11-MPL/AAA mutant or the first 202 amino acids of Nxf1. MBP-CF Im25 and MBP-TEV-HA-CF Im25 were expressed in BL21(DE3)LysS and purified over an amylose resin (New England Biolabs), followed by the removal of the MBP-tag from MBP-TEV-HA-CF Im25 by digestion of the fusion protein with ProTev protease (Promega). The purified GST-fusions were coupled to glutathione sepharose 4B beads (Amersham Biosciences) and incubated with baculovirus produced hexahistidine-tagged CF Im68 or bacterially produced HA-tagged CF Im25. The beads and proteins were incubated in IIP150-NP40 (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40) at 4°C for 2 h with gentle agitation on a wheel. Subsequently, the beads were washed with IIP150-NP40 and the bound and input materials were resolved on 12% high-TEMED SDS–polyacrylamide gels (28), analysed by western blots with appropriate antibodies (see above) and developed by the enhanced chemiluminescence method (Amersham).
Li-Cor western blots
Western blots were performed essentially as described above. Bands were visualized with secondary antibody donkey anti rabbit IRDye800CW (Figure 5 F). Bands were quantified with an Odyssey Infrared Imaging system (LI-COR Biosciences).
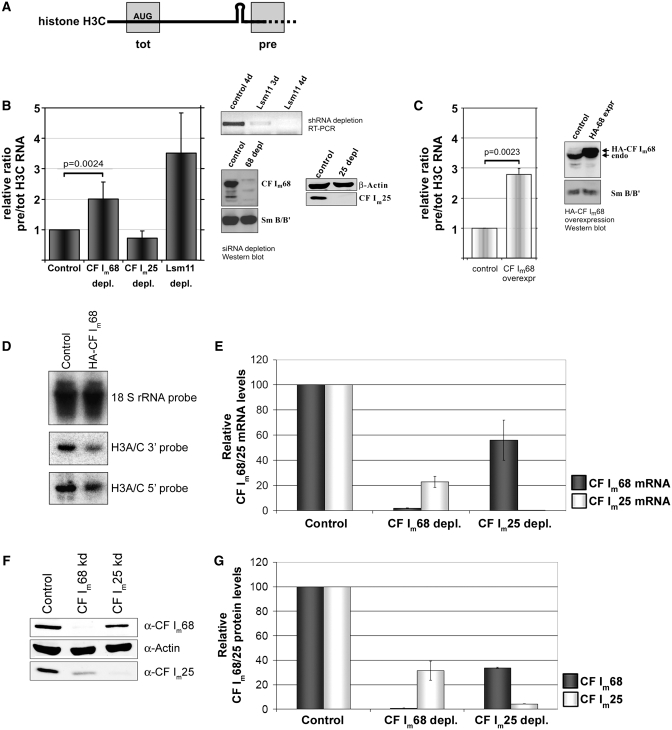
Figure 5.
Effects of CF Im68 depletion/overexpression or CF Im25 depletion on apparent histone RNA 3′-end processing in vivo. (A) Schematic representation of quantitative (real-time) reverse transcription–PCR (qRT–PCR) assay. Total mRNA (tot) of histone H3 C is measured with a primer-probe set spanning the translation start codon. To measure the unprocessed precursors (pre), the primer-probe set spans the 3′-cleavage site. See ‘Materials and Methods’ section for further details. (B) H3 C RNA 3′-end processing efficiency in Hela cells depleted of CF Im68, CF Im25 or Lsm11. The western blots show endogenous CF Im68 or CF Im25 detected with corresponding antibodies. SmB/B′ and β-actin served as loading controls. The RT–PCR shows Lsm11 mRNA detected in total RNA. (C) H3 C RNA 3′-end processing efficiency in Hela cells overexpressing CF Im68. The western blots show endogenous and HA-tagged CF Im68 detected with anti-CF Im68 antibody. SmB/B′ served as loading control. The relative in vivo processing efficiency was calculated as ratio of pre-mRNA to total mRNA and then normalized with respect to the ratio obtained in cells treated with either control siRNA, Tcrβ-specific shRNA (B) or pcDNA-puro (C). The data shown represent means ± standard deviations. Number of independent measurements (CF Im68 depletion, 5; CF Im25 depletion, 3; Lsm11 depletion, 3; CF Im68 overexpression, 4) p, significance values determined by Student’s t-test. (D) Northern blot detecting H3 C mRNA. Nuclear RNA from control cells or cells overexpressing HA-tagged CF Im68 was separated by polyacrylamide electrophoresis and transferred to positively charged nylon membranes. Histone H3 C mRNA was detected with probes either hybridizing to the 5′ or the 3′-end of the mRNA. 18 S rRNA served as loading control. (E) Quantitation of mRNA levels (relative to U6 snRNA) for CF Im68 (dark columns) or CF Im25 (light columns) from cells depleted of CF Im68 or CF Im25 (number of depletions: 3). (F) Western blot detecting endogenous CF Im68 or CF Im25 from cells transfected with a control siRNA or depleted of CF Im68 or CF Im25. β-actin served as loading control. (G) Quantitation of CF Im68 (dark columns) and CF Im25 (light columns) protein levels, relative to β-actin, in cells depleted of CF Im68 or CF Im25 (number of evaluated blots: 2).
BiFc detection
HeLa cells that had been incubated for 24 h after transfection were washed with PBS (130 mM NaCl, 20 mM potassium phosphate, pH 7.4), fixed for 10 min with PBS containing 4% (w/v) paraformaldehyde and mounted in Fluorsave Reagent (Calbiochem).
For measurement of Venus Fluorescent Protein fluorescence after complementation, the YFP beam-path was used as described (29). Complementation of the C-terminal YFP fragment VC155 with the N-terminal YFP fragment VN173 generates a fluorophore with an excitation maximum at 515 nm and an emission maximum at 528 nm. Therefore, the fluorescence resulting from complementation was recorded through the Leica laser scanning acquisition system by using the argon laser line of excitation.
For immunofluorescence, cells fixed as described above were washed with PBS, permeabilized in CKS solution (HEPES 20 mM, sucrose 300 mM, NaCl 50 mM, MgCl2 3 mM, Triton-X-100 0.2%) for 5 min at 4°C, blocked in PBS containing 10% foetal bovine serum for 1 h at room temperature, and incubated with the primary antibodies diluted in PBS containing 0.2% bovine serum albumin (BSA) for 1 h at 37°C. Subsequently, cells were washed with PBS (three times for 5 min) and incubated with the secondary antibody diluted in PBS/0.2% BSA for 45 min at room temperature. The cells were again washed with PBS (three times for 5 min) and mounted in Fluorsave Reagent (Calbiochem).
Images were collected with a confocal microscope Leica TCS SP2 AOBS, by using LSC software. Fluorophores were excited by using standard laser lines.
Quantitative reverse transcription PCR
Usually 400–1000 ng RNA were reverse transcribed in 50 μl Stratascript 6.0 RT buffer in the presence of 4 mM of all four desoxy nucleoside triphosphates, 300 ng random hexamers, 40 U RNasIn (Promega) and 1 μl Stratascript 6.0 reverse transcriptase (Stratagene). For real-time PCR, reverse-transcribed material corresponding to 40 ng RNA was amplified in 25 μl Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems) with specific primers, and TaqMan probes by using the ABI SDS7000 Sequence Detection System. The TaqMan assays for endogenous histone H3 RNA are specific for the histone H3 A/C genes (Homo sapiens histone cluster 2, H3 A and H3 C; the sequences of the two transcripts are identical and will henceforth be referred to as H3 C). The assays use probes spanning the start codon and the 3′-cleavage site to measure total RNA and pre-mRNA, respectively (Supplementary Table S1).
Northern blot
Samples of 10 μg nuclear RNA, isolated as described (24), were separated on a 3.6% polyacrylamide gel containing 7 M urea in 1× TBE and transferred onto positively charged Nylon membranes (Roche Applied Sciences) in 0.5× TBE at 400 mA by using a TE 77 ECL Semi-dry transfer unit (GE Healthcare). RNA was cross-linked to the membrane in a UV Stratalinker® 2400 (Stratagene) by application of 120 mJ ultraviolet light (254 nm). Hybridization was carried out in ULTRAhyb Ultrasensitive Hybridization Buffer (Applied Biosystems) at 68°C. The histone H3 C 5′ and 3′-probes were generated by run-off transcription incorporating α-[32 P]-UTP (Hartmann Analytic GmbH). They hybridize to nt 1–130 and 448–507, respectively, of Histone H3 C (NM_021059). After detection of the histone mRNA signals, the blot was rehybdrized with a [32P]-labelled oligonucleotide complementary to 18 S rRNA (5′-CTTTCGCTCTGGTCCGTCTTGCGCCGGTCC-3′) in ULTRAhyb-Oligo Hybridization buffer (Applied Biosystems) at 42°C. Excess of probe or oligo were removed by following the instructions of the ULTRAhyb protocol. Signals were detected with a Phosphoimager Storm 820 (810-UNV) scanner.
In vitro histone RNA 3′-end processing
Processing reactions were carried out with 24 μl of HeLa nuclear extract and 10 000 cpm (1–3 fmol) 12/12 RNA template (30) in a total volume of 40 μl and incubated for 90 min at 30°C, unless stated otherwise. To the processing reactions different amounts of either BSA or hexahistidine-tagged CF Im68 in Buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 100 mM KCl, 0.2 mM EDTA and 0.5 mM DTT) were added. After incubation at 30°C, 1 ml TRI reagent was added followed by subsequent purification of the RNA. In some experiments, ∼1.2 µg rabbit polyclonal anti-HA (Y-11 probe, Santa Cruz), rabbit polyclonal anti-β-actin (20–33, Sigma Aldrich), rabbit polyclonal anti-CDC73 (A300-170 A, Bethyl) or rabbit polyclonal anti-CFIm68 were added to the processing reaction mix without substrate and incubated for 10 min on ice. After addition of the processing substrate, the reaction was incubated for 1 h 45 min at 30°C. The samples were resolved by 8% denaturing polyacrylamide gel electrophoresis (7 M urea). The gel was dried and revealed on a Storm820 Phosphoimager (Amersham). The band intensities were quantitated by using Aida software V3.11 (Raytest Isotopenmessgeräte GmbH, Straubenhardt).
RNA Immunoprecipitation
RNA immunoprecipitation was performed essentially as described (31). Briefly, 2.5 × 107 cells were fixed in 1% formaldehyde for 30 min at room temperature, lysed in FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors) containing RNase inhibitor [RNAsin (Promega); 80 U/500 μl buffer], and then nucleic acids were fragmented by sonication. Before immunoprecipitation, the extract was treated with DNase [400–500 units RNase-free DNase I (Sigma) per 500 μl extract] in 25 mM MgCl2, 5 mM CaCl2, 3 μl RNAsin for 10 min at 37°C. DNase I digestion was stopped by the addition of EDTA to 20 mM. Reactions were cleared by centrifugation at 16 100g in a microcentrifuge for 15 min. For input RNA, all following steps up to proteinase K digestion were omitted. Immunoprecipitation was performed by adding 30 μg anti-CF Im68, 30 μg BSA (negative control), 50 μg anti-CF Im25 or 50 μg anti-β-actin (negative control) to the DNase-treated extract and incubating head over tail at 4°C overnight. Then 67 μl of protein G slurry (equilibrated in FA lysis buffer containing RNasin) were added to each tube and incubated for 2 h at 4°C. All subsequent precipitate washes and the final elution were performed as described in (31). To the eluate fractions, NaCl was added to a final concentration of 200 mM before addition of 80 μg proteinase K and incubation at 42°C for 1 h. Crosslinks were reversed by heating at 65°C for 5 h. The reactions were extracted with an equal volume of acid-equilibrated (pH 4.8) phenol:chloroform 5:1. Then, 1/10 volume of 3 M sodium acetate (pH 5.5), 20 μg glycogen, and 2.5 volumes of ice-cold absolute ethanol were added to the aqueous layer before precipitation at −80°C. The reaction was centrifuged at maximum speed in a microcentrifuge for 15 min and the precipitate washed twice with ice-cold 70% ethanol. To remove contaminating DNA, RNA was treated with Turbo DNase (Ambion) according to the manufacturer’s protocol.
RESULTS
Identification of CF Im68 as an interaction partner of the U7 snRNP
We previously reported the purification of U7 snRNPs and the characterization of some of the associated proteins including the U7-specific protein Lsm10 (5). Apart from the subunits already described, two additional bands of ∼70 and ∼50 kDa, respectively, were present in our most highly purified U7 snRNP preparation [marked with asterisks in Figure 1 C of (5)]. Microsequencing of the 50 kDa band later led to the identification of Lsm11 (6). The band of ∼70 kDa was also subjected to microsequencing by nano-electrospray tandem mass spectrometry coupled to the Sequest database search algorithm (32). This analysis revealed several peptides from bovine serum albumin (theoretical, unmodified molecular mass: 69 293 Da) and human cytokeratin 10 (59 511 Da) that we considered to be contaminants. Additionally, the peptides GFALVGVGSEASSK, AISSSAISR and GPPPTDPYGRPPPYDR were indicative of the 68 kDa subunit of cleavage factor I (CF Im68; 59 210 Da). The location of the peptides is indicated in Figure 1 as bars below the schematic domain organization of CF Im68.
Because it was not clear whether this association with highly purified U7 snRNPs might be due to a contamination or reflect a real interaction, we immunopreciptated U7 snRNPs from total cell extract of 293-T cells expressing HA-tagged CF Im68. Separate precipitations were carried out with antibodies specific for Lsm10 (5) or Lsm11 (6), and co-precipitation of CF Im68 was analysed by western blotting with anti-HA antibody. Indeed, HA-CF Im68 was efficiently co-precipitated by both antibodies, but not by beads alone (Figure 2 A). Since the extracts had been treated with RNAse A, the interaction of CF Im68 with the U7 snRNP-specific proteins Lsm10 and Lsm11 was apparently not mediated by RNA but rather due to a protein–protein interaction.
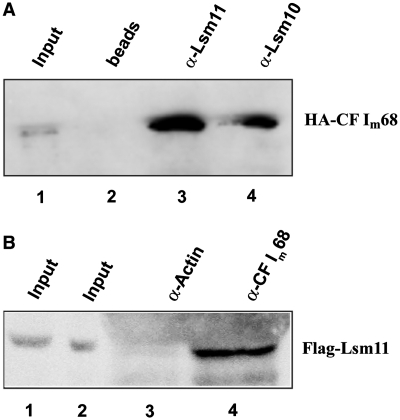
Figure 2.
CF Im68 interacts with Lsm10 and Lsm11. (A) HA-tagged CF Im68 was expressed in human 293-T cells and its ability to interact with Lsm11 and Lsm10 was assessed by immunoprecipitation of whole cell extracts with antibodies specific for Lsm11 (lane 3) and Lsm10 (lane 4). Negative control (lane 2), beads incubated with bovine serum albumin. Input, 1/30 of the amount used in the co-immunoprecipitation. (B) Flag-tagged Lsm11 was expressed in HeLa cells and its ability to interact with CF Im68 was assessed by immunoprecipitation of small scale nuclear extracts with an antibody specific for CF Im68. An antibody specific for β-actin served as negative control (lane 3). In both cases, the extracts had been treated with RNAse A. Lanes 1 and 2 are 1/10 and 1/15 of the amount used in the co-immunoprecipitation, respectively.
In an inverse experiment, protein G sepharose beads coupled with anti-β-actin (negative control) or anti-CF Im68 were used. However, in this case, nuclear extract from HeLa cells expressing Flag-tagged Lsm11 was used. The western blot was probed with anti-Flag antibody, and the results indeed showed that the anti-CF Im68 antibody can co-precipitate Flag-Lsm11 (Figure 2B). Note that we could not probe for a co-precipitation of endogenous Lsm11, because its abundance is below the detection limit of western blots (data not shown).
As CF Im68 is usually found in a complex with CF Im25 (19), we tested whether the small 25 kDa subunit of CF Im is also associated with Lsm11. However, we could not detect any interaction, by trying to precipitate either Flag-tagged Lsm11 with anti-CF Im25 antibody or CF Im25 with anti-Flag antibody from cells expressing Flag-Lsm11 (Supplementary Figure S1).
CF Im68 interacts directly with the N-terminus of Lsm11
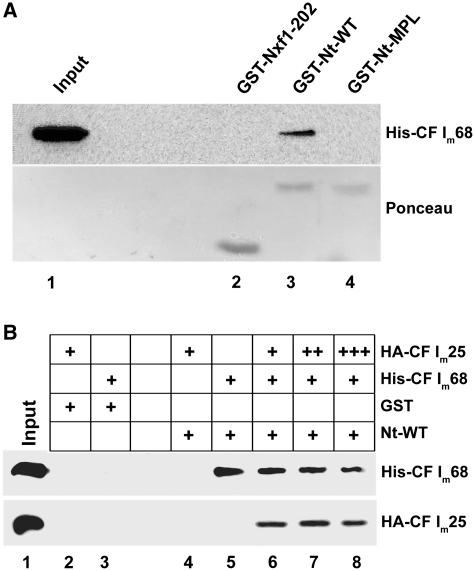
The N-terminus of the U7-specific Lsm11 protein and, more specifically, an evolutionarily conserved amino acid motif, NVLTRMPLH, are essential for histone RNA 3′-end processing but not required for incorporation of Lsm11 into U7 snRNPs (6,23,33). To investigate if CF Im68 directly interacts with this part of Lsm11, we coupled purified, recombinant, GST-tagged N-terminal fragment of Lsm11 to glutathione beads and used these to precipitate hexahistidine-tagged CF Im68 purified from a baculovirus expression system. Indeed, CF Im68 interacted with the wild-type N-terminus of Lsm11, but not with a mutant in which the three amino acids MPL of the mentioned motif had been replaced by alanines (23) (Figure 3 A, lanes 3 and 4). Additionally, no binding was observed to beads loaded with a GST fusion of the first 202 amino acids of the nuclear export factor Nxf1 (Figure 3 A, lane 2) (24). These interaction data were confirmed by GST-pulldowns using in vitro translated HA-tagged CF Im68, and we also found that the GST-fusion of the N-terminus of Lsm11 could bind CF Im68 from an extract of mammalian cells (data not shown).
Figure 3.
The interaction with CF Im68 is specific for the wild-type N-terminus of Lsm11. (A) Analysis of the binding of hexahistidine-tagged CF Im68 to glutathione beads containing GST-NXF1-202 (lane 2) or GST fused to the first 157 amino acids of wild-type mouse Lsm11 (lane 3) or of a processing-deficient mutant thereof in which the amino acids 149–151 (MPL) are replaced by three alanines (23) (lane 4). The beads were washed, and the bound material was analysed by western blotting (upper panel). To confirm that the absence of CF Im68 in lane 4 is not due to an absence of GST-Nt-MPL, the blot was stained with Ponceau S (lower panel). Input (lane 1), one-third the amount used in the binding assays was analysed directly. (B) Analysis of the binding of hexahistidine-tagged CF Im68 and HA-tagged CF Im25 to glutathione beads containing GST fused to the first 157 amino acids of wild-type Lsm11. (Lane 1) input representing one-third the amount used in the binding assays, (lanes 2 and 3) negative controls in which equimolar amounts of His-CF Im68 (600 ng) or HA-CF Im25 (200 ng), repectively, were incubated with GST. (Lanes 4–8) glutathione beads containing GST fused to the first 157 amino acids of wild-type mouse Lsm11 that was incubated with HA-CF Im25 (lane 4; 200 ng), His-CF Im68 (lane 5; 600 ng), or both subunits with increasing amounts of HA-CF Im25 (lanes 6–8; 200, 600 1200 ng). The bound material was analysed by western blotting by using anti-His and anti-HA antibodies, respectively.
CF Im68 is normally in a complex with CF Im25, but the small subunit was neither found in the U7 snRNP purification, nor in Lsm11 co-precipitations. However, we wondered whether at least in vitro assembled CF Im and the N-terminus of Lsm11 can form a trimeric complex or whether CF Im25 competes with Lsm11 for the binding to CF Im68. To adress this question we repeated the GST pulldown experiments, but this time adding recombinant HA-tagged CF Im25 to the binding reaction between the GST-tagged Lsm11 N-terminal fragment and CF Im68. As can be seen in Figure 3B, CF Im25 does not interact directly with Lsm11 (lane 4) but can form a trimeric complex in vitro by interaction with CF Im68 (lanes 6–8). When higher molar ratios of CF Im25 in respect to CF Im68 are added to the binding reaction, the amount of precipitated CF Im25 does not increase, showing that the co-precipitated amount of CF Im25 depends on the precipitated amount of CF Im68. This experiment was also performed with MBP-CF Im25 yielding the same results (data not shown).
In vivo interaction of CF Im68 with Lsm11 but not the MPL mutant
To analyse whether the interaction between CF Im68 and the U7 snRNP or Lsm11 can be detected in unperturbed cells in vivo, we used the recently developed method of BiFC (25,34). In this method, fluorescent proteins are split into two non-fluorescent halves, which then tag the proteins under study. The affinity of the fragments for each other is very low and therefore fluorescence complementation increases considerably when an interaction between the respective fusion partners brings them into close proximity. BiFC detects stable interactions: for complementation to occur, the halves have to interact for several seconds (34).
Full length wild-type Lsm11, its MPL mutant mentioned above, as well as CF Im25 were fused between a Flag tag and the N-terminal 173 amino acids of the YFP mutant Venus (Flag-Lsm11-VN, Flag-MPL-VN and Flag-CF Im25-VN, respectively), while CF Im68 and CF Im25 open reading frames were cloned between a HA tag and the C-terminal 155 Venus residues (HA-CF Im68-VC and HA-CF Im25-VC, respectively). The use of Venus allows for BiFC analysis under physiological conditions and increases the signal output and specificity (25). First, we characterized the intracellular distribution of the fusion proteins in transiently transfected HeLa cells. While HA-CF Im68-VC, like endogenous CF Im68, was concentrated in the nucleus, with exclusion of the nucleoli (21) (Figure 4 A), Flag-Lsm11-VN, Flag-MPL-VN, HA-CF Im25-VC and Flag-CF Im25-VN, were distributed both in the cytoplasm and in the nucleus (Figure 4B–E). In contrast, endogenous Lsm11 was found to be similarly distributed as U7 snRNA detected by fluorescent in situ hybridization (35) and Lsm10 (5), i.e. predominantly in Cajal bodies with some background nucleoplasmic staining (Supplementary Figure S2). HA-CF Im25-VC was also mostly cytoplasmic when co-expressed with Flag-Lsm11-VN, but the presence of the green BiFC signal (see below) prevented a double labelling of the same cells with anti-HA and anti-Flag antibodies (data not shown). Therefore, to analyse the distribution of both the HA- and the Flag-tagged proteins in the same cell, we coexpressed Flag-CF Im25-VN with HA-CF Im68, HA-Lsm11 or HA-MPL, all of which lack the Venus C-terminal fragment, and analysed these cells by double immunostaining. As can be seen in Supplementary Figure S3, HA-CF Im68 was predominatly nuclear, whereas all three other proteins showed a mostly cytoplasmic localization.
The observation that Flag-Lsm11-VN or the shorter HA-Lsm11 showed a stronger cytoplasmic staining than the endogenous protein could be explained by the fact that, upon overexpression, some of the recombinant protein does not get assembled into U7 snRNPs and therefore remains in the cytoplasm. The same could be true for HA-CF Im25-VC or Flag-CF Im25-VN. In fact, we previously showed that nuclear localization of exogenous CF Im25 is strongly increased by the concomitant expression of the 68 kDa subunit, indicating that the nuclear import of the small subunit of CF Im requires association with one of the larger subunits (24).
Importantly, as shown in Figure 4 (A row 2 and B row 2), a strong complementation signal was detected between HA-CF Im68-VC and Flag-Lsm11-VN. This signal which we observed in >90% of the cotransfected cells was predominantly nuclear with exclusion of the nucleoli and was characterized by a patchy appearance. However, no BiFC colocalization was observed between the MPL mutant of Lsm11 and CF Im68 (Figure 4 A row 3 and 4 C row 2).
While CF Im25, CF Im68, and Lsm11 can form a trimeric complex in vitro, in vivo there seems to be no interaction between the small subunit of CF Im and Lsm11 in the nucleus, where these proteins normally reside (Figure 4D, row 2). The cytoplasmic fluorescence complementation observed between Flag-Lsm11-VN and HA-CF Im25-VC could be an artefact of the high cytoplasmic level of both proteins that are not efficiently imported into the nucleus. The failure of the tagged CF Im25 to yield a nuclear BiFC signal with Lsm11 was not due to a general deficiency of the CF Im25 open reading frame, as was shown by the nuclear BiFC signal obtained between Flag-CF Im25-VN and HA-CF Im68-VC (Figure 4E). Control western blot analyses indicated that the different Venus fusion proteins were expressed at similar levels in the cells (Supplementary Figure S4).
Taken together, these experiments clearly demonstrate that the tagged Lsm11 and CF Im68 colocalize in the cell nucleus in a speckled pattern. In contrast, the small subunit CF Im25 does not efficiently associate with Lsm11 in the nucleus.
Altering CF Im68 levels affects apparent histone RNA 3′-end processing in vivo
Since we found CF Im68 to interact with Lsm11, but not with the processing-defective MPL mutant, we wanted to investigate the importance of this association for histone pre-mRNA 3′-end processing. For this purpose, we established an assay to study a possible dependence of the histone RNA processing reaction on CF Im68 in vivo. The measurements are based on quantitative real-time reverse transcription PCR (qRT–PCR) for histone H3 C total RNA and pre-RNA (‘Materials and Methods’ section). To ensure that both primer-probe sets are specific for H3 C, the one for the total mRNA spans the AUG initiation codon (Figure 5 A), thereby excluding several non-allelic H3 genes which are very similar in the coding sequences but differ in the 5′-untranslated region. The primer-probe set for the pre-mRNA spans the 3′-processing site.
HeLa cells were depleted of CF Im68 or CF Im25 by transfection of siRNAs. Depletion of Lsm11 by plasmids encoding shRNAs (‘Materials and Methods’ section), which was expected to reduce 3′-end processing of histone mRNA, was used as positive control. For all three proteins, the depletion was almost complete within 4 days, as detected by western blot for CF Im68 and CF Im25 or by RT–PCR for Lsm11, since in this case the endogenous protein can not be detected by western blot due to its very low abundance (Figure 5B). As reference for the normalization of qRT–PCR, we used HeLa cells similarly transfected with a control siRNA or with two control plasmids encoding shRNAs specific for human T-cell receptor β subunit (TcRβ) mRNA that is not expressed in this cell line. Please also note that, because of the toxicity of the Lsm11 depletion, the cells had sometimes to be harvested at different times, in order to have enough live cells for the analysis. Moreover, in agreement with data published by others (36), Lsm11 depletion changed the cell cycle distribution of the cells, i.e. it increased the G1 fraction and decreased the S and G2 fractions (Supplementary Figure S5). In contrast, neither depletion nor overexpression of CF Im68 appeared to induce any cell cycle changes.
In the cells transfected with control siRNAs or shRNA-encoding plasmids, H3 C pre-mRNA constituted 0.5–2% of total H3 C mRNA (data not shown). As expected, the depletion of Lsm11 affected the 3′-processing of histone H3 RNA. Compared to TcRβ-depleted cells, the apparent in vivo processing efficiency was reduced, as reflected by a 3.5-fold increase in the ratio of pre to tot RNA (Figure 5B). Most importantly, depletion of CF Im68 reduced the level of total histone H3 C RNA to 92%, and increased the level of histone H3 pre-mRNA to 174% in comparison to the control cells, resulting in an overall 2-fold decrease in the apparent in vivo processing efficiency (i.e. the ratio of pre to tot RNA). In contrast, CF Im25 depletion had no significant effect on the apparent processing efficiency (Figure 5B), although it affected β-actin and c-myc pre-mRNA processing in a similar assay (Supplementary Figure S6).
In addition, overexpression of HA-tagged CF Im68 decreased total H3 C RNA and increased H3 C pre-RNA, resulting in an overall 3.1-fold decrease in the apparent in vivo processing activity (Figure 5 C). This processing deficiency was additionally reflected by a 2.5- and 3-fold decrease of the cytoplasmic levels of histone H3 and histone H1 mRNAs, respectively (data not shown). A possible explanation for this observation might be that exogenous CF Im68 sequesters other 3′-end formation factors into non-functional complexes.
To obtain an independent read-out and to ascertain that the changes in CF Im68 levels did not alter the transcript structure (e.g. by favouring the use of potential downstream processing sites), we performed a northern blot on nuclear RNA and probed it for histone H3 C by using a combination of two hybridization probes (‘Materials and Methods’ section). By this approach no alternative H3 C transcripts could be detected, but HA-CF Im68 overexpression caused a reduction of the mature H3 C (Figure 5D), in agreement with the results obtained by qRT–PCR.
The differences between the control siRNA and CF Im68 siRNA as well as between control or HA-CF Im68 transfected cells were highly significant as evaluated by Student’s t-test (Figure 5B and C). Taken together, these experiments therefore indicate a significant quantitative contribution of CF Im68 to histone mRNA 3′-end processing in vivo.
In the course of these experiments, we made the observation that CF Im68 depletion induces a partial depletion of CF Im25 and vice versa (Figure 5 E and F). Interestingly, this co-depletion did not only affect the protein but also the RNA levels of the respective partner subunit, indicating that the two CF Im subunits must affect each other’s expression at some step of RNA metabolism.
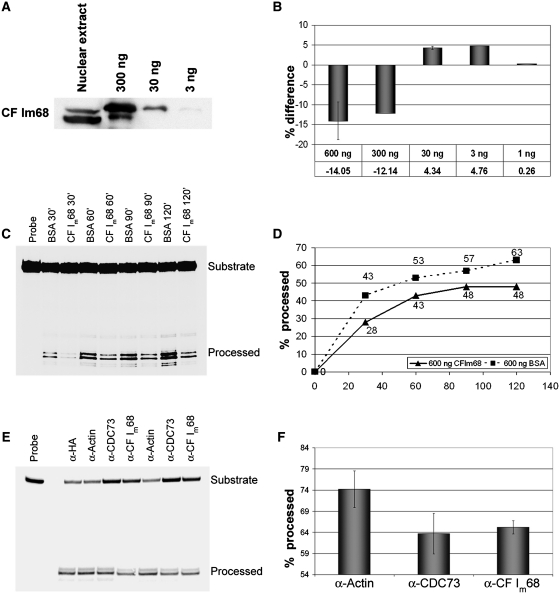
CF Im68 is involved in histone RNA 3′-end processing in vitro
To further clarify the role of CF Im68 in histone RNA processing, we performed in vitro cleavage assays in the presence of recombinant CF Im68. We titrated different amounts of histidine-tagged CF Im68 and analysed its effect on 3′-end formation. Interestingly, the addition of an excess of CF Im68 affected 3′-end formation (Figure 6B) leading to a >15% decrease of the processing efficiency, whereas more physiological amounts slightly stimulated 3′-end formation. Although the positive effect of the lower concentrations was very small, it was reproducible. Presumably, if CF Im68 contributes to histone RNA processing, it is not strongly limiting in the nuclear extracts used. The stronger, but inhibitory effect of adding an excess of 600 ng of His-CF Im68 could be seen during an entire reaction time course (Figures 6 C and D). Presumably, in this experiment, the excess CF Im68 acted by sequestering other 3′end formation factors. As an additional test for an involvement of CF Im68 in histone RNA processing, we added ∼1.2 µg of different antibodies to the processing reactions. Indeed, antibodies against CF Im68 or against the known histone RNA processing factor CDC73/parafibromin (37) inhibited the processing in comparison to the negative control in which an antibody against β-actin was used (Figures 6 E and F).
Figure 6.
In vitro processing. (A) Western blot detecting CF Im68 in 24 μl nuclear extract and a dilution series of hexahistidine-tagged recombinant CF Im68. (B) Graph showing the effect on the processing efficiency after 90 min upon addition of recombinant His-CF Im68 in comparison to the addition of the same amount of BSA. Error bars represent standard deviations for concentrations tested in multiple experiments (number of experiments: three for 600 ng, two for 30 ng). (C) Autoradiography of an in vitro processing time course with the addition of 600 ng BSA or 600 ng His-CF Im68 to the processing reaction. (D) Graph showing the accumulation of processed substrate from the experiment shown in (C). The time course was performed three times. One representative experiment is shown. (E) Effect of adding antibodies against CF Im68, the known histone processing factor CDC73/parafibromin, or β-actin (negative control) to processing reactions incubated for 105 min. (F) Quantitation of the results. Error bars represent standard deviations (number of experiments: three).
CF Im68 is associated with both unprocessed and processed histone pre-mRNA
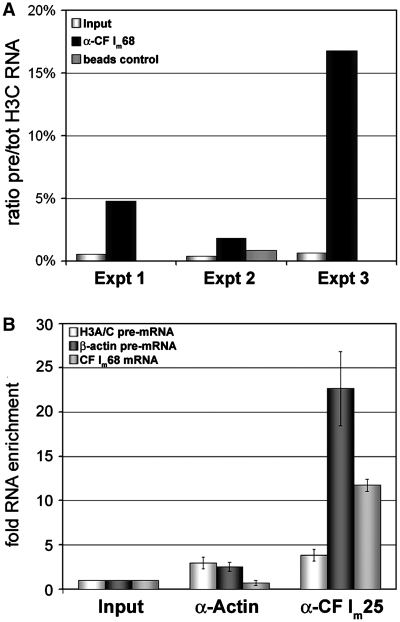
Because we could demonstrate an interaction between Lsm11 and CF Im68 in vitro as well as in vivo and because both CF Im68 depletion and overexpression significantly influenced the 3′-processing efficiency, we asked if an association of CF Im68 with histone pre-mRNAs or total RNAs can be demonstrated in vivo. Alternatively, an interaction of CF Im68 with mature mRNAs could be interesting in the light of a recent study of our two groups that demonstrated a novel role of CF Im68 in mRNA export from the nucleus to the cytoplasm (24). To address these questions, we performed RNA immunoprecipitations.
For this analysis, HeLa cells were fixed with formaldehyde, and whole cell lysates, supplemented with either anti-CF Im68 antibody or bovine serum albumin (as negative control), were subjected to immunoprecipitation. After washing, the formaldehyde fixation was reversed by incubation with proteinase K and heat treatment, RNA was isolated, reverse transcribed and the amounts of precipitated histone H3 C total RNA and pre-RNA were measured by quantitative real-time reverse transcription PCR (qRT–PCR) as already described. The results indicated that, in these experiments, the H3 C pre-RNA represented 0.53 ± 0.11% of the total H3 C RNA. In the immunoprecipitated sample, this proportion was increased to 7. 8 ± 8.0%. Despite this high standard variation which resulted from a very high pre/tot ratio in one of the immunoprecipitates, the pre/tot ratios were at least 4-fold higher in the precipitates than in the controls in all experiments (Figure 7 A). These results clearly indicate an interaction of CF Im68 with both unprocessed histone precursors and mature histone RNA, but they also show that the immunoprecipitation enriched the histone pre-mRNA ∼10-fold, strongly suggesting that CF Im68 is present in the 3′-processing complex and that CF Im68 remains associated with some of the processed mRNA.
Figure 7.
(A) Co-immunoprecipitation of histone H3 C pre-mRNA and total RNA with CF Im68. The graph shows the ratio between pre-mRNA and total histone H3 C RNAs either in the input sample (light columns) or after co-immunprecipitation with anti-CF Im68 (dark columns) as determined in three independent experiments. In Experiments 1 and 3 no RNA contamination was detected in the negative controls (BSA-coated beads), whereas for Experiment 2 the contamination is shown as grey column. (B) Co-immunoprecipitation of histone H3 C pre-mRNA (light columns), β-actin pre-mRNA (dark columns) and CF Im68 mRNA with CF Im25. An antibody against β-actin served as negative control. The cells were fixed with formaldehyde prior to total cell extract preparation and immunprecipitation. All precipitated transcripts were quantitated by qRT–PCR (number of experiments: two).
Despite the fact that the in vivo depletion did not reveal an involvement of CF Im25 in histone RNA processing (Figure 5B), we wanted to know whether CF Im25 might be associated with histone pre-mRNA, similar to CF Im68. Therefore cell extracts were subjected to immunoprecipitation with antibodies against CF Im25 or β-actin (negative control). The precipitated RNA was then analysed for the presence of pre-mRNAs for histone H3 C or β-actin, or for the presence of CF Im68 mRNA by qRT–PCR. In this experiment, H3 C pre-mRNA was not enriched in the CF Im25 immunoprecipitate compared to the β-actin precipitate (Figure 7B). In contrast, both β-actin pre-mRNA and CF Im68 mRNA were precipitated significantly more efficiently than by the control antibody. Note that CF Im68 mRNA has three binding sites for CF Im (38). Importantly, the lack of association of CF Im25 with histone pre-mRNA strongly supports the notion that the small CF Im subunit does not participate in the function of CF Im68 in histone RNA processing.
DISCUSSION
Our observations provide strong evidence for a new role of CF Im68 in histone RNA 3′-end processing. Although several other components of the cleavage and polyadenylation machinery, including all five CPSF subunits, symplekin, as well as two CStF subunits (CStF-64 and -77) have been found to constitute the heat-labile histone RNA processing factor (HLF), other components, including CF Im, have not been detected in that complex (16), and have also not been reported to contribute to histone RNA processing before. Here, we have shown that CF Im68 can be found in highly purified U7 snRNP fractions, and that it interacts with the U7-specific Sm-like protein Lsm11. This interaction was revealed in cell extracts by co-immunoprecipitation (Figure 2), in vitro by GST-pulldown experiments with recombinant proteins (Figure 3) as well as in vivo by the BiFC technique (Figure 4). Interestingly, this interaction was not observed with a mutant of Lsm11 (MPL mutant) which, when assembled with U7 snRNA, produces U7 snRNPs that are non-functional in histone RNA processing (23).
These findings suggested that CF Im68 might play a role in histone RNA 3′-end processing. In line with this idea, we found that both depletion and overexpression of CF Im68 caused significant decreases in the apparent in vivo processing efficiency (Figure 5B and C). Although this inibition might also have been mediated through reduced mRNA levels for a short-lived processing factor we consider this possibility unlikely, because depletion of the small subunit CF Im25, which is known to affect cleavage/polyadenylation (39) and which inhibited apparent in vivo 3′-end processing of β-actin and c-myc mRNAs in our hands (Supplementary Figure S6), did not affect the histone processing efficiency in vivo (Figure 5B). Moreover, the reduction of apparent processing measured by qRT–PCR did not appear to be due to an aberrant transcript structure, e.g. the use of an alternative processing site, as was revealed by northern blot analysis (Figure 5D). The notion that CF Im68 participates in histone RNA 3′-end processing was further substantiated by the finding that the addition of His-tagged CF Im68 can stimulate or inhibit in vitro processing, dependent on the amount added, and that the processing is specifically inhibited by the presence of anti-CF Im68 antibody (Figure 6). Finally, we also found that histone mRNA precursors are enriched ∼14-fold by immunoprecipitation of CF Im68 (Figure 7 A).
Interestingly, while the small CF Im, subunit, CF Im25, can form trimeric complexes with CF Im68 and Lsm11 in vitro (Figure 2) and can associate with Lsm11 in the cytoplasm when both proteins are overexpressed in the same cell (Figure 4D), it does not appear to interact with Lsm11 in vivo (Supplementary Figure S2). Its depletion also does not affect histone RNA 3′-end processing, as already mentioned above (Figure 5B), and it is not found associated with histone pre-mRNA (Figure 7B). All these results strongly suggest that CF Im68 may exist in two different states: one, associated with CF Im25, that is involved in cleavage/polyadenylation and the other, free of CF Im25, which participates in histone mRNA metabolism. How these two states of CF Im68 are controlled will be an interesting topic for future investigations.
In this context, it is noteworthy that we found the two subunits of CF Im to be co-regulated (Figure 6 E–G). The depletion of CF Im68 led to a reduction of CF Im25 mRNA and protein levels and vice versa, but not to a complete depletion. The fact that, in both cases, the reduction in mRNA level of the co-regulated gene was commensurate with that of the protein indicates that the two proteins affect each other’s expression at some step of RNA metabolism. Although we presently have no experimental data, the most likely steps to be affected are cleavage/polyadenylation, mRNA export or mRNA stability. Such a mutual cross-regulation makes sense for two proteins which depend on each other for most of their functions. In particular, it may be important for the cell not to allow CF Im25, which carries the main RNA binding activity of CF Im, to accumulate to high levels or to enter the nucleus. Its accumulation can be prevented by the cross-regulation mechanism discovered here, whereas the previous discovery that CF Im25 requires one of the larger CF Im subunits to efficiently enter the nucleus (24) would ensure that free CF Im25 does not massively enter the nucleus with possible negative consequences for the expression of many genes. It will be interesting to analyse how this co-regulation works at the molecular level and to see whether other multi-subunit 3′-end formation factors are subjected to a similar regulation.
Although a histone 3′-processing signal by itself may be efficiently recognized in an in vitro processing system, it is not certain whether the same signal is sufficiently well defined to achieve efficient cleavage in vivo. Aided by a local concentration within histone processing bodies (40), HBP and the U7 snRNP, with the help of ZFP100, may bind efficiently to nascent histone pre-mRNAs. However, the interaction of these cleavage site recognition components with HLF has not been characterized and may require additional helpers in vivo. Along these lines, it is interesting that an RNA element in the coding sequence of histone genes was shown to act as a binding site for U2 snRNPs and that this binding could stimulate U7-dependent RNA 3′-cleavage (17). It has not yet been clarified how this stimulation works, but, based on a study on 3′-terminal exon definition in polyadenylation substrates (18), the mechanism may involve an interaction between the U2 snRNP and CPSF. Additionally to the U2 snRNP, also SR proteins such as SRp20 and 9G8 can bind to the sequence element in the coding region of histone mRNAs, and this interaction was found to stimulate histone mRNA export (41). Since CF Im68 is known to bind to these SR proteins via its C-terminal alternating charge domain (21), it is also possible that it links the U7 snRNP to this mRNA internal element. Alternatively or additionally, CF Im68 might contribute to histone processing by helping the U7 snRNP to attract the cleavage effector HLF to the cleavage site that has already been defined by the U7 snRNP, HBP and ZFP100. Together with the supporting function of the U2 snRNP, this could result in an enhancement of 3′-end cleavage not unlike the one seen in 3′-terminal exon definition. To underline these parallels between polyadenylation substrates and metazoan histone pre-mRNAs, one should perhaps refer to them as ‘3′-cleavage exons’.
An interaction between CF Im68 and SR proteins bound to the mRNA internal element could also have implications for the nucleo-cytoplasmic transport of histone mRNA. Our observation that not only histone pre-mRNAs, but also a fraction of mature mRNAs, are immunoprecipitated along with CF Im68 (Figure 7 A) indicates that an interaction exists at least for a subpopulation of cleaved histone mRNAs. Important in this respect, we have recently described a function of CF Im68 in promoting the export of polyadenylated mRNAs from the nucleus (24). A similar role for the export of histone mRNAs therefore appears possible, but it could not be tested with the experiments described here, because of the effect that CF Im68 has on in vivo processing. Further experiments more specifically measuring nucleo-cytoplasmic transport will be required to explore this possibility.
In conclusion, our data indicate that CF Im68 plays one or more important role(s) in the expression of histone mRNAs. It increases the efficiency of 3′-end processing in vivo, presumably by helping to attract HLF to the factors directly recognizing the processing substrate and thereby improving the definition of the ‘3′-cleavage exon’. Moreover, it may also stimulate histone mRNA export.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canton Bern and Swiss National Science Foundation grants 3100A0-065225, -105547 and -120064 (to D.S.); Ministero dell’Istruzione, dell’Università e della Ricerca-Progetti di ricerca di interesse nazionale (MUIR-PRIN) 2006 and the Cariplo Foundation (to S.M.L.B.). Funding for open access charge: University of Bern, Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Walter Keller and Georges Martin (Biozentrum Basel) for generous gifts of clones, antibodies, and baculovirus hexahistidine-tagged CF Im68, Oliver Mühlemann (ICB Bern) for the donkey-anti-rabbit IRDye800CW antibody, Chang-Deng Hu (Purdue University) for the BiFC plasmids, Elmar Wolf, Reinhard Lührmann (MPI Göttingen) and Assad Alhaboub (ICB Bern) for HeLa nuclear extracts and Karin Schranz (ICB Bern) for excellent technical support.
REFERENCES
- 1.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schümperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ZF, Whitfield ML, Ingledue TC, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 8.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;15:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski Z, Erkmann JA, Yang X, Sanchez R, Marzluff WF. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gick O, Krämer A, Vasserot A, Birnstiel ML. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl Acad. Sci. USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lüscher B, Schümperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller B, Schümperli D. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 1997;8:567–576. doi: 10.1006/scdb.1997.0182. [DOI] [PubMed] [Google Scholar]
- 14.Furger A, Schaller A, Schümperli D. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA. 1998;4:246–256. [PMC free article] [PubMed] [Google Scholar]
- 15.Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol. Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Rüegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 20.Rüegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor I(m) involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 21.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SML. Distinct sequence motifs within the 68-kDa subunit of cleavage factor im mediate RNA binding, protein-protein interactions, and Subcellular Localization. J. Biol. Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 22.Cardinale S, Cisterna B, Bonetti P, Aringhieri C, Biggiogera M, Barabino SML. Subnuclear localization and dynamics of the pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol. Biol. Cell. 2007;18:1282–1292. doi: 10.1091/mbc.E06-09-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzouz TN, Gruber A, Schümperli D. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids. Res. 2005;33:2106–2117. doi: 10.1093/nar/gki516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruepp MD, Aringhieri C, Vivarelli S, Cardinale S, Paro S, Schumperli D, Barabino SML. Mammalian pre-mRNA 3′ end processing factor CF Im68 functions in mRNA export. Mol. Biol. Cell. 2009;20:5211–5223. doi: 10.1091/mbc.E09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 26.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 27.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Will CL, Kastner B, Lührmann R. Analysis of ribonucleoprotein interactions. In: Higgins SJ, Hames BD, editors. RNA Processing. Vol. 1. Oxford: Oxford University Press; 1994. pp. 141–177. [Google Scholar]
- 29.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotech. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spycher C, Streit A, Stefanovic B, Albrecht D, Wittop Koning TH, Schümperli D. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 32.Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 33.Azzouz TN, Schümperli D. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA. 2003;9:1532–1541. doi: 10.1261/rna.5143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 35.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells [published erratum appears in Proc. Natl Acad. Sci. USA, 1995,92:8532] Proc. Natl Acad. Sci. USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol. Cell. Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farber LJ, Kort EJ, Wang P, Chen J, Teh BT. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol. Carcinog. 2010;49:215–223. doi: 10.1002/mc.20591. [DOI] [PubMed] [Google Scholar]
- 38.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 39.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.