Abstract
Aptamers are short single-stranded nucleic acids with high affinity to target molecules and are applicable to therapeutics and diagnostics. Regardless of an increasing number of reported aptamers, the structural basis of the interaction of RNA aptamer with proteins is poorly understood. Here, we determined the 2.15 Å crystal structure of the Fc fragment of human IgG1 (hFc1) complexed with an anti-Fc RNA aptamer. The aptamer adopts a characteristic structure fit to hFc1 that is stabilized by a calcium ion, and the binding activity of the aptamer can be controlled many times by calcium chelation and addition. Importantly, the aptamer–hFc1 interaction involves mainly van der Waals contacts and hydrogen bonds rather than electrostatic forces, in contrast to other known aptamer–protein complexes. Moreover, the aptamer–hFc1 interaction involves human IgG-specific amino acids, rendering the aptamer specific to human IgGs, and not crossreactive to other species IgGs. Hence, the aptamer is a potent alternative for protein A affinity purification of Fc-fusion proteins and therapeutic antibodies. These results demonstrate, from a structural viewpoint, that conformational plasticity and selectivity of an RNA aptamer is achieved by multiple interactions other than electrostatic forces, which is applicable to many protein targets of low or no affinity to nucleic acids.
INTRODUCTION
Aptamers are folded single-stranded nucleic acids that can target given molecules. The concept is based on the ability of short (20–80 mer) sequences to fold, in the presence of a target, into unique 3D structures, which allow the aptamer to bind target molecules with high affinity and specificity (1–3). Therefore, aptamers can be thought of as nucleic-acid analogs to antibodies. RNA aptamers are selected from a combinatorial library of randomized sequences (4–8) by in vitro selection, known as SELEX (systematic evolution of ligands by exponential enrichment), and the target molecules can be nucleic acids, proteins, or small organic compounds. Aptamers have therefore many potential uses in medicine and technology. The first aptamer-based therapeutic, Pegaptanib (Macugen), which targets vascular endothelial growth factor, was approved by the FDA in 2004 for the treatment of age related macular degeneration (AMD) (9,10). Considering the basic principles of aptamer selection, the high potential of RNA to create a vast set of tertiary structures, which we referred to as ‘RNA plasticity’ (11), is conceivable from both the ‘RNA world’ hypothesis (12) and the concept of ‘molecular mimicry’ between RNA and protein (13).
Whereas RNA aptamers have been examined for their 3D structures by X-ray crystallography or NMR spectroscopy (14), only three high resolution structures of RNA aptamers in complex with target proteins have been reported: RNA aptamers in complex with NF-κB solved at 2.45 Å (15), with bacteriophage MS2 capsid at 2.8 Å (16) and with thrombin at 1.9 Å resolutions (17). NF-κB and bacteriophage MS2 capsid naturally bind to nucleic acids. The crystal structures of RNA aptamers in complex with NF-κB and bacteriophage MS2 capsid indicate that the aptamers bind to the respective nucleic-acid-binding sites and mimic naturally occurring electrostatic interactions (16,18). Thrombin does not naturally bind to nucleic acid, but bears an extensive positively charged surface responsible for heparin binding. The crystal structure of an RNA aptamer in complex with thrombin demonstrated that the RNA aptamer binds to the positively charged surface required for heparin binding (17). Thus, the crystal structures determined to date have suggested that RNA aptamers bind target proteins predominantly through electrostatic forces.
Using SELEX, we have previously identified an RNA aptamer containing 2′-fluoro pyrimidine nucleotides that binds to the Fc portion of human IgG1 (hFc1) in the presence of Ca2+ and Mg2+ (19). A unique characteristic of the Fc fragments, including hFc1, is the absence of an extensive positively charged molecular surface (20); it is thus tempting to speculate that the RNA aptamer may interact with the hFc1 via non-electrostatic forces. The aptamer’s high specificity to human IgGs and its requirement of divalent cations for binding activity are additional distinctive characteristics (19). This specificity provides us with an alternative reagent for the mass purification of therapeutic antibodies (19), as described earlier. Protein A affinity chromatography is currently the most common procedure used for purification of humanized or chimeric antibodies (21,22). This process requires an acidic elution step, which may sometime cause unexpected aggregation or denaturing of antibodies (22–24). Aptamer bound IgGs are instead easily released from the aptamer resin under neutral pH conditions using simple elution buffers, such as an EDTA solution (19). This is a potential advantage of aptamers for affinity purification. In this study, to understand the structural basis of the hyperspecificity and high affinity of the anti-hFc1 aptamer as well as prompt release of bound IgG by EDTA, we solved the crystal structure of the aptamer–hFc1 complex at the 2.15-Å resolution.
MATERIALS AND METHODS
Crystallization and data collection
Human IgG1 was purchased from Calbiochem (USA). To produce Fc fragments, human IgG1 was digested with papain (Wako Pure Chemical Industries Ltd, Japan) at 37°C for 1 h in 100 mM sodium phosphate (pH 7.2), 10 mM l-cysteine and 2 mM EDTA (molar ratio of IgG1:papain is 17:1).The reaction was stopped by the addition of protease inhibitor. The Fc fragment was purified with Protein A column (GE Healthcare) (25). The chemically synthesized RNA aptamer containing 2′-fluoropyrimidines (Figure 1a) was purchased from GeneDesign Inc. (Japan) and purified as described previously (25). The previous NMR study indicated a 1:2 stoichiometry of the Fc dimer:aptamer complex (19). Thus, the aptamer was mixed with the Fc dimer in a molar ratio of 1:2.2 (Fc dimer:aptamer) for crystallization. The final concentration of the Fc fragment was 7 mg ml−1 and it was stored at 193 K (25). Crystals were grown at 293 K by the sitting-drop vapor-diffusion method by mixing the sample with an equal volume of reservoir solution containing 0.1 M Tris–HCl buffer (pH 8.0), 20% (w/v) PEG 1000 and 0.2 M calcium acetate. The solution stirring technique with a rotary shaker (26–28) produced the best diffracting crystals.
Figure 1.
Structure of the aptamer and the aptamer–hFc1 complex. (a) M-fold predicted secondary structure of anti-hFc1 aptamer. Nucleotide symbols: N (black), RNA; and fN (blue), 2′-fluoro modification. Note that it differs from the RNA structure in the aptamer–hFc1 complex in Figure 2. (b) 2Fo–Fc electron density contoured at 1σ, corresponding to RNA aptamer. (c) Stereo view of the structure of the aptamer–hFc1 complex. The aptamers and hFc1 are shown in orange and green, respectively. The loop connecting CH2 and CH3 domains are shown in yellow. Ca2+ ions bound to the aptamers are indicated by green spheres. The side chains of Lys340 and Gln342, and carbohydrates are shown as stick models. The areas in contact with the symmetry-related molecules in the crystal lattice are shown in red. (d) Structural comparison of the residues at the aptamer–hFc1 interface. All the residues of hFc1 responsible for binding to the aptamer are shown as green stick models. The residues of uncomplexed form of hFc1 (PDB entry: 1FC1), which are superimposed onto the corresponding residues of aptamer-bound hFc1, are shown as yellow stick model. The black arrow indicates the induced fit of Lys340.
The crystals were soaked briefly in Paratone-N oil and then frozen by rapidly submerging them in liquid nitrogen. Diffraction of data sets was collected at the beam line BL41XU in SPring-8. Crystals belonged to the space group P21212 with the cell parameters a = 83.59, b = 107.24, c = 79.46 Å. Diffraction data were processed with the program HKL2000 (29). Data statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Data Collection | |
| Space group | P21212 |
| Cell dimensions (Å, °) | a = 83.7, b = 107.2, c = 79.0 |
| Resolution (Å) | 30.5–2.15 (2.23–2.15) |
| Rmerge (%)a | 8.1 (43.3)b |
| Completeness (%) | 98.3 (89.1)b |
| No. of measured reflections | 195 721 |
| No. of unique reflections | 38 726 |
| I/σ(I) | 7.8 (2.4)b |
| Redundancy | 5.1 (5.1) |
| Refinement | |
| Resolution (Å) | 30.5–2.15 |
| No. of reflections | 36 041 |
| Rwork / Rfree (%) | 23.7/27.8 |
| No. of atoms | |
| Protein | 3082 |
| Ligand/ion | 1072/2 |
| Water | 253 |
| B-factors | |
| Protein | 48.5 |
| Ligand/ion | 75.4/68.3 |
| Water | 44.5 |
| Rmsd | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.2 |
aRmerge = Σ|I(k)-I|/ΣI(k), where I(k) is value of the kth measurement of the intensity of a reflection, I is the mean value of the intensity of that reflection and the summation is the over all measurements.
bValues in parentheses are for the highest resolution shell (2.23–2.15 Å).
Structural determination and refinement
The structure of hFc1 in complex with RNA aptamer was determined by the molecular replacement method using the program MOLREP (30) with the structure of the uncomplexed form of hFc1 (PDB entry: 1FC1) as a search model. Refinement procedure for IgG complexed with the aptamer was carried out using the program CNS (31) without non-crystallographic symmetry (NCS) restrains/constraints. The structures were visualized and modified using the programs O (Alwyn Jones, Uppsala University, Sweden), and COOT (32).
The stereochemical qualities of the final structures were assessed with programs PROCHECK (33). Contact area values were calculated with the program ICM Pro (Molsoft), in which all of the parameters were used default. Figures were generated by PyMOL (www.pymol.org), and structural comparisons were carried out using the programs Chimera (34) and LSQKAB (35).
SPR analysis of base substitution derivatives
3′-end poly(dA) 16-tailed Apt8-2 (a functional derivative of the aptamer in this study) and mutant RNAs were prepared by chemical synthesis (Gene Design Inc.), and the binding activity was measured by SPR using the BIAcore2000 instrument (GE Healthcare). 5′-biotinylated poly(dT) 16 was first bound to a streptavidin sensor chip (BIAcore), and poly(dA) 16-tailed RNAs were immobilized to the sensor chip (∼1000 RUs) via hybridization to the poly(dT) 16 oligomer. Human IgG1 solution (Calbiochem, 100–500 nM) was injected, and complex formation was monitored via the sensorgram coupled with the BIAevaluation 3.0 software analysis (BIAcore). The running buffer was a mixture of 145 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2 and 20 mM Tris–HCl (pH 7.6). A 6 M urea solution was used for regeneration.
Repeated unfolding/refolding of anti-hFc1 aptamer resin
The refolding experiments were conducted with BIAcore2000. The 5′-biotinylated Apt8-2 was immobilized on a streptavidin sensor chip (BIAcore, 1000RU). Human IgG1, dissolved in binding buffer (Calbiochem, 400 nM), was injected. The composition of binding buffer was the same as the running buffer shown above. The aptamer was inactivated by injection of unfolding buffer (10 mM EDTA and 10 mM Tris–HCl pH 7.6) and the released IgG was washed out. Injections of the IgG solution and the unfolding buffer were repeated 10 times.
RESULTS
Overall structure
To clarify the structural basis for the high specificity and characteristic properties of the RNA aptamer, we have determined the 2.15 Å crystal structure of the RNA aptamer in complex with hFc1 (Figure 1). The final model, an asymmetric unit including homodimeric hFc1 residues 241–262, 272–296, 302–321 and 334–443 for chain A and 237–444 for chain B, two RNA aptamer molecules (G1-C11 and G16-C23 and G1-A24), two carbohydrate chains (1–8, 3–7), two Ca2+ ions, and 254 water molecules, of which we observe well-defined electron density. The monomeric hFc1 is composed of two globular domains (referred to as CH2 and CH3 domains) (Figure 1).
The superimposition of two hFc1 molecules in the asymmetric unit reveals a minor difference in their conformation (rmsd of 1.2 Å over 170 aligned Cα atoms). The difference is very likely caused by crystal packing, because residues 268 and 298–300 of chain B make van der Waals contacts with residues 289–292 of a symmetrically related chain A, while the corresponding regions of chain A does not (Figure 1 c). Ca2+ ions, which were contained in the crystallization solution, are observed in the structure of the aptamers (discussed in detail later). The bound carbohydrate chains are structurally identical to those observed in the uncomplexed structure of dimeric hFc1 (PDB entry: 1FC1). The model, which has been refined to an Rfree of 27.7%, displays good geometrical parameters (Table 1). The overall structure of the aptamer–hFc1 complex is represented in Figure 1 c.
The structure of the aptamer-bound hFc1 can be superimposed on the uncomplexed form of the structure of hFc1 (20) (PDB entry: 1FC1) with rmsd of 1.1 Å over 357 aligned Cα atoms. This suggests that aptamer binding caused no significant structural changes to the backbone of hFc1. This deviation value is also comparable to the rmsd value between two hFc1 molecules within the aptamer complex. Except for Lys340, the side chains at the aptamer–hFc1 interface adopt similar conformations that are observed in the uncomplexed form of hFc1 (Figure 1 d) (rmsd of 1.0 Å over 82 nonhydrogen atoms of residues 339–344, 373, 374, 398, 402 and 404). Upon aptamer binding, the side chain of Lys340 swings ∼4 Å to interact with the phosphate backbone of RNA (Figure 1 d).
RNA structure
Figure 2a shows the overall structure of the RNA aptamer. The atomic coordinates of the apical loop in one aptamer could not be determined due to low electron density, while the apical loop in another aptamer adopts a GAAA tetraloop. Although G1:C23 and G2:C22 are base paired in one of the aptamers, the two base pairs in the other aptamer are not formed due to crystal packing. The structures of the binding region (A3-C11 and G16-U21) to hFc1 for both aptamers are almost identical (rmsd of 0.43 Å over all nonhydrogen atoms). The global fold of the aptamer adapts a distorted hairpin structure with a GAAA tetraloop, an internal loop, and a terminal A-form helix (Figure 2 a). The characteristic structure of the binding region to hFc1 was predominantly formed by the internal loop, as we describe below.
Figure 2.
Overall structure of the aptamer within the aptamer–hFc1 complex. (a) Tertiary (left) and secondary (right) structures of the bound aptamer. Ca2+ is shown as green sphere. The nucleotide sequences are color coded as follows: adenine, red; guanine, cyan; 2′-fluoro uridine, green; and 2′-fluoro cytidine, yellow. (b) U6:A18:U9 base triple. The interactions are indicated with thin dashed lines and the distances (Å) are given in black. RNA atoms are colored, red, blue, light blue and orange for oxygen, nitrogen, fluorine and phosphorus, respectively. The color coding of the carbon is the same as in Figure 2 a. (c) Interaction between the H8 of G7 and the 2′-fluoro of U6. The H8 atom is shown at the calculated ideal positions. (d) Functional annotation of base substitutions and ribose 2′ modifications in the anti-hFc1 aptamer. Base substitutions described previously (19) and those newly manipulated in this study are indicated in the M-fold structure of Apt8-2 (19), a functional derivative of the aptamer used for crystallization. Each oligonucleotide with the indicated substitutions were chemically synthesized and examined for affinity to hIgG1 by SPR. Nucleotide symbols: N (black), RNA; n (green), DNA; fN (blue), 2′-fluoro modification; and mN (orange), 2′ O-methyl modification. All substitutions shown here completely abrogated binding activity.
The aptamer contains six Watson–Crick type base pairs, two non-canonical base pairs, and one base triple. One non-canonical base pair is found in the GAAA tetraloop (G12:A15 base pair), and the other is a G5:A19 base pair in the internal loop. The latter is categorized to be Trans-Sugar Edge/Hoogsteen GA (36,37). In the internal loop, a U6:A18:U9 base triple is formed (Figure 2 b). In the base triple, U9 and A18 form a cis-Watson–Crick/Watson–Crick base pair, while U6 and A18 form a Trans-Watson–Crick/Hoogsteen base pair (38,39). Bases are continuously stacked in G1-U6, G7-G12 and A13-C23, whereas the flipping bases of U6 and G7 are not stacked. Torsion angles ζ for U6 and α, β, γ for G7 adopt non-standard values (−160°, 85°, −155° and 175°, respectively) for the base flipping between U6 and G7. Our aptamer contains 2′-fluoro substitutions on pyrimidine residues for ribonuclease resistance. It is known that 2′-fluoro substitutions in RNA increase the propensity of the sugar to adopt a C3′-endo conformation which results in a stabilized A-form helix (40,41). Consistent with this propensity, the sugars of all nucleotides including the 2′-fluoro pyrimidines adopt a C3′-endo conformation. The 2′-fluoro of U6 is crucial for binding to Fc fragment (19). The conformation of the G7 base is stabilized by the van der Waals interaction between the H8 of G7 and the 2′-fluoro of U6 (Figure 2 c), which also participates in the U6:A19:U9 base triple.
To investigate the structural and functional contribution of each nucleotide, mutagenesis experiments of RNA were further conducted in addition to those described previously (19) (Figure 2 d). When the UAU base triple was destroyed by mutation, such as fU6-to-fC, a18-to-t, or fU9:a18-to-fC:G, the aptamer completely lost its affinity. Similarly, destruction of the interaction between fU6 and G7 by mutation, such as fU6-to-U/mU/u and G7-to-A/fU, also caused a loss of affinity. These data suggest the functional importance of the interaction between H8 of G7 and 2′-fluoro of U6 and UAU base triple, as observed in the crystal structure. In addition, the mutation of G5-to-A or fC lost the affinity, which also suggests the importance of maintaining the G5:A19 non-canonical base pair.
Role of the divalent cations
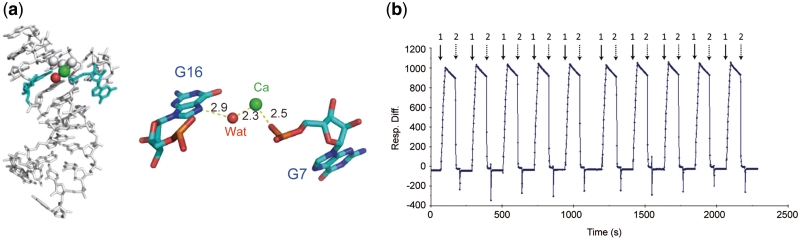
In a difference electron density map calculated using a model lacking metals, a strong peak (at a contour level of 5σ above the mean value of the electron density) was found in the RNA major groove. These densities cannot be accounted for by water molecules but rather as heavier metals. On the basis of an average metal-to-atoms distance, geometry of the coordination sphere, an atomic temperature factor and an atomic occupancy (Table 2), the metal ions were identified as Ca2+, which were present in the crystallization solution. The hydrated Ca2+ in the RNA major groove coordinates with no protein ligands (Figure 3a), but rather binds to non-bridging oxygen atoms of the G7 phosphates in the RNA. Therefore, Ca2+ also may help to maintain the distinct conformation of G7, which is one of the hFc1-interacting residues (described later). This site might also be occupied by Mg2+, since the aptamer could bind to hFc1 in the presence of Ca2+ or Mg2+.
Table 2.
Calculated atomic occupancy of the hypothetical metal ions
| Peaka | Average B-factorb | Calculated atomic occupancyc |
|||
|---|---|---|---|---|---|
| Ca2+ | Na+ | Mg2+ | Water | ||
| 1 | 65.17 | 0.86 | 1.52 | 1.41 | 2.50 |
| 2 | 54.05 | 1.01 | 1.77 | 1.68 | 2.68 |
aThe peak corresponds to the electron density peak. Peaks 1 and 2 are located in the major groove of the aptamer, while peaks 3 and 4 are observed at the aptamer–hFc1 interface.
bThe average B-factors were calculated and averaged from the atoms within a 5 Å radius of the peak (left column).
cThe atomic occupancy refinements were carried out by CNS (31). Before the refinement, the cations (Ca2+, Na+, Mg2+, and water) were put at the peak of the electron density; then, the temperature factors of the atoms were set to the average B-factor (centre columns). Peaks 1 and 2 are likely to be equivalent to Ca2+ because the calculated values of the atomic occupancy are close to 1.
Figure 3.
(a) Coordination spheres of Ca2+. The interactions are indicated with thin dashed lines and selected nucleotides are labeled. Ca2+ is bound in a distorted octahedral coordination environment with the phosphate backbone and five water molecules (left). In the enlarged view (right), only one water molecule involved in the aptamer binding is shown for clarity. The Ca2+ and the water molecule are shown with green and red spheres, respectively. (b) A SPR sensorgram of IgG-aptamer interactions. Folding and unfolding the aptamer was repeated 10 times. Arrows: 1, injection of hIgG dissolved in binding buffer; 2, injection of unfolding buffer. Experimental procedures and conditions are described in Supplementary Notes online.
To investigate the importance of Ca2+ for the affinity, surface plasmon resonance (SPR) experiments were conducted with different running buffer conditions. Under a condition without divalent cations, such as PBS or saline, the aptamer could not bind to human IgG (hIgG). Addition of EDTA, a chelating agent, caused loss of affinity and the affinity was regained with addition of Ca2+ (Figure 3 b). These biochemical assays indicate the importance of divalent cations for the aptamer activity. The reversible binding suggests reversible folding of the aptamer, achieved by the presence of the divalent cation. The SPR analysis demonstrates that this conformational change is reversible and rapid, and therefore can also be envisioned as a sensor for divalent cations. In cells, such cation-specific sensors are also known as naturally occurring riboswitches (42–44).
Interaction between the aptamer and hFc1
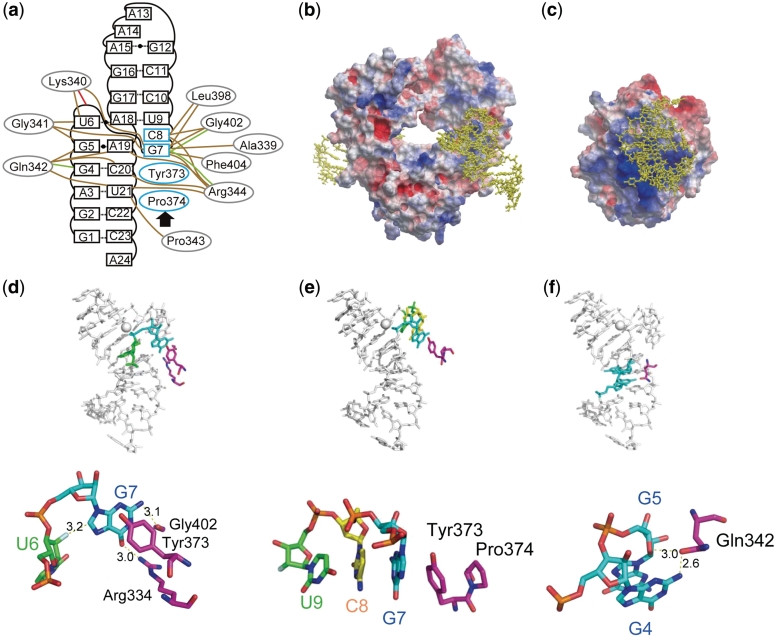
The interaction between the aptamer and hFc1 is summarized in Figure 4a. The interaction involves hydrogen bonding and van der Waals forces, and only one ion-pair was observed between the backbone phosphate of G7 and the side chain of Lys340. Analysis of the surface electrostatic potential in hFc1 demonstrates that the aptamer-bound area consists of a less-positively charged surface (Figure 4 b). This feature is significantly different from aptamer complexes with NF-κB (15) and bacteriophage MS2 capsid (16), which also naturally bind nucleic acids (Supplementary Figure S1), or to the aptamer complex with thrombin, which does not naturally bind nucleic acid (17) (Figure 4 c).
Figure 4.
Interactions of the aptamer with target proteins. (a) Interactions between hFc1 and the aptamer are color-typed: red, ion pair; green, hydrogen bond; brown, van der Waals contact; blue, stacking interaction. (b) Overall structure of aptamer–hFc1 complex with the electrostatic surface potential of hFc1. The RNA aptamer is shown as a yellow ball-and-stick model. (c) Structure of aptamer–thrombin complex with the electrostatic surface potential of thrombin (17) with the RNA aptamer. (d) The detailed interaction of the base of G7 with the RNA aptamer and protein portions. (e) The continuous stacking interaction between hFc1 and RNA aptamer. (f) The detailed interaction of the side chain of Gln342. Images of the electrostatic surface potential were produced by ICM Pro (Molsoft Inc.) using default setting: the potential scale used was 5.
The interaction between hFc1 and aptamer covers 580 Å2 of surface area per Fc fragment, and this area is relatively small to the area covered by the other RNA aptamer interactions (ca. 1000 Å2) (13–15). The average contact area of all amino acids that interact with the aptamer is 30 Å2. The contact area of the aptamer’s G7 to hFc1 is the largest and covers 110 Å2, whereas the average contact area of all interacting nucleotides is 35 Å2. As shown in Figure 4 d, the flipped base of G7 is stabilized by the following interactions: the stacking interaction between the G7 base and the side chain of Tyr373; the hydrogen bonds between N2 of the G7 base and the carbonyl oxygen of Gly402, as well as the O6 of the G7 base and the side chain of Arg344; and van der Waals contacts between the G7 base and 2′-fluoro of U6 ribose. Bases G7-G12 continuously stack on Tyr373 and Pro374 of hFc1 (Figure 4 e), revealing structural adaptations that create a characteristic interaction. In addition, the 2′-fluoro groups of the C8 and the C20 potentially stabilize the aptamer–hFc1 interaction. The 2′-fluoro group of the C8 makes van der Waals contacts with the side chain of Leu398 and the 2′-fluoro group of the C20 contacts the side chains of Gln342 and Arg344. The previous mutagenesis studies have indicated that 2′-fluoro groups of C8 and C20 could be replaced by hydroxyl groups (19).
DISCUSSION
The aptamer binds to four subtypes of human IgGs, IgG1 through IgG4, but not to IgGs from other species, including mouse, rat, rabbit, bovine, feline and canine (19). It is likely that Gln342 and Phe404, conserved in human IgG1-4, play key roles for the specificity as they interact directly with the aptamer. The largest contact area (75 Å2) in hFc1 is occupied by Gln342, which is located in the loop connecting the CH2 and CH3 domains. The side chain of Gln342 interacts with N2 of the base G4, O2 of the base C20 and O4′ of the ribose in U21 (Figure 4f). Gln342 is also conserved in rabbit and feline IgG. Therefore, Gln342 is not the only residue crucial for the specificity. The other key residue seems to be Phe404, which directly interacts with G7 (Figure 4 a). At this position, Phe is substituted by Tyr in the other species, with the exception of rat IgG, and this substitution would likely lead to steric hindrance with G7. Since the presence of both of these residues is common only to human IgG1-4, we conclude that the two residues are most important for the specificity of the aptamer.
RNA generally binds to proteins via an electrostatic interaction between the negatively charged phosphate backbones and the positively charged surface of proteins. However, the stacking interaction between G7 and Tyr373 was one of the major interactions in the aptamer–hFc1 complex. As shown in Figure 4 b, the aptamer obviously interacts through the weaker forces supported by van der Waals contacts and hydrogen bonds. This study strongly suggests that SELEX technology can select for not only molecules that interact through predominantly electrostatic forces (13–15), but also high specificity molecules that interact through weaker forces such as van der Waals contacts and hydrogen bonds. The nature of the latter resembles that of protein-protein interactions, rather than that of nucleic acids-protein interactions.
The crystal structure revealed that oxygen atoms of the phosphate of G7 is bound to a divalent ion, which is located in the major groove of the aptamer but not located in the interface between the aptamer and hFc1. Thus, it is suggested that the divalent ion is important for folding the active structure of the aptamer, and could be quickly removed by a chelating agent. Indeed, the SPR analysis demonstrated that the active structure of the aptamer is destabilized with EDTA solution, but quickly refolds by adding the divalent cations again. It is noteworthy that hFc1 undergoes no conformational change upon binding to the aptamer. This, in turn, emphasizes conformational plasticity of aptamer RNA. Consequently, this aptamer could be a novel ligand for affinity purification of therapeutic antibodies and can substitute for protein A to avoid possible loss-of-quality during the acid elution process.
In summary, the most intriguing findings in the current study are related to the multiple weaker interactions, rather than electrostatic forces, that contribute to the highly selective and strong binding to the target protein, and to the fact that the hFc1 conformation is the least affected by aptamer binding. These findings demonstrate excellent conformational plasticity of RNA molecules, and therefore suggest that RNA aptamers may be applicable to a wider range of targets than previously expected.
ACCESSION NUMBER
The final atomic coordinates and structure factor amplitudes (PDB entry 3AGV) have been deposited in the Worldwide Protein Data Bank (wwPDB; http://www.wwpdb.org), Protein Data Bank Japan at the Institute for Protein Research in Osaka University (PDBj; http://www.pdbj.org).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT); Core Research for Evolution Science and Technology (CREST) grants (to Y.N. and Y.M.); Science and Technology Incubation Program from the Japan Science and Technology Agency (to H.M.). Funding for open access charge: CREST.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The synchrotron radiation experiments were performed at SPring-8 with the Institute for Protein Research, Osaka University (2008A6806).
REFERENCES
- 1.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y. RNA aptamers to initiation factor 4 A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyakawa S, Oguro A, Ohtsu T, Imataka H, Sonenberg N, Nakamura Y. RNA aptamers to mammalian initiation factor 4 G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA. 2006;12:1825–1834. doi: 10.1261/rna.2169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohuchi SP, Ohtsu T, Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface. Biochimie. 2006;88:897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Klussmann S. The Aptamer Handbook. Germany: WILEY-VCH, Weinheim; 2006. [Google Scholar]
- 8.Keefe AD, Schaub RG. Aptamers as candidate therapeutics for cardiovascular indications. Curr. Opin. Pharmacol. 2008;8:147–152. doi: 10.1016/j.coph.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, Wang B. Pegaptanib for the treatment of age-related macular degeneration. Exp. Eye. Res. 2006;83:615–619. doi: 10.1016/j.exer.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura Y, Endo K, Adachi H, Ishiguro A. RNA aptamers to translational components. In: Hershey JWB, editor. Translational Control in Health and Disease. Amsterdam: Elsevier Press; 2009. pp. 369–395. [DOI] [PubMed] [Google Scholar]
- 12.Gesteland RF, Cech TR, Atkins JF. The RNA World. New York: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 13.Nakamura Y. Molecular mimicry between protein and tRNA. J. Mol. Evil. 2001;53:282–289. doi: 10.1007/s002390010218. [DOI] [PubMed] [Google Scholar]
- 14.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 15.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd, Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl Acad. Sci. USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn WT, Convery MA, Stonehouse NJ, Adams CJ, Liljas L, Phillips SE, Stockley PG. The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions. RNA. 2004;10:1776–1782. doi: 10.1261/rna.7710304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh G, Huang DB, Huxford T. Molecular mimicry of the NF-kappaB DNA target site by a selected RNA aptamer. Curr. Opin. Struct. Biol. 2004;14:21–27. doi: 10.1016/j.sbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Miyakawa S, Nomura Y, Sakamoto T, Yamaguchi Y, Kato K, Yamazaki S, Nakamura Y. Structural and molecular basis for hyperspecificity of RNA aptamer to human immunoglobulin G. RNA. 2008;14:1154–1163. doi: 10.1261/rna.1005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 21.Fahrner RL, Knudsen HL, Basey CD, Galan W, Feuerhelm D, Vanderlaan M, Blank GS. Industrial purification of pharmaceutical antibodies: development, operation, and validation of chromatography processes. Biotechnol. Genet. Eng. Rev. 2001;18:301–327. doi: 10.1080/02648725.2001.10648017. [DOI] [PubMed] [Google Scholar]
- 22.Ghose S, Allen M, Hubbard B, Brooks C, Cramer SM. Antibody variable region interactions with Protein A: implications for the development of generic purification processes. Biotechnol. Bioeng. 2005;92:665–673. doi: 10.1002/bit.20729. [DOI] [PubMed] [Google Scholar]
- 23.Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004;20:1301–1308. doi: 10.1021/bp0498793. [DOI] [PubMed] [Google Scholar]
- 24.Cromwell ME, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8:E572–E579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiyama S, Nomura Y, Sakamoto T, Kitatani T, Kobayashi A, Miyakawa S, Takahashi Y, Adachi H, Takano K, Murakami S, et al. Crystallization and preliminary X-ray diffraction studies of an RNA aptamer in complex with the human IgG Fc fragment. Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 2008;64:942–944. doi: 10.1107/S1744309108028236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adachi H, Matsumura H, Niino A, Takano K, Kinoshita T, Warizaya M, Inoue T, Mori Y, Sasaki T. Improving the quality of protein crystals using stirring crystallization. Jpn. J. Appl. Phys. 2004;43:L522–L525. [Google Scholar]
- 27.Adachi H, Takano K, Matsumura H, Inoue T, Mori Y, Sasaki T. Protein crystal growth with a two-liquid system and stirring solution. J. Synchrotron Rad. 2004;11:121–124. doi: 10.1107/s0909049503026980. [DOI] [PubMed] [Google Scholar]
- 28.Adachi H, Takano K, Niino A, Matsumura H, Kinoshita T, Warizaya M, Inoue T, Mori Y, Sasaki T. Solution stirring initiates nucleation and improves the quality of adenosine deaminase crystals. Acta Crystallogr. D Biol. Crystallogr. 2005;61:759–762. doi: 10.1107/S0907444905013466. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Data Collection and Processing: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Warrington: Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 31.Adams PD, Pannu NS, Read RJ, Brunger AT. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl Acad. Sci. USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 37.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 38.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 40.Guschlbauer W, Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980;8:1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westhof E, Patel DJ. Nucleic acids. From self-assembly to induced-fit recognition. Curr. Opin. Struct. Biol. 1997;7:305–309. doi: 10.1016/s0959-440x(97)80044-9. [DOI] [PubMed] [Google Scholar]
- 43.DeRose VJ. Sensing cellular magnesium with RNA. Nat. Chem. Biol. 2007;3:693–694. doi: 10.1038/nchembio1107-693. [DOI] [PubMed] [Google Scholar]
- 44.Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.