Abstract
Sequence-specific transcription factors (TFs) are important to genetic regulation in all organisms because they recognize and directly bind to regulatory regions on DNA. Here, we survey and summarize the TF resources available. We outline the organisms for which TF annotation is provided, and discuss the criteria and methods used to annotate TFs by different databases. By using genomic TF repertoires from ∼700 genomes across the tree of life, covering Bacteria, Archaea and Eukaryota, we review TF abundance with respect to the number of genes, as well as their structural complexity in diverse lineages. While typical eukaryotic TFs are longer than the average eukaryotic proteins, the inverse is true for prokaryotes. Only in eukaryotes does the same family of DNA-binding domain (DBD) occur multiple times within one polypeptide chain. This potentially increases the length and diversity of DNA-recognition sequence by reusing DBDs from the same family. We examined the increase in TF abundance with the number of genes in genomes, using the largest set of prokaryotic and eukaryotic genomes to date. As pointed out before, prokaryotic TFs increase faster than linearly. We further observe a similar relationship in eukaryotic genomes with a slower increase in TFs.
INTRODUCTION
Regulation of gene expression has always been one of the most prominent areas in the field of genetics. The mechanism of genetic regulation was unveiled for the first time, when Jacob and Monod (1) uncovered the gene regulation apparatus of the lac operon in Escherichia coli. Since then, numerous studies [e.g. (2–4)] have shown that regulation of gene expression is essential to determining organismal complexity and morphological diversity in different species across the tree of life. Transcriptional regulation is a crucial step in gene expression regulation because the genetic information is directly read from DNA by sequence-specific transcription factors (TFs). The unique role of TFs is highlighted by several studies demonstrating their abilities to reprogramme fibroblasts into embryonic stem cells (5,6).
Numerous studies have provided a great deal of insight into the conserved and specific DNA-binding TFs in different lineages, though they tended to concentrate on particular phylogenetic groups. The DNA-binding domains (DBDs), evolutionary components of sequence-specific TFs that mediate the specificity of the TF–DNA interaction, are often used to represent TF families, which is appropriate from functional as well as evolutionary points of view. Despite their importance, the global DBD repertoire was only once reviewed from a structural perspective a decade ago (7).
Being aware of the importance of TFs on genetic regulation, the community has put a great amount of effort into the development of resources for the systematic collection and classification of annotated TFs in genomes from diverse lineages. Here, we summarize key publications of genome-wide studies of TFs and survey TF databases currently available, as well as discuss the criteria and the methods used to obtain TF catalogues. A better understanding of global TF repertoires in species from diverse and related lineages will not only serve as a starting point for experimental design of high-throughput studies for determining the binding sites of TFs in different model organisms (8–12), but will also offer an insight into the evolution of TFs in conjunction with the remainder of the proteins in genomes that they regulate. To summarize our current knowledge on the genomic repertoires of TFs across the tree of life (from Bacteria, Archaea and Eukaryota superkingdoms), we used TFs annotated by the DBD database (13) in ∼700 organisms as representatives to review their abundance with respect to the number of genes in different genomes, as well as their complexity in terms of DBD and other protein domain components in diverse phylogenetic branches.
GENOME-WIDE STUDIES OF TFs IN DIFFERENT PHYLOGENETIC GROUPS
Since the first whole-genome sequencing was completed in 1995 on a pathogenic bacterium Haemophilus influenza (14), the number of completely sequenced prokaryotic genomes has been increasing rapidly, with a doubling time of ∼20 months for bacteria and ∼34 months for archaea (15). Due to the abundance of publicly available prokaryotic genomes, a large number of whole genome TF studies have focused on these organisms. Aravind and Koonin (16) published one of the earlier analyses on the repertoire of TF families in four complete archaeal genomes. Using sequence profile methods in conjunction with protein structure information, they presented the intriguing finding that the majority of archaeal DBDs had helix-turn-helix (HTH) structures similar to bacterial HTH domains. This contrasts with the core components of the archaeal transcriptional machinery, such as basal TFs and RNA polymerases, which are more closely related to eukaryotic systems. A more recent study by Coulson and coworkers confirmed this finding (17). Since then, similar types of analysis were conducted by different groups with larger sets of prokaryotic species.
Perez-Rueda et al. (18) addressed the distribution of 75 TF families across 90 prokaryotes based on the well-characterized set of TFs in E. coli K12. Because the reference TFs were taken from one bacterial species, the predicted TFs were restricted to close homologues of TFs found in E. coli. Similarly, Minezaki et al. (19) classified TFs from 154 complete prokaryotic genomes into 52 TF families. Their TF families were collected from TFs found in eight different archaea and bacteria, with additional DBDs documented in Pfam (20). Thus, this reference TF set was likely to detect additional varieties of TF homologues across prokaryotic proteins. Different criteria for constructing the reference TFs notwithstanding, both studies consolidated the predominance of HTH DBDs in prokaryotes, especially the winged-HTHs. They also demonstrated a significant depletion of TF families in intra-cellular pathogenic and endosymbiotic bacteria including Mycoplasma and Chlamydophila. These pathogenic life forms normally inhabit hosts whose environment lacks selective pressure to maintain the specific genes to respond to environmental stress. Other groups considered more restricted lineages of bacteria including Moreno-Campuzano et al. (21) and Brune et al. (22). Their studies provided comprehensive lists of TF repertoires in firmicutes and corynebacteria, respectively.
Baker’s yeast Saccharomyces cerevisiae was the first eukaryotic species to have its genome completely sequenced. The paper describing the whole-genome sequencing of baker’s yeast (23) was published in 1996, only slightly after the first prokaryotic genome H. influenza. The number of completely sequenced eukaryotic genomes, however, increases significantly more slowly than that of prokaryotic genomes. This is likely due to the combination of larger average size of eukaryotic genomes, and the difficulty in assembling and annotating the genomes that contain a great amount of repetitive and non-coding elements (24). Nonetheless, an increasing number of studies on the genomic TF repertoires are being conducted using complete eukaryotic genomes.
Riechmann et al. (25) surveyed specific TF families occurring in four eukaryotic genomes: Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster and S. cerevisiae. They demonstrated that a number of DBD families are shared across all three major eukaryotic kingdoms, i.e. Metazoa (animal), Fungi and Viridiplantae (plant), but the domain combinations of DBDs and other domains in TFs are highly kingdom-specific. According to Coulson and Ouzounis (26), each eukaryotic kingdom possesses not only the families common to all eukaryotes, but also a number of kingdom-specific transcriptional regulators, which possibly participate in kingdom-specific processes. Other studies focused on particular eukaryotic kingdoms. In plant, Shiu et al. (27) pointed out that not only were the TF families more diverse compared with fungi and animals, but the expansion and duplication rates in plants were also considerably greater. This suggests a more frequent adaptive response to selection pressure among plants since they do not have mobility to avoid stress stimuli in the same way as other eukaryotes. More recent work by Shelest (28) concentrated on TFs in fungi, reporting 37 TF families in 62 fungal species, of which only six families were fungal specific. Being phylogenetically distant from animals, fungi and plants, the genomes of parasitic protists such as apicomplexans and ciliates are known to be substantially divergent from the current model eukaryotic genomes and thus less well-understood. Iyer and coworkers (29) were the first to investigate the repertoires of TFs and chromatin proteins in these parasitic unicellular eukaryotes.
In the Metazoa (animal) kingdom, TFs are particularly essential to the morphological development of animals’ organ systems. Messina et al. (30) compiled one of the first lists of metazoan TFs by focusing on human. They aimed to produce a starting point for array experiments across species. By taking known TFs from TRANSFAC (31), InterPro (32) and FlyBase (33) as seed sequences, additional human TFs were discovered using hidden Markov model (HMM) searches, followed by manual curation. As part of the initiative to characterize the transcription regulatory network in mammalian cells, the International Regulome Consortium (IRC) have put together a comprehensive list of mouse TFs by mapping cDNA sequences from several libraries to the NCBI mouse genome. More recently, Vaquerizas et al. (34) have manually compiled a human TF repertoire and analyzed their expression patterns and evolutionary conservation. These studies on mammalian TFs will contribute to a better understanding of gene expression control in higher organisms.
In summary, several key publications mentioned here highlight the importance of TFs in the development and maintenance of cellular phenotypes in different kinds of organisms. These genome-wide studies provide a starting point for a systematic comparative analysis of genomic TF repertoires in both closely and distantly related genomes.
TF RESOURCES FOR ORGANISMS FROM DIVERSE LINEAGES
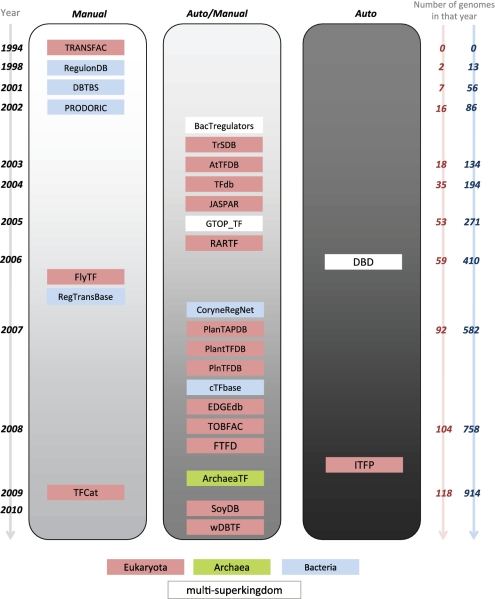
In this section, we survey the TF resources available to date and summarize them in Table 1. The resources are categorized according to the organisms for which the TF annotations are provided. We also illustrate the list of these resources on a timeline (Figure 1), which indicates the year in which different resources were first developed. The figure shows the trends in methods used to annotate TFs, with respect to the number of complete genomes available over the past 16 years according to the Genome OnLine Database (35).
Table 1.
TF resources currently available
| Database | Annotation | Organism | Link | Comment | |
|---|---|---|---|---|---|
| I | GTOP_TF | A/M | Prokaryotes | http://spock.genes.nig.ac.jp/∼gtop_tf/index2.html/ | Covers over 150 prokaryotic genomes |
| BacTregulators* | A/M | Prokaryotes | http://www.bactregulators.org/ | AraC-Xyls and TetR transcription regulator families. Last updated 2004 | |
| PRODORIC | M | Bacteria | http://prodoric.tu-bs.de | Contains protein–DNA interaction information | |
| RegTransBase | M | Bacteria | http://regtransbase.lbl.gov | Contains protein–DNA interaction information | |
| ArchaeaTF | A/M | Archaea | http://bioinformatics.zj.cn/archaeatf/ | Covers 37 archaeal genomes | |
| CoryneRegNet | A/M | Corynebacteria | http://www.coryneregnet.de/ | Contains protein–DNA interaction information | |
| cTFbase | A/M | Cyanobacteria | http://cegwz.com/ | Covers 21 cyanobacterial genomes | |
| DBTBS | M | Bacillus subtilis | http://dbtbs.hgc.jp/ | Contains other literature-curated information for B. subtilis | |
| RegulonDB | M | E. coli K-12 | http://regulondb.ccg.unam.mx/ | Contains other literature-curated information for E. coli K-12 | |
| II | TRANSFAC | M | Eukaryotes | http://www.gene-regulation.com/pub/databases.html/ | Partially commercial. Licence required to access some restricted areas |
| JASPAR | A/M | Eukaryotes | http://jaspar.cgb.ki.se/ | Contains collections of experimentally defined TF binding sites | |
| TrSDB* | A/M | Eukaryotes | http://bioinf.uab.es/cgi-bin/trsdb/trsdb.pl/ | Covers nine eukaryotic proteomes. Last updated 2004 | |
| ITFP | A | Mammals | http://itfp.biosino.org/itfp/ | Contains TFs and target genes from human, mouse and rat | |
| TFcat | M | Mammals | http://www.tfcat.ca/ | Contains manually curated TFs from human, mouse | |
| TFdb* | A/M | Mouse | http://genome.gsc.riken.jp/TFdb/ | Based on LocusLink and GO annotations. Last update: 2004 | |
| FTFD | A/M | Fungi | http://ftfd.snu.ac.kr/ | Covers 69 fungal and three oomycete genomes | |
| PlanTAPDB | A/M | Plants | http://www.cosmoss.org/bm/plantapdb/ | Contains taxonomic information of transcription associated protein families | |
| PlantTFDB | A/M | Plants | http://planttfdb.cbi.pku.edu.cn/ | Integrates other plant databases: DPTF (poplar), DRTF (rice), DATF (Arabidopsis) | |
| PlnTFDB | A/M | Plants | http://plntfdb.bio.uni-potsdam.de/v2.0/ | Covers five model plant genomes | |
| RARTF | A/M | A. thaliana (thale cress) | http://rarge.gsc.riken.jp/rartf/ | TF database devoted to A. thaliana | |
| AtTFDB* | A/M | A. thaliana | http://arabidopsis.med.ohio-state.edu/AtTFDB/ | Sister database, AtsicDB, contains cis-regulatory data. Last updated 2004 | |
| SoyDB | A/M | Glycine Max (soybean) | http://casp.rnet.missouri.edu/soydb/ | Predicts TFs using InterProScan | |
| wDBTF | A/M | Triticum aestivum T (wheat) | http://wwwappli.nantes.inra.fr:8180/wDBFT/ | Predicts TFs from wheat Expressed Sequence Tags (ESTs) and mRNA. | |
| TOBFAC | A/M | Nicotiana tabacum (Tobacco) | http://compsysbio.achs.virginia.edu/tobfac/ | Predicts TFs from tobacco gene-space sequence reads (GSRs) | |
| FlyTF | M | D. melanogaster (fruit fly) | http://flytf.org/ | TF database devoted to D. melanogaster | |
| EDGEdb | A/M | C. elegans (worm) | http://edgedb.umassmed.edu/ | Contains protein–DNA interaction information | |
| III | DBD | A | Cellular organisms | http://www.transcriptionfactor.org/ | Contains TF predictions of more than 1000 cellular organisms |
The databases can be divided into three categories: (I) prokaryotic TF databases; (II) Eukaryotic TF databases; (III) databases that provide TF annotations in genomes from different superkingdoms. The databases which have ceased to be developed or not been updated since 2004 are marked with asterisks. The years of the latest update are included in the comment field. Annotation methods are indicated as A (Automated) and M (Manually curated).
Figure 1.
Historical timeline of TF resources. The timeline to the left shows the years of the first publications describing the databases (not to scale). The panel on the right shows how the number of completely sequenced eukaryotic and bacterial genomes has increased according to the Genome OnLine Database (35). The TF resources are grouped according to their main annotation methods (manual curation, automatic plus manual curation or automatic). They are colored according to the organisms the resources annotate (blue for Bacteria, green for Archaea, red for Eukaryota and white if the resource covers two or three superkingdoms).
Prokaryotic TF resources
It has been thought that the level of transcription in prokaryotes is largely governed by the binding strengths of RNA polymerase and TFs to DNA (36). In addition, prokaryotic genomes are typically smaller, with fewer repeats and lower fractions of non-coding DNA, compared to eukaryotes. Consequently, the current knowledge on prokaryotic transcriptional mechanisms is believed to be more complete. Early established databases for prokaryotes serve as integrated resources of transcriptional machineries for specific model organisms, instead of focusing on TF annotation per se. RegulonDB (37) was one of the first TF databases to be established. The database provides high-quality information relating to the transcriptional regulatory network in the Gram-negative bacterium E. coli K12. Apart from the literature curated TFs, RegulonDB contains other molecular categories such as small RNAs and operon structures. Similarly, DBTBS (38) provides TFs as well as other transcriptional regulation-related knowledge such as recognition sequences and target genes for the Gram-positive bacterium Bacillus subtilis.
Due to the continuous development of sequencing technology, large number of completely sequenced genomes can be generated within shorter periods of time. This undoubtedly facilitates TF annotation and thus has resulted in a number of TF databases with larger and wider ranging prokaryotic taxonomic groups. In addition to RegulonDB and DBTBS, there are other resources, such as PRODORIC (39) and RegTransBase (40), that provide different aspects of data related to genetic regulation including lists of manually curated TFs and regulatory sites for multiple bacterial species. Other databases, such as the GTOP_TF database (19) provide a comparative insight into TF repertoires in >150 species from taxonomically diverse prokaryotic groups. Similarly, BacTregulators (41) is another resource providing TF collections for 123 genomes from archaea and bacteria, although the annotations are restricted to only two TF families: AraC-XylS and TetR. CoryneRegNet (42) integrates data on TFs and gene regulatory networks of eight corynebacteria, two mycobacteria and E. coli K12. cTFbase (43) is a database devoted to TF prediction in cyanobacteria, an ancient group of Gram-negative bacteria which reside in diverse environments. They obtain energy through photosynthesis and are believed to be the origin of eukaryotic chloroplasts according to the endosymbiotic theory (44). ArchaeaTF (45) is a unique database which is dedicated to archaea. Among the three main superkingdoms, Archaea are the least studied lineage. By providing TF prediction for 37 archaeal species, Archaea TF serves as an important platform for understanding the genetic regulation mechanisms in these life forms.
Eukaryotic TF resources
Transcription mechanisms in eukaryotes are known to be sophisticated but less well-understood, compared with the prokaryotes. Among the early eukaryotic TF resources is TRANSFAC (31). The database contains literature-curated information on sequence-specific TFs together with their binding sites, nucleotide distribution matrices as well as composite elements. While TRANSFAC is semi-commercial and some parts of the database require registration to access, JASPAR (46) is an open-access database which also mainly focuses on eukaryotic TF-binding sites. It also contains useful information on structural classes of TFs that share binding properties.
Owing to the continuous improvement of eukaryotic genome assembly and annotation, the number of databases containing TF annotation for eukaryotic species has increased rapidly in the past 5 years. A great deal of attention has been focused on plants in particular, possibly due to their importance as model organisms (e.g. Arabidopsis), food (e.g. rice, potato, wheat) and alternative energy sources (e.g. corn, sugarcane). Consequently, databases have been created to concentrate solely on TFs from groups of plant species: PlanTAPDB (47), PlantTFDB (48) and PlnTFDB (49). In addition, there are many databases devoted to single plant species including RARTF (50) and AtTFDB (51) for A. thaliana (thale cress), SoyDB (52) for Glycine max (soybean), wDBTF (53) for Triticum aestivum (wheat) and TOBFAC (54) for Nicotiana tabacum (tobacco).
The only resource focusing on TF annotation in fungi is FTFD (55). The database has been developed to provide TF predictions for >70 fungal species. For animals, the currently available databases contain TF collections for specific model animal species but not for larger taxonomic groups. These databases include FlyTF (56) which provides a manually curated list of D. melanogaster TFs. EDGEdb (57) contains protein–DNA interaction data in addition to a comprehensive collection of TFs in C. elegans. A number of databases which provide TF catalogues specifically for vertebrates include TFdb (58) (mouse), ITFP (59) (human, mouse and rat) and TFcat (60) (human and mouse).
Among the currently available TF resources, the DBD TF database (13,61) is one of the most comprehensive and unique TF collections in terms of number and variety of species available. The current version of the DBD database contains TF predictions for >1000 cellular organisms from the three major superkingdoms (Bacterial, Archaea and Eukaryota).
Although our survey is by no means exhaustive, it is the most comprehensive compilation of DNA-binding TF resources to date. Apart from the databases in Table 1, there are relevant databases that concentrate on TF-binding sites but also provide some TF predictions, e.g. MATCH (62) and MAPPER (63). In addition, we also noted that there are general resources for specific genomes that contain literature-curated gene annotations, as well as listings of known TFs such as FlyBase (33) and FlyMine (64), both for Drosophilids. Because these two groups of databases focus more on the TF-DNA interactions and particular genomes, respectively, and not specifically TF annotations, we did not include them in our table.
In our survey, we collected >25 TF databases that are available to date. Almost all databases concentrate on model organisms or specific taxonomic groups. Evidently, we still lack a universal platform that systematically integrates and validates the TFs annotated using criteria from a variety of resources.
VARIETY OF TF ANNOTATION METHODS FOR COMPILING TF CATALOGUES
A variety of TF identification methods have been used by different authors. The databases launched before the year 2000 rely on manual literature curation (Figure 1). These databases include RegulonDB (E. coli), DBTBS (B. subtilis) and TRANSFAC (eukaryotes). The other resources that also exclusively contain a compilation of manually curated TFs but were published more recently are PRODORIC and RegTransBase (both for bacteria), FlyTF (fruit fly) and TFCat (human and mouse).
Computational approaches became more popular after the year 2000 when a large number of fully sequenced genomes became available. This is because automated methods allow scientists to identify putative TF sets from completely sequenced genomes within a short period of time. Sequence-based pair-wise alignment applications such as BLAST (65) were widely used to detect homologues of known DBDs from numerous protein sequences. TFs can then be annotated based on the presence of DBDs in the protein chains. Due to its low sensitivity in remote homologue detection, the technique has become less popular and most of the second-generation databases relying on pair-wise comparisons have now ceased to be developed further (indicated by asterisks in Table 1, last update in 2004 or earlier).
In contrast, most regularly updated databases use profile-based searches such as HMM and PSI-BLAST (66) as opposed to conventional pair-wise alignments such as BLAST. The profile-based approaches have a number of advantages, including the superior sensitivity and specificity in recognizing remote homologues of sequence-specific DBDs (61). These third-generation TF databases include GTOP_TF, RATTF, DBD, PlanTAPDB, PlantTFDB, PlnTFDB, cTFbase and ArchaeaTF. The sources of reference query sets and refinement processes after the profile searches vary across the databases. Reference libraries can either be taken directly from existing projects including Pfam (20), SUPERFAMILY (67), PROSITE (68) or newly compiled from the literature and text mining. More recently, a support vector machine (SVM) was used in the ITFP database (59) instead of the profile-based searches for detecting DBDs. Apart from the methods involving sequence comparisons, structural alignment has also been introduced as an alternative for TF DBD classification (69).
Since most databases focus on a small number of organisms from specific taxonomic groups, manual refinements are plausible after putative TFs have been identified computationally. The automatic searches are often followed by literature curation and/or benchmarking against other databases, which can be performed manually or by automatic pair-wise sequence comparison. During this step, false negatives and true positives are removed and added, respectively, according to experimentally verified evidence. These additional curation processes generally refine the annotated TF collection and improve the specificity of the databases. The DBD database, on the contrary, is unique compared to other TF resources because it contains TF predictions for >1000 species, which is more than any other TF database to date. The post-automatic search curation is both time- and resource-inefficient so manual refinement is performed at the beginning of the process where the DBD HMM list is manually curated.
GENOMIC TF REPERTOIRES IN EUKARYOTES AND PROKARYOTES
The TF resources discussed in the previous sections not only serve as a starting point for experimental design of TF–DNA interaction studies in different model organisms (8–12), but can also provide an insight into the evolution of TFs in conjunction with the rest of the proteins in genomes that they regulate.
Cross-lineage analysis of TF repertoires based on the DBD database predictions
To demonstrate our current knowledge on the genomic repertoires of TFs across the tree of life, we extracted TF predictions for 683 non-redundant genomes (449 bacteria; 49 archaea; 185 eukaryotes) from the DBD database. To obtain the non-redundant genome set, we minimized the bias due to well-characterized species by manually excluding multiple strains of pathogenic bacteria and fungi, according to species name. These genomes are important from medical and agricultural points of view and thus have been intensively studied. Only the well-studied strains are included. For instance, E. coli K12 and Candida albicans SC5314 were used to represent E. coli and C. albicans, respectively. For eukaryotic genomes, only the longest transcript of each gene is included in this study. We noted that a survey on splice variants across multiple genomes is confounded by the heterogeneity of the data available for different organisms. For instance, mouse is extremely well-characterized, while chimpanzee is not. As a result, alternative splicing was excluded entirely from this study. This also allows the numbers of eukaryotic TFs to be compared with the bacterial TFs, which do not contain splice variants.
We chose TF predictions from the DBD database to illustrate the genome-wide TF repertoires in related and diverse species mainly because all the TF annotation in all the species was performed using a single platform. Although single-species databases such as RegulonDB (E. coli K12) and FlyTF (fruit fly) are more comprehensively and thoroughly curated, inconsistent criteria and methods used by different resources hinder an inter-species comparison. The DBD database is thus most suitable for a cross-lineage TF repertoire comparison. The initial benchmark of the DBD database against the proteins from PDB (70) and UniProt (71) classified by Gene Ontology (72) as TFs showed 95–99% accuracy and 66–67% coverage (61). More recent databases that benchmark their TF predictions against the DBD database include FlyTF, cTFbase and TFcat. Note that, in many cases, the annotation of TFs in one database relies on the annotated TFs from other databases as a starting point. Furthermore, many are based on similar sets of HMMs from Pfam or SUPERFAMILY. That is, TF annotations in different resources are not entirely independent. Consequently, the benchmarks and comparisons of TFs annotated by different databases need to be treated with care.
The TF prediction in the DBD database was performed based on the presence of DBDs (DBDs), scored by two HMM libraries: SUPERFAMILY (67) and Pfam (20). The DBD lists were manually curated and undergo occasional refinement. For SUPERFAMILY, the HMM models were designed to identify members of superfamilies, based on the domain definitions from the Structural Classification of Proteins (SCOP) (73). Since protein domain members in SCOP superfamilies tend to be functionally diverse, manual curation in the DBD database was done at the SCOP family level instead (61). Moreover, it has been shown that many SCOP families have homologous connections to Pfam families (74). For these reasons, we performed our analysis based on the Pfam and SCOP family DBDs. Although here we only discuss the number of domains and DBDs, assigned to proteins and TFs based on the presence of the Pfam HMMs, it is worth noting that the same observations also hold for SCOP families (see Supplementary Data for SCOP family results).
Table 2 describes the median values of various structural features of proteins and TFs in the three superkingdoms, predicted based on Pfam domain assignment to all proteins in each genome. We observed large variations among the eukaryotic species so we further divided them into three major kingdoms: Metazoa (animal), Fungi and Viridiplantae (plant) in Table 3. These results will be discussed in the next section. The means with standard deviations, SCOP family results and a table containing the numbers of TFs annotated and DBDs in the 683 genomes are available in Supplementary Data.
Table 2.
TF repertoires in the three main superkingdoms of life: Bacteria, Archaea, and Eukaryota
| Bacteria | Archaea | Eukaryota | Cellular organisms | |
|---|---|---|---|---|
| Proteins | ||||
| Proteins per species | 3140 | 1966 | 14 141 | 3885 |
| Length of all proteins (residues) | 322 | 289 | 465 | 328 |
| Domains assigned per protein | 1.41 | 1.30 | 1.53 | 1.42 |
| Distinct domain families per protein | 1.33 | 1.25 | 1.29 | 1.32 |
| Length of protein domains (residues) | 180 | 171 | 161 | 177 |
| TFs | ||||
| TFs per species | 131 | 60 | 325 | 155 |
| Distinct architectures per species | 39 | 19 | 45 | 39 |
| Length of TFs (residues) | 242 | 196 | 560 | 253 |
| DBDs per TF | 1.04 | 1.00 | 1.41 | 1.05 |
| Distinct DBD families per TF | 1.00 | 1.00 | 1.01 | 1.00 |
| Length of DBDs (residues) | 62 | 60 | 64 | 62 |
| Partner domains per TF | 0.58 | 0.25 | 0.24 | 0.49 |
| Distinct partner domain families per TF | 0.57 | 0.24 | 0.21 | 0.48 |
| Length of partner domains (residues) | 153 | 97 | 85 | 139 |
| TF content in genome (%) | 4.39 | 2.94 | 2.91 | 3.59 |
| DBDs | ||||
| DBD families | 61 | 15 | 77 | 131 |
| Superkingdom-specific DBD families | 43 | 0 | 69 | |
| Partner domain families | 228 | 55 | 795 | 938 |
| Superkingdom-specific partner domain families | 116 | 12 | 693 | |
| Distinct domain architectures | 605 | 118 | 2209 | 2779 |
| DBDs per species | 109 | 42 | 206 | 122 |
| Distinct DBD families per species | 23 | 12 | 27 | 24 |
Domain assignments are from Pfam. Median values of all species in each lineage are displayed. Mean values and their SDs for each property are described in ‘Supplementary Data’.
Table 3.
TF repertoires in three major eukaryotic kingdoms: Viridiplantae (plants), Fungi, and Metazoa (animals), plus all eukaryotes combined
| Viridiplantae | Fungi | Metazoa | Eukaryota | |
|---|---|---|---|---|
| Proteins | ||||
| Proteins per species | 27 235 | 9997 | 16 371 | 14 141 |
| Length of all proteins (residues) | 387 | 466 | 479 | 465 |
| Domains assigned per protein | 1.48 | 1.47 | 2.00 | 1.53 |
| Distinct domain families per protein | 1.24 | 1.29 | 1.37 | 1.29 |
| Length of protein domains (residues) | 158 | 185 | 150 | 161 |
| TFs | ||||
| TFs per species | 591 | 203 | 806 | 325 |
| Distinct architectures per species | 77 | 38 | 160.5 | 45 |
| Length of TFs (residues) | 375 | 604 | 545 | 560 |
| DBDs per TF | 1.13 | 1.36 | 2.75 | 1.41 |
| Distinct DBD families per TF | 1.01 | 1.00 | 1.03 | 1.01 |
| Length of DBDs (residues) | 73 | 65 | 56 | 64 |
| Partner domains per TF | 0.23 | 0.14 | 0.40 | 0.24 |
| Distinct partner domain families per TF | 0.20 | 0.13 | 0.35 | 0.21 |
| Length of partner domains (residues) | 90 | 83 | 85 | 85 |
| TF content in genome (%) | 2.12 | 2.53 | 4.65 | 2.91 |
| DBDs | ||||
| DBD families | 38 | 34 | 58 | 78 |
| Kingdom specific DBD families | 12 | 6 | 26 | |
| DBDs per species | 602 | 152 | 2151 | 206 |
| Distinct DBD families per species | 37 | 25 | 53 | 27 |
The domain assignments are Pfam families. Median values of all species in each lineage are displayed.
TF and DBD repertoires in the major superkingdoms of life
According to Table 2, eukaryotic species have much larger protein repertoires that contain longer average peptide sequences than bacteria, the superkingdom that dominates the prokaryotic group (medians of 465 and 322 residues, respectively). Note that the P-values (P), calculated using a non-parametric test (Mann–Whitney), are <10−15 for all comparisons discussed here unless specified otherwise. The longer eukaryotic proteins also contain more domains per protein chain than bacteria (1.53 versus 1.41 domains per protein). This might allow protein sequences to possess more functionality such as enzymatic properties, DNA binding, as well as binding to other proteins.
The average length of TFs is greater than the average length of all proteins in most eukaryotic species (Table 2). Within the Eukaryota superkingdom, the average length of TFs versus all proteins is 545 versus 479 residues in animals (P = 10−6), and 604 versus 466 in fungi. Interestingly, the length of TFs in plants are not significantly different from other proteins on average, 375 versus 387 (P = 0.4) (Table 3). In contrast, TFs in bacteria are significantly shorter than their average proteins. One possible explanation for longer TFs compared to all proteins in animals and fungi is a high fraction of intrinsically disordered (ID) regions, which are absent in bacterial TFs (75,76). These ID segments in proteins are naturally unfolded and unstructured but may serve as flexible linkers that aid protein interactivity (77). Through promoting protein–protein interaction, these long-ID regions aid formation of composite regulatory protein elements in eukaryotes.
A single eukaryotic TF typically contains 1.41 DBDs on average but these DBDs only belong to ∼1 distinct DBD family per TF. This suggests that many eukaryotic TFs include >1 repeated DBD from the same family. Among the eukaryotic species, the number of DBD repeats is greatest in the animals where DBDs from the same family can occur almost three times on average (2.75 DBDs, median) in a single TF chain; while fungi (1.36) and plants (1.13) possess significantly fewer DBD repeats per TF. Zinc fingers are among the DBD families that occur multiple times within a single polypeptide. By increasing the number of DBD repeats, eukaryotes can boost the length of recognition sequence and thus overcome the DNA-binding site length limitation of a single DBD (78). On the other hand, a single DBD seems sufficient for most prokaryotic TFs to recognize their binding elements. The only exception is the HTH_AraC family (arabinose operon regulatory), a bacterial DBD that occurs more than once in the same TF chain.
In both eukaryotic and prokaryotic groups, the number of distinct DBD families per TF is close to 1.00. Indeed the combination of DBDs from more than one family in the same TF chain is extremely rare and restricted to certain phylogenetic groups. For instance, we observed two bacterial DBD families, HTH_AraC and AraC_N, that occur in the same TF but this combination is restricted to proteobacteria. In animals, the HLH DBD mainly appears in single domain TFs but can also combine with two other DBD families, Myc_N and Basic, to form a TF. These are examples of only eight combinations in total (five in eukaryotes and three in bacteria) of multiple DBD families occurring in the same TF chain (79). One possible advantage of having few TFs with multiple DBD families is the minimization of crosstalk between two or more distinct DBDs on the same TF, and a large number of possible binding sites of different DBD families on DNA.
Apart from DBDs, TFs may contain non-DBD domains of different functions, which we hereby call ‘partner domains’. In both prokaryotes and eukaryotes, repeats of the same partner domains within one TF chain are rare. The average number of total domains (DBDs plus partner domains) in TFs is greater than in other proteins on average: 1.65 versus 1.53 for eukaryotes (P = 0.002), and 1.62 versus 1.41 for bacteria. However, only in eukaryotes is the number of distinct families per protein fewer in TFs than in other proteins, i.e. 1.22 versus 1.29 (P = 10−6), suggesting a higher rate of domain repeats in TFs than in eukaryotic proteins in general.
One might think that the greater number of domains in eukaryotic TFs could be a probable reason that explains why they are longer in sequence than non-TF proteins. Nonetheless, we have shown that this is not the case because bacterial TFs also contain more domains than other proteins on average, but are shorter in length. An alternative explanation would be that the protein domains present in TFs (DBDs and partner domains) are longer than other domains found in non-TF proteins. Interestingly, we demonstrate here, for the first time, that the average length of DBDs does not vary by much across different superkingdoms (60–64 amino acid residues, medians). Furthermore, they are significantly shorter than other protein domain families. Thus, the average length of domains, as well as their number of occurrence, cannot explain the longer eukaryotic TFs compared to other proteins. Instead, this is more likely to be due to long stretches of intrinsically disordered regions detected in eukaryotic TFs but not in bacteria as we mentioned earlier.
The average fraction of TFs in genomes (TF content) is highest in animal groups where ∼4.7% of proteins are TFs. Fungi and plant genomes possess significantly smaller TF contents of between 2–2.5%, which surprisingly are less than the average TF fraction in bacteria of 4.2%. This is because the DBD repertoires in different eukaryotic kingdoms are highly lineage-specific (79), while plant and fungal TF repertoires are less well-characterized than animals. The difference within the animal kingdom is most apparent between vertebrates (dominated by Chordata) and invertebrates (dominated by Arthropoda). The average number of proteins and TFs per species, as well as TF contents are significantly greater in Chordata than in Arthropoda. This could be a result of whole genome duplication events, a greater rate of segmental duplication in vertebrates, or simply due to better characterized TF catalogues in Chordata.
Unicellular obligate parasites such as apicomplexa and euglenozoa, e.g. Plasmodium falciparum (malaria apicomplexan) and Trypanosoma brucei (sleeping sickness euglenozoa), contain surprisingly small TF fractions of their genomes. Their entire protein repertoires typically contain only 0.5% TFs. To illustrate the point, as many as 6% of human proteins are classified as TFs, as opposed to only 0.3% in P. falciparum. These parasitic organisms have different lifestyles from the other eukaryotes considered here, as they only survive or replicate in a relatively stable environment inside their hosts. The low fraction of predicted TFs in these life forms is likely to be due to their reduced number of proteins and regulatory components, as well as their less well-characterized TF repertoire (29,80).
Bacteria make use of 61 Pfam DBDs in total, which combine with 228 partner domains and give rise to 605 distinct domain architectures (Table 2). When considering them individually, each bacterial organism possesses 131 TFs on average but only 39 distinct architectures (all averages are medians). This corresponds to the previous finding that the majority of bacterial TFs have arisen through gene duplication events (18,81). Although there are fewer complete genomes available, the eukaryotic superkingdom possesses a large DBD repertoire of 77 distinct families. They combine with 795 partner domain families and form 2209 distinct domain architectures. Besides the larger DBD repertoire, a greater number of partner domains utilized by eukaryotes also plays a part in creating more diverse architectures.
Within the eukaryotic genomes, the Metazoa kingdom possess a considerably larger DBD repertoire than the Fungi and Viridiplantae kingdoms (Table 3). This reflects the greater morphological complexity and number of body structures in animals, as well as a potential bias towards the study of animal model organisms. On average, eukaryotic species possess 325 TFs per genome but make use of only 45 distinct arrangements (medians). This suggests that a large fraction of eukaryotic TFs also emerged through gene duplication (82), possibly even at a higher rate than in bacteria. The result is in accordance with previous work suggesting that as many as 90% of eukaryotic genes have arisen by duplication (83).
The wealth of completely sequenced genomes and automatic TF annotation allow us to predict the TF sets from entire genomes and analyze at a global level the genomic TF repertoires in species from diverse phylogenetic groups. We observe distinct features of TFs and DBDs such as TF length, TF content in genome, number of DBD families and number of DBD repeats per TF in different lineages. During the course of evolution, as genomes expand via gene duplication, a greater number of TFs are required to orchestrate the expression of these expanded genes. In the next section, we will investigate whether or not the TF expansion with respect to the total number of genes is also lineage-specific.
TF ABUNDANCE FOLLOWS A POWER LAW INCREASE WITH NUMBER OF GENES
As morphological complexity increases, organisms require a greater proportion of TFs for gene expression control. As well as TFs, cell adhesion molecules and proteins involved in extra-cellular processes have been shown to be greatly expanded in animals (84,85). A power law increase in TF numbers with gene numbers has previously been observed in several bacterial genomes and a very limited number of eukaryotic genomes (15,86–90). In accordance with these previous studies, we not only confirm a linear trend of TF abundance with the number of genes on the log–log scale using the TFs obtained from the DBD database for a large set of bacteria (449 genomes), but also extend this analysis to eukaryotes (185 genomes). This implies a power law relationship between the two variables in both prokaryotic and eukaryotic genomes.
Power law relationship between TFs and number of genes
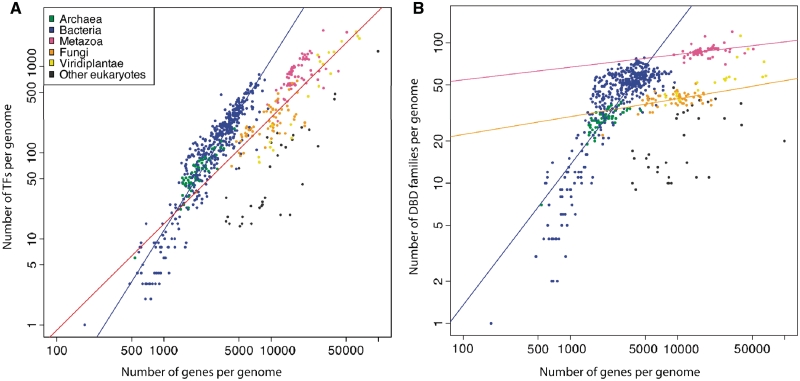
In bacteria, as the number of genes becomes larger, the TF expansion strictly follows a power law increase with an exponent close to 2, which infers a quadratic increase (power law exponent of 1.98, coefficient of determination, R2 of 0.87, Figure 2A). It is worth noting that sigma factors and other non-sequence-specific TFs were not included in this dataset. We observe similar exponents when the numbers of TFs from different bacterial phyla are correlated separately with their number of genes (Supplementary Figure S1). A similar exponential TF expansion can also be seen in eukaryotic genomes but with a slower increase (power law exponent of 1.23) and less fitting quality (lower R2 of 0.61). The exponents >1 observed in both cell types mean that the TF repertoire expands faster than linearly for every gene added to the genome.
Figure 2.
(A) TF abundance against number of genes per genome in different lineages across the tree of life. Each colored dot represents a genome. Different colors are used to highlight genomes from different phylogenetic groups. According to the linear model fit on a log–log scale, TF expansion in bacteria strictly follows a power law increase, with an exponent close to quadratic (logT = 1.98logG – 4.84 with R2 = 0.87 where T is number of predicted TFs, G is number of genes and R2 is coefficient of determination). The TF increase in eukaryotes has a lower exponent as well as degree of correlation (logT = 1.23logG – 2.53 with R2 = 0.61). (B) The number of unique DBD families increases linearly with the total number of proteins in bacteria (power law exponent = 1.00, R2 = 0.71). In contrast, the number of families is independent of the number of genes in metazoans (pink, exponent = 0.09, R2 = 0.11) and fungi (orange, exponent = 0.13, R2 = 0.23). Grey dots in the figures represent other eukaryotic species that do not belong to the main kingdoms such as apicomplexan and euglenozoa.
Two possible implications of this power law relationship were proposed separately in the context of metabolic networks (89) and microeconomics (88). From the metabolic network point of view, when organisms evolve to explore a new environment, a new set of TFs are required to monitor new tasks necessary to adapt to different conditions. On the other hand, some of the metabolic enzymes can be reused and fewer new ones are required to regulate each new task. This may explain why the number of new tasks and their regulators increase faster than linearly with the number of genes encoding enzymes (89).
The necessity of a sharper TF increase with number of genes in bacteria might be linked to the absence of a nucleus and other eukaryote-specific transcriptional mechanisms that might hinder the organisms from having a larger genome. This observation corresponds with a previous study (88) where a microeconomic model was used to speculate that bacteria already have a maximal number of genes, given their transcriptional mechanisms. A further increase in number of genes would be ‘economically’ ineffective since the average cost to regulate a gene becomes prohibitively expensive.
Eukaryotes employ more complex mechanisms for gene expression control compared to bacterial systems that may partly explain the slower TF increase with the number of genes observed. For instance, the high degree of combinatorial regulation in eukaryotes (91) means that TFs are involved in many different multi-protein transcription complexes. The greater fraction of non-coding DNA in the eukaryotic genomes has an important role in producing small RNAs that provide an additional layer of gene regulation. This large amount of non-coding DNA also harbours cis-regulatory sequences with more complex-binding site architecture than in prokaryotes (92–94). Eukaryotic DNA is packaged into chromatin repressed in the transcriptional ground state and the promoter is only accessible in the presence of chromatin remodelling proteins (2). This system also acts as an extra switch for expression control. Although some bacterial chromosome packaging has been observed (95), the system is less well-characterized. Tissue-specific regulatory circuits are another way multi-cellular eukaryotes utilize the same transcriptional associated elements to temporally and spatially control gene expression (96,97). The existence of splice variants in eukaryotes is another possible explanation for the slower TF increase (98). This is, however, not considered here as only the longest transcript per gene is included, due to the heterogeneity in splice variant datasets as discussed earlier. These machineries unique to eukaryotes together enhance genetic regulation beyond the context of TFs and their target genes.
All eukaryotic obligate parasites have less than 50 TFs (grey dots below the red line in Figure 2A). They are known to be divergent in sequence as well as structure from other eukaryotes (29,80) and less well-characterized. Even when such parasitic species were excluded from the model fitting, the degree of correlation of the linear model in eukaryotes is drastically weaker than in bacteria. The poor correlation could be due to a greater organismal complexity in multi-cellular eukaryotes, which cannot be captured by the total number of proteins alone.
Slow increase in the number of DBD families reveals TF evolution via gene duplication
In addition to the total number of TFs per genome, in Figure 2B we illustrate the repertoires of distinct DBD families in each species. Bacterial organisms with larger numbers of genes contain more distinct DBD families. As opposed to the quadratic increase in the total number of TFs with respect to number of genes (power law exponent of 1.98), the increase in number of distinct DBD families recruited by larger prokaryotic organisms is close to linear (power law exponent of 1.00, R2 = 0.71). From this finding, we can infer that the number of TFs per DBD family gradually increases as the total number of genes grows larger. Most likely the TFs belonging to the same DBD family have arisen through multiple gene duplication events followed by a series of protein sequence divergence and domain re-combination events. This has also been shown previously (18,81).
In contrast to bacteria, we did not observe any clear correlation between the number of distinct DBD families and the number of genes per genome in eukaryotes. When we performed a linear regression separately for different eukaryotic kingdoms, we found that the number of unique eukaryotic DBD families per species is relatively conserved in all animals, regardless of the number of genes, and the same is true for fungi (power law exponents are 0.09 and 0.13 for animals and fungi, respectively). This suggests there might be a minimal requirement of DBD diversity in animals, and similarly in fungi. Evidently, there seem to be at least two major bursts of DBD family expansion: the first when eukaryotes branched off from prokaryotes and the second at the common ancestral node of animals and fungi. This gives rise to the unique set of DBD repertoires in the eukaryotic kingdoms, which reflects the organismal complexity and morphological diversity of the two lineages. As discussed previously, some eukaryotic DBD families can occur repeatedly within the same TF chain. This may also allow eukaryotes to boost the length and diversity of DNA-recognition sequence without recruiting additional DBD families.
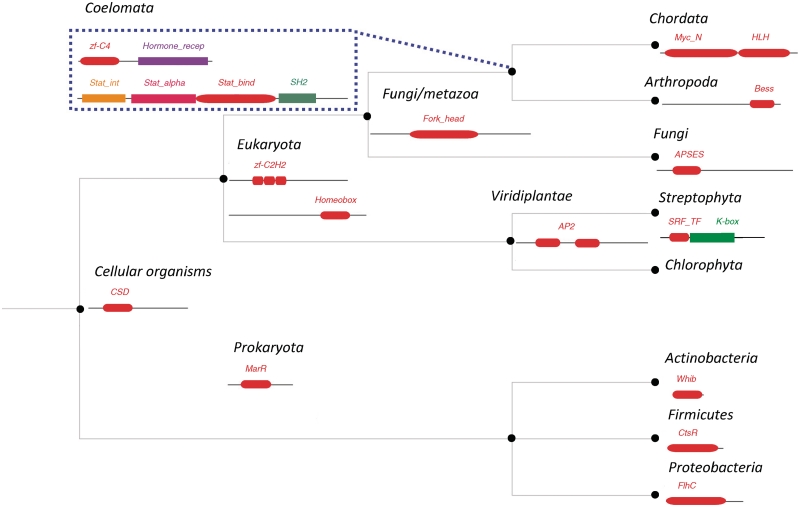
Apart from the lineage-specific increase in TFs with the total number of genes, as well as the distinct structural features of TFs and DBDs in different lineages we discussed in the previous section, we also noted lineage-specific presence and absence of DBD families, and their domain combinations with partner domains. We discuss this analysis in greater detail in a separate article (79). Examples of the most frequently observed lineage-specific Pfam DBD families and their architectures in TFs are shown in Figure 3.
Figure 3.
Examples of lineage-specific DBDs and domain architectures of TFs across the tree of life. Commonly found DBDs and TF architectures in different taxonomic species are projected onto the simplified NCBI taxonomic tree. DBDs and their architectures in TFs at different taxonomic nodes are unique to their descendent branches. DBDs are represented by red oblongs, and other protein domains occurring within the same TFs (partner domains) are represented by colored rectangles.
CONCLUSIONS AND PERSPECTIVE
Sequence-specific TFs are a vital class of proteins because they directly bind to DNA and thus regulate differential expression of genes. Importantly, they determine physiological diversity of organisms in different lineages across the tree of life (99). We survey and summarize TF resources currently available, as well as discuss the criteria and methods used to annotate TFs by different authors. Comprehensive and high quality TF catalogues serve as a starting point for the experimental design of high-throughput studies on TF–DNA interactions (8–12), as well as being a platform for protein engineering such as in the emerging field of synthetic biology, e.g. engineered zinc finger nucleases (100,101).
As we are moving into the post-genomic era, computational tools have been employed more often to help TF prediction for a large number of completely sequenced genomes. Exclusive literature curation of entire genomes is still available for a small number of model organisms, e.g. FlyTF for fly, and TFcat for human and mouse. Over 25 databases providing genomic TF catalogues have become available over the past 15 years, however, most of them focus on a small number of model organisms or specific taxonomic groups. A universal platform that systematically integrates and validates TFs annotated using different criteria from different TF databases would be of great benefit to the community, in a similar way to InterPro (32) that integrates protein families, domains and functional sites from other protein databases such as Pfam (20), SUPERFAMILY (67) and PROSITE (68).
We used TF annotations obtained from the DBD database to demonstrate the current knowledge on the global TF repertoire in ∼700 genomes across the tree of life. TF catalogues of different species in the DBD database are automatically annotated based on consistent criteria and this eliminates the biases due to different methods of TF annotation. Although the biases due to different levels of knowledge of TF repertoires in diverse lineages remain (model organisms are better studied), the datasets we use can serve as representative examples for summarizing the community’s current understanding of the genomic TF abundance and structural complexity.
We observe several features of TF families and their protein domain architectures unique to specific lineages, most apparently between prokaryotic and eukaryotic genomes. Firstly, the eukaryotic TFs are significantly longer than eukaryotic proteins of other functions while this relationship is reversed in prokaryotes. This could be due to the presence of long intrinsic disordered segments in eukaryotic TFs that are required to leverage the formation of multi-protein transcription complexes (76). Second, repeats of the same DBD family in one polypeptide chain are common only in eukaryotes. This has been suggested as one mechanism used by eukaryotes to increase the length and diversity of DNA-binding recognition sequence from a limited number of DBD families (78). It also potentially explains why the number of unique DBD families keeps increasing when the total number of prokaryotic genes grows larger, while the abundance of DBD family repertoires seem to be relatively conserved in animals and fungi.
We not only confirmed the quadratic increase in TFs with the number of genes in prokaryotes observed by previous studies (92–94), but also extended the model fitting to a large group of eukaryotic species. We observe a similar exponential TF expansion in eukaryotic genomes but with a lower exponent and fitting quality than bacteria. We speculate that this may be due to the complex mechanisms for gene expression control utilized only by eukaryotes such as a greater fraction of regulatory non-coding DNA, combinatorial regulation of multiple TFs and chromatin repressed transcriptional ground state.
We demonstrate the lineage-specific structural features, distinct rates of increase with respect to the total number of genes of TFs and DBD families in different lineages. In addition, we observe distinct patterns of DBD family expansion and their domain combinations with partner domains in diverse phylogenetic groups. The lineage-specific characteristic of DBD families and TF architectures can be used as signatures for the genetic regulatory circuits, which can improve methods for remote homology detection and thus the discovery of new TFs in genomes. Coin and coworkers (102,103) have shown that techniques along these lines can be used to enhance protein domain discovery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council, and a Royal Thai Government Scholarship (to V.C.). Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Daniel Hebenstreit, Joseph Marsh, Anuphon Laohavisit, as well as anonymous referees for critical commentary on the manuscript.
REFERENCES
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 3.Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. doi: 10.1016/s0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 4.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 7.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallikas O, Taipale J. High-throughput assay for determining specificity and affinity of protein-DNA binding interactions. Nat. Protoc. 2006;1:215–222. doi: 10.1038/nprot.2006.33. [DOI] [PubMed] [Google Scholar]
- 10.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 11.Meng X, Wolfe SA. Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system. Nat. Protoc. 2006;1:30–45. doi: 10.1038/nprot.2006.6. [DOI] [PubMed] [Google Scholar]
- 12.Deplancke B, Dupuy D, Vidal M, Walhout AJ. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD–taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulson RM, Touboul N, Ouzounis CA. Lineage-specific partitions in archaeal transcription. Archaea. 2007;2:117–125. doi: 10.1155/2006/629868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Rueda E, Collado-Vides J, Segovia L. Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput. Biol. Chem. 2004;28:341–350. doi: 10.1016/j.compbiolchem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Minezaki Y, Homma K, Nishikawa K. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res. 2005;12:269–280. doi: 10.1093/dnares/dsi016. [DOI] [PubMed] [Google Scholar]
- 20.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Campuzano S, Janga SC, Perez-Rueda E. Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes–a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brune I, Brinkrolf K, Kalinowski J, Puhler A, Tauch A. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics. 2005;6:86. doi: 10.1186/1471-2164-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 24.Stein L. Genome annotation: from sequence to biology. Nat. Rev. Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 25.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 26.Coulson RM, Ouzounis CA. The phylogenetic diversity of eukaryotic transcription. Nucleic Acids Res. 2003;31:653–660. doi: 10.1093/nar/gkg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiu SH, Shih MC, Li WH. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 2005;139:18–26. doi: 10.1104/pp.105.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelest E. Transcription factors in fungi. FEMS Microbiol Lett. 2008;286:145–151. doi: 10.1111/j.1574-6968.2008.01293.x. [DOI] [PubMed] [Google Scholar]
- 29.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Messina DN, Glasscock J, Gish W, Lovett M. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 2004;14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 35.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grote A, Klein J, Retter I, Haddad I, Behling S, Bunk B, Biegler I, Yarmolinetz S, Jahn D, Munch R. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res. 2009;37:D61–D65. doi: 10.1093/nar/gkn837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase–a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Bueno M, Molina-Henares AJ, Pareja E, Ramos JL, Tobes R. BacTregulators: a database of transcriptional regulators in bacteria and archaea. Bioinformatics. 2004;20:2787–2791. doi: 10.1093/bioinformatics/bth330. [DOI] [PubMed] [Google Scholar]
- 42.Baumbach J. CoryneRegNet 4.0 - A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Zhao F, Wang S, Deng G, Wang J, Bai J, Lu J, Qu J, Bao Q. cTFbase: a database for comparative genomics of transcription factors in cyanobacteria. BMC Genomics. 2007;8:104. doi: 10.1186/1471-2164-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raven JA, Allen JF. Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol. 2003;4:209. doi: 10.1186/gb-2003-4-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J, Wang S, Bai J, Shi L, Li D, Xu Z, Niu Y, Lu J, Bao Q. ArchaeaTF: an integrated database of putative transcription factors in Archaea. Genomics. 2008;91:102–107. doi: 10.1016/j.ygeno.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardt S, Lang D, Reski R, Frank W, Rensing SA. PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiol. 2007;143:1452–1466. doi: 10.1104/pp.107.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005;12:247–256. doi: 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- 51.Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Libault M, Joshi T, Valliyodan B, Nguyen HT, Xu D, Stacey G, Cheng J. SoyDB: a knowledge database of soybean transcription factors. BMC Plant Biol. 2010;10:14. doi: 10.1186/1471-2229-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romeuf I, Tessier D, Dardevet M, Branlard G, Charmet G, Ravel C. wDBTF: an integrated database resource for studying wheat transcription factor families. BMC Genomics. 2010;11:185. doi: 10.1186/1471-2164-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rushton PJ, Bokowiec MT, Laudeman TW, Brannock JF, Chen X, Timko MP. TOBFAC: the database of tobacco transcription factors. BMC Bioinformatics. 2008;9:53. doi: 10.1186/1471-2105-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Park B, Jung K, et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24:1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 56.Pfreundt U, James DP, Tweedie S, Wilson D, Teichmann SA, Adryan B. FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 38:D443–D447. doi: 10.1093/nar/gkp910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrasa MI, Vaglio P, Cavasino F, Jacotot L, Walhout AJ. EDGEdb: a transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanamori M, Konno H, Osato N, Kawai J, Hayashizaki Y, Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem. Biophys. Res. Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 59.Zheng G, Tu K, Yang Q, Xiong Y, Wei C, Xie L, Zhu Y, Li Y. ITFP: an integrated platform of mammalian transcription factors. Bioinformatics. 2008;24:2416–2417. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 60.Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY–sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponomarenko JV, Bourne PE, Shindyalov IN. Building an automated classification of DNA-binding protein domains. Bioinformatics. 2002;18(Suppl. 2):S192–S201. doi: 10.1093/bioinformatics/18.suppl_2.s192. [DOI] [PubMed] [Google Scholar]
- 70.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 71.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Chandonia JM, Ding C, Holbrook SR. Comparative mapping of sequence-based and structure-based protein domains. BMC Bioinformatics. 2005;6:77. doi: 10.1186/1471-2105-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. J. Mol. Biol. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 76.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 78.Itzkovitz S, Tlusty T, Alon U. Coding limits on the number of transcription factors. BMC Genomics. 2006;7:239. doi: 10.1186/1471-2164-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Charoensawan V., Wilson D., Teichmann S.A. Lineage-specific expansion of DNA-binding transcription factor families. Trends in Genetics. 2010;26:388–393. doi: 10.1016/j.tig.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madan Babu M, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 84.Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tordai H, Nagy A, Farkas K, Banyai L, Patthy L. Modules, multidomain proteins and organismic complexity. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 86.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 87.Ranea JA, Buchan DW, Thornton JM, Orengo CA. Evolution of protein superfamilies and bacterial genome size. J. Mol. Biol. 2004;336:871–887. doi: 10.1016/j.jmb.2003.12.044. [DOI] [PubMed] [Google Scholar]
- 88.Ranea JA, Grant A, Thornton JM, Orengo CA. Microeconomic principles explain an optimal genome size in bacteria. Trends Genet. 2005;21:21–25. doi: 10.1016/j.tig.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 89.Maslov S, Krishna S, Pang TY, Sneppen K. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc. Natl Acad. Sci. USA. 2009;106:9743–9748. doi: 10.1073/pnas.0903206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cases I, de Lorenzo V, Ouzounis CA. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003;11:248–253. doi: 10.1016/s0966-842x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- 91.Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J. Mol. Biol. 2006;360:213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 92.Mattick JS. A new paradigm for developmental biology. J. Exp. Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 93.Ahnert SE, Fink TM, Zinovyev A. How much non-coding DNA do eukaryotes require? J. Theor. Biol. 2008;252:587–592. doi: 10.1016/j.jtbi.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 94.Harafuji N, Keys DN, Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl Acad. Sci. USA. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Travers A, Muskhelishvili G. Bacterial chromatin. Curr. Opin. Genet. Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 96.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- 98.Taneri B, Snyder B, Novoradovsky A, Gaasterland T. Alternative splicing of mouse transcription factors affects their DNA-binding domain architecture and is tissue specific. Genome Biol. 2004;5:R75. doi: 10.1186/gb-2004-5-10-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McClintock B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 100.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug. Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 101.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coin L, Bateman A, Durbin R. Enhanced protein domain discovery by using language modeling techniques from speech recognition. Proc. Natl Acad. Sci. USA. 2003;100:4516–4520. doi: 10.1073/pnas.0737502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coin L., Bateman A., Durbin R. Enhanced protein domain discovery using taxonomy. BMC Bioinformatics. 2004;5:56. doi: 10.1186/1471-2105-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.