Abstract
The post-genomic era presents many new challenges for the field of bioinformatics. Novel computational approaches are now being developed to handle the large, complex and noisy datasets produced by high throughput technologies. Objective evaluation of these methods is essential (i) to assure high quality, (ii) to identify strong and weak points of the algorithms, (iii) to measure the improvements introduced by new methods and (iv) to enable non-specialists to choose an appropriate tool. Here, we discuss the development of formal benchmarks, designed to represent the current problems encountered in the bioinformatics field. We consider several criteria for building good benchmarks and the advantages to be gained when they are used intelligently. To illustrate these principles, we present a more detailed discussion of benchmarks for multiple alignments of protein sequences. As in many other domains, significant progress has been achieved in the multiple alignment field and the datasets have become progressively more challenging as the existing algorithms have evolved. Finally, we propose directions for future developments that will ensure that the bioinformatics benchmarks correspond to the challenges posed by the high throughput data.
INTRODUCTION
Bioinformatics challenges in the high throughput era
Today, high throughput technologies are transforming the biology data landscape. The 1000 Genomes Project (www.1000genomes.org) and the next generation of deep sequencing platforms (1) are providing unprecedented amounts of DNA sequence data. Other large-scale data resources are also emerging from high-throughput experimental technologies, such as gene expression data sets, protein 3D structures, protein–protein interactions, etc. But, the scale of the data is not the only issue. The quality of the high throughput data is also notoriously variable, with relatively high error rates and low reproducibility. This flood of complex, heterogeneous and inherently noisy data has stimulated the development of novel algorithms and software analysis tools to organize and explore the raw data and to extract the hidden knowledge. Applications include the more traditional tasks, such as identifying genes in DNA sequences and determining the 3D structure of proteins, as well as new subfields of computational biology such as expression data analysis, metabolic and regulatory network analysis, proteomics, functional analysis of regulatory non-coding RNAs, etc. In all these domains, new computational methodologies are being developed based on statistical analyses, learning and data mining algorithms, mathematical modelling and simulation.
The wide variety of the analysis tools available today means that it is often difficult for the non-specialist to choose an appropriate tool for his specific problem. Comparative evaluation of the different methods has become a crucial task, in order to select the most suitable method for a particular problem (e.g. more efficient, more correct, more scalable), to evaluate the improvements obtained when new methods are introduced, and to identify the strong and weak points of the different algorithms. The goal of this review is to discuss general good practices in benchmarking development and the performance of objective comparative studies. We briefly mention some examples of benchmarking studies that illustrate the widespread applications of benchmarking. We then study benchmarking in the field of protein sequence alignment in more detail. Finally, we discuss a number of important considerations that will need to be addressed in the future.
Benchmarking and objective, comparative studies
In computer science, comparative evaluations of computer systems, compilers, databases and many other technologies are performed using standard benchmarks. One of the simplest experiments involves running specific programs, e.g. the Standard Performance Evaluation Corporation CPU2000 suite (www.spec.org), on different computer systems, in order to compare the time required to complete the test. The use of a standard benchmark reduces variability in the test results and allows an objective, quantitative comparison of the different tools and techniques. Another advantage of benchmarking is that replication is built into the method. Since the materials are designed to be used by different laboratories, the evaluation can be performed repeatedly on various tools and techniques. In addition, many benchmarks can be automated so that the computer does the work of executing the tests, gathering the data, and producing the performance measures. Benchmarking can also be used to compare the accuracy of the results obtained from alternative software tools or approaches. Here, the benchmark represents a tool that is used to measure the ability of a software system to meet the requirements of the user. In this case, the benchmark requires two components: (i) the tests designed to compare the qualities of the alternative software tools and (ii) some means of evaluating the fitness for purpose. The evaluation of a given tool can be based on independent fitness scores; more often, though, the output of the program is compared to the ‘correct’ solution, known as the gold standard, specified by the benchmark.
Benefits for the user community
Within a scientific discipline, a benchmark captures the community consensus on which problems are worthy of study, and determines what are scientifically acceptable solutions. For example, standard datasets, such as the Caltech series (http://www.vision.caltech.edu/Image_Datasets), have been widely applied in the field of image recognition and classification. Caltech contains a large number of images, divided into a number of distinct object categories (faces, watches, ants, pianos, etc.), handpicked by the authors to represent a wide variety of natural and artificial objects. The benchmark provides three different measures: (i) the accuracy with which objects are identified, (ii) the ability to differentiate between the objects of interest and uninteresting background, (iii) the level of differentiation between similar objects. In practice, the level of differentiation depends on the needs of the user, ranging from fine discrimination, e.g. the exact identification of a particular species of mammal, to a more inclusive classifier that would be better at discriminating between mammals and other animals. Since their introduction, these datasets have become progressively more challenging as existing algorithms consistently saturated performance. The latest version, Caltech-256, contains over 30 000 images in 256 object categories and is designed to challenge today’s state of the art classification algorithms.
Benefits for the developer community
The benchmarking process is also beneficial for the software developers. A benchmark is usually formal, in the sense that it is created intentionally and applied with a specific intent. During deployment, the results from different technologies are compared, which requires researchers to look at each other’s contributions. Thus, researchers become more aware of one another’s work and ties between researchers with similar interests are strengthened. The resulting evaluations also allow developers to determine where they need to improve and to incorporate new features in their programs, with the aim of increasing specific aspects of performance. As a consequence, the creation and widespread use of a benchmark within a research area is frequently accompanied by rapid technical progress (2–4).
BENCHMARKING IN BIOINFORMATICS
In the early days of the bioinformatics field, when relatively little data were available, new methods were evaluated using a small number of test cases, that were chosen by the developers of the software. Different test sets were used each time, making comparisons difficult. Some small scale comparative studies were performed; for example in 1994, McClure et al. (5) compared several multiple sequence alignment methods based on four test sets and concluded that global methods generally performed better than local ones. However, the number of suitable test sets available at that time was somewhat limited and this was therefore not a comprehensive test. In 1995, Pearson (6) used 134 query sequences from 67 protein superfamilies to compare tools for searching sequence databases. In 1996, a larger set of 570 protein coding genes was used to evaluate gene finder programs (7), where a decrease in accuracy was observed when the programs were confronted with sequences showing little similarity to the training sequences.
With the widespread use of high throughput technologies and the corresponding growth of the bioinformatics databases, larger test sets can be built and recently, a number of benchmarks have been designed specifically for particular fields or applications. The field of bioinformatics benchmarking is now huge, ranging from traditional methods, such as sequence alignment, structure prediction, to more recent applications, such as protein–protein interaction prediction and protein docking, pharmacophore and drug development, etc. Here, we will mention a few recent examples, although this is clearly not intended to be an exhaustive list.
Multiple sequence alignment plays a central role in most bioinformatics analyses and provides a framework for the analysis of evolution, a major driving force in biology. The first large scale benchmark, BAliBASE (8) specifically designed for protein alignment was introduced in 1999. Other benchmarks for protein sequence alignment will be discussed in more detail below. Benchmarks have also been designed to evaluate the alignment of RNA sequences, for example, BRAliBASE (9,10). Similarly, benchmarks have been developed for the alignment of DNA sequences, including protein-coding regions based on ‘correct’ alignments inferred from 3D structural data (11) and non-coding regions, where simulations are typically used to generate synthetic data sets (12,13).
Protein 3D structure prediction, including both automatic and manually-assisted approaches, have been evaluated since 1994 in a biannual competition known as CASP (Critical Assessment of Techniques for Protein Structure Prediction). The methods are evaluated in blind experiments and the results are made available at predictioncenter.org. Other aspects of structure modeling, including 3D contact identification, and prediction of disorder, ligand binding sites or model quality, have also been assessed. Fully automatic methods are also evaluated in the CAFASP (www.cs.bgu.ac.il/∼dfischer/) competitions and the EVA (cubic.bioc.columbia.edu/eva) evaluation server. Protein–protein docking methods are compared in the CAPRI experiments (www.ebi.ac.uk/msd-srv/capri/). These continuous evaluations of methods have clearly led to significant progress (14), although, as the protein targets change each year, it can be difficult to directly compare the results of each experiment. Therefore, other benchmarks have been developed that address specific aspects of protein structure prediction. An evaluation of the sequence alignments produced by 3D structure superposition methods was performed in 2005 (15) using a test set of 2930 protein domains selected from the CATH database (16). Another benchmark based on experimentally determined structures for 124 protein–ligand complexes (17) is widely used for the development and testing of protein–protein docking algorithms.
Protein function annotation often involves the classification of an unknown protein as a member of a known family. In this domain, a protein classification benchmark (18) was constructed containing a total of 6405 classification tasks, selected to represent various degrees of difficulty. The tests involving closely related sequences within a group and relatively weak similarities between the groups are relatively easy. More difficult tests involve low (or high) similarities both within and between groups. Today, function annotation is evolving beyond the basic biochemical role, to include other features such as post-translational modifications, cellular localization, binding site localization and interactions with other molecules. Benchmarks are now being developed, e.g. focusing specifically on the problem of predicting involvement in biological processes, such as DNA damage repair in Saccharomyces cerevisiae (19). Unfortunately, this benchmark focuses on a single biological process in a specific organism, and it is not clear whether the methodology can be generalized for other systems.
Gene-expression analysis has now become a widespread tool in diverse areas of biological and biomedical research. Processing of the datasets ranges from the normalization of the data to the identification of genes that are expressed differently under various conditions. The most well-known benchmark for Affymetrix data analysis is probably Affycomp (20), consisting of a dilution study and a spike-in study. Because the truth is known for these data, they can be used to evaluate normalization and summarization methods. For the identification of differential expression, the Golden Spike data set (21) includes two conditions (control and sample) with 2535 probesets known to be differentially expressed, and 1331 probesets known to be not differentially expressed. The final step in the analysis pipeline is often the identification of over-represented functional classes in the detected gene lists. A benchmark system has been proposed recently (22) for this step, which takes a large set of genes with known Gene Ontology (GO) classifications and systematically generates gene lists with a given number of independent over-represented classes.
Image analysis: The rapid growth in microscope technologies and high throughput bioimaging has led to the development of numerous image processing methods capable of performing quantitative analyses, e.g. image segmentation and tracking. This motivated the construction of the biosegmentation benchmark infrastructure (23) that provides representative datasets of microbiological structures ranging from the subcellular level (nanometers) to the tissue level (micrometers). The datasets were obtained through collaborations with domain scientists and highlight many of the current challenges in image analysis. Applications are evaluated by comparing their results to the manually verified benchmark annotations, including segmentation, cell counting and tracking data.
BENCHMARK DESIGN AND DEVELOPMENT
With the proliferation of these benchmarks and their increasing acceptance by the community, it is important to consider what makes a good benchmark. The process of constructing a benchmark implies the rigorous definition of both what is to be measured (e.g. the quality of a solution or the time required to produce it) and how it should be measured. A number of requirements for successful benchmarks have been identified previously (e.g. 13), which can be used as design goals when creating a benchmark or as dimensions for evaluating an existing one.
Relevance: Benchmarks should be adapted to the application. The tasks set out in the benchmark should be representative of ones that the system is reasonably expected to handle in a real world (i.e. not artificial or synthetic) setting and the performance measure used should be pertinent to the comparisons being made. The tasks should also adequately reflect the scope of the problems currently encountered in the field, without over-representation of one particular task.
Solvability. It should be possible to complete the task sample and to produce a good solution. The tasks should be neither too hard nor too easy. If a task is too easy, the testing process becomes an exercise in tuning existing algorithms. If a task is too hard, it is beyond the ability of existing algorithmic techniques. The results from the test are poor and yield little data to support comparisons. A task that is achievable, but not trivial, provides an opportunity for systems to show their capabilities and their shortcomings.
Scalability: The benchmark tasks should scale to work with tools or techniques at different levels of maturity. This property influences the size of the task: it should be sufficiently large to demonstrate the advantages of the more mature techniques, but not too large to test techniques currently being researched.
Accessibility: The benchmark needs to be easy to obtain and easy to use. The test materials and results need to be publicly available, so that anyone can apply the benchmark to a tool or technique and compare their results with others.
Independence: The methods or approaches to be evaluated should not be used to construct the gold standard tests. Otherwise, the developers could be accused of ‘cheating’, i.e. designing the benchmark to suit the software. Ideally, independent information from other techniques or from human experts should be used to evaluate the correctness of the results.
Evolution: Continued evolution of the benchmark is necessary to prevent researchers from making changes to optimize the performance of their contributions on a particular set of tests. Too much effort spent on such optimizations indicates stagnation, suggesting that the benchmark should be changed or replaced.
Benchmarks that are designed according to these conditions will lead to a number of benefits, including a stronger consensus on the community’s research goals, greater collaboration between laboratories, more rigorous examination of research results, and faster technical progress. To achieve this, the benchmark tests must be planned and thought out ahead of time. It is essential to decide such things as what exactly is to be tested, the way the test is going to be run and applied, what steps are required, etc. First, the specific features to be tested should be defined as accurately as possible. If only one aspect of the test subject is to be tested, this limitation should be made clear and the results should be interpreted accordingly. Next, a benchmark test is usually based on some kind of understanding of what the correct result should be, and a specific definition of what ‘correct’ means is crucial. A misunderstood or inadequately planned test can waste time and provide bad information, because the interpretation of the test results will be flawed and misleading.
These issues will be discussed in more detail in the next section, for the specific case of multiple sequence alignment benchmarking. The criteria for good benchmarks listed above are indicated by italics.
A BENCHMARK CASE STUDY: MULTIPLE SEQUENCE ALIGNMENT
The multiple sequence alignment field represents an ideal case study to discuss the development and evolution of good benchmarking practice and to understand how benchmarking studies can be used to benefit both users and developers.
Multiple sequence alignment is one of the most fundamental tools in molecular biology. It is used not only in evolutionary studies to define the phylogenetic relationships between organisms, but also in numerous other tasks ranging from comparative multiple genome analysis to detailed structural analyses of gene products and the characterization of the molecular and cellular functions of the protein. The accuracy and reliability of all these applications depend critically on the quality of the underlying multiple alignments. Consequently, a vast array of multiple alignment programs have been developed based on diverse algorithms, from multi-dimensional dynamic programming, via progressive, tree-based programs to the more recent methods combining several complementary algorithms and/or 3D structural information (24).
Multiple sequence alignment benchmarks
Several benchmarks are now available, whose primary goal is to assess the quality of the different multiple sequence alignment programs (Table 1). A brief overview of each benchmark is provided below:
HOMSTRAD (25) is a database of protein domains, clustered on the basis of sequence and structural similarity. Although HOMSTRAD was not specifically designed as a benchmark database, it has been employed as such by a number of authors. The database provides combined protein sequence and structure information extracted from the PDB (26) and other databases, including Pfam (27) and SCOP (28). The latest version of the database contains 1032 domain families (from 2 to 41 sequences in each family) and 9602 single-member families.
BAliBASE (8,29–31) was the first large scale benchmark specifically designed for multiple sequence alignment. The alignment test cases are based on 3D structural superpositions that are manually refined to ensure the correct alignment of conserved residues. The 217 alignments in the current version of BAliBASE contain from 4 to 142 sequences and are organized into nine reference sets representing many of the problems encountered by multiple alignment methods, from a small number of divergent sequences, via sequences with large N/C-terminal extensions or internal insertions, to the particular problems posed by transmembrane regions, repeated or inverted domains, and eukaryotic linear motifs.
OXBench (32) provides multiple alignments of proteins that are built automatically using structure and sequence alignment methods. The benchmark is divided into three data sets. The master set currently consists of 673 alignments of protein domains of known 3D structure, with from 2 to 122 sequences in each alignment. The extended data set is constructed from the master set by including sequences of unknown structure. Finally, the full-length data set includes the full-length sequences for the domains in the master data set.
PREFAB (33) was also constructed using a fully automatic protocol and currently contains 1932 multiple alignments. Pairs of sequences with known 3D structures were selected and aligned using two different 3D structure superposition methods. A multiple alignment was then constructed for each pair of structures, by including 50 homologous sequences detected by sequence database searches. The automatic construction means that a large number of tests can be included. A disadvantage of this benchmark is that multiple alignment accuracy is inferred only from the alignment of the initial pair of sequences of known 3D structure.
SABmark (34) contains reference sets of sequences derived from the SCOP protein structure classification, divided into two sets, twilight zone (Blast E-value ≥1) and superfamilies (residue identity ≤50%). Pairs of sequences in each reference set are then aligned based on a consensus of two different 3D structure superposition programs. Again, the benchmark only provides ‘gold standard’ alignments for pairs of sequences. Although the sequences are grouped into families, with at most 25 sequences in each family, no consistent multiple alignment solution is provided.
IRMBASE (35) is an example of an alignment benchmark that uses synthetic datasets. The alignments consist of simulated conserved motifs implanted in non-related random sequences. The tests are divided into subsets containing 1, 2 or 3 conserved motifs per sequence, and 4, 8 or 16 sequences. The benchmark was thus designed specifically to test local multiple alignment methods. Only the alignment of the conserved motifs is taken into account when assessing the quality of an alignment program.
Table 1.
Widely used multiple sequence alignment benchmarks
| Sequence type | Test alignments | No. of test alignments | Core block annotation | No. of subsets | |
|---|---|---|---|---|---|
| HOMSTRAD | Protein | Multiple | |||
| BAliBASE | Protein | Multiple | 217 | Yes | 9 |
| Oxbench | Protein | Multiple | 673 | Yes | 3 |
| Prefab | Protein | Pairwise | 1932 | Yes | 3 |
| SABmark | Protein | Pairwise | 634 | No | 2 |
| IRMbase | Synthetic | Multiple | 180 | yes | 3 |
Each benchmark consists of a set of ‘gold standard’ test alignments. The sequences are either real protein sequences or produced by computer simulations in order to exhibit specific properties. The test alignments contain either two sequences (pairwise alignments) or multiple sequences and are divided into a number of subsets representing different alignment problems. Reliably aligned regions (core blocks) in the alignments may be annotated.
A previous study by Blackshields and coworkers (36) evaluated the relative performance of various alignment algorithms on each of these benchmark sets. The authors showed that the predicted alignment quality was more dependent on the chosen benchmark than on the alignment algorithm and that the ability of all the programs to accurately align sequences was largely dependent on the diversity of the sequences. Although there has been some argument against the quality of BAliBASE as a benchmark (37), the study by Blackshields et al., showed that program ranking was similar across all the benchmarks tested. In addition, BAliBASE was identified as one of the most useful benchmarks available thanks to its reference set architecture and its explicit coverage of distinct alignment problems.
Benchmark design
There are three main issues involved in the design of a multiple alignment benchmark. First, what is the ‘gold standard’, or correct alignment, for the sequences included in the tests? Second, which alignment problems should be covered by the benchmark, and how many test cases are needed? Third, what measure should be used to evaluate the alignments produced by different programs? These three problems are discussed in the following sections, particularly in relation to the requirements for successful benchmarks described earlier.
Correct alignments: the gold standard
The goal of a multiple sequence alignment is to identify equivalent residues in nucleic acid or protein molecules that have evolved from a common ancestor. Some authors have used probabilistic models of evolution, including insertion, deletion and substitution of characters, to construct benchmarks based on families of artificial sequences (35,38). In this case, the correct alignment solution is known, although it may not be totally independent of the sequence alignment methods to be tested. Some alignment methods may incorporate certain features of the evolutionary model used to construct the gold standard. Furthermore, the recent availability of high throughput ‘omics’ data and the advent of systems biology has revealed unexpected correlations between protein evolution and a variety of genomic features, including structure, function, network interactions and expression levels. It is now becoming clear that the evolution of most real world proteins is also affected by other processes such as gene and genome duplications, recombinations and deletions (39). As a consequence, benchmarks containing true protein sequences are more relevant, in that they provide a better representation of the problems encountered in real world situations.
Given the current status of our knowledge of protein evolutionary mechanisms, an alternative gold standard for sequence alignment is needed. The higher order tertiary structure of a protein is generally more conserved during evolution than its primary sequence. Structural similarity thus represents an objective criterion for protein sequence alignment and most benchmarks incorporate 3D structural information to some extent. First, the sequences to be included in an alignment test set are often selected from protein families in 3D structure classification databases, such as CATH (16) or SCOP (28). Second, the superposition of similar secondary structure elements is widely considered to be a good indication of an accurate sequence alignment. In themselves, these are clearly relevant factors that should be taken into account when building reference alignments, but they are not enough as a ‘gold standard’. They are themselves the result of a computer program and are not always reliable or consistent.
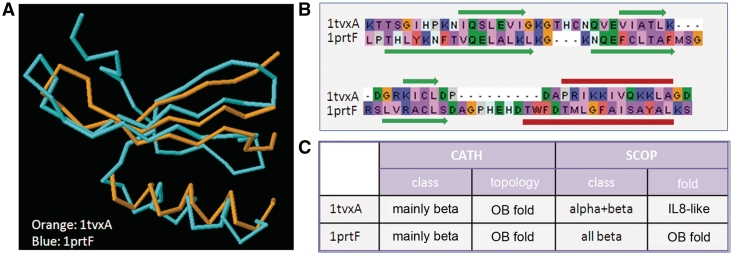
At the global similarity level, structural classifications such as the CATH or SCOP databases use additional information, including sequence or functional similarities based on a combination of computational algorithms and expert analysis. These hierarchical classifications of domains into ‘folds’ and ‘superfamilies’ are clearly useful and have contributed to important scientific progress. However, the classifications are not always unambiguous or consistent between databases. For example, 1tvxA and 1prtF (Figure 1) are both classified in CATH in the same fold family, the OB fold. In SCOP, 1prtF is again classified as an OB fold, but 1tvxA is classified as an interleukin 8-like fold. In this hierarchical view, it is implicitly assumed that protein structure space is discrete, in the sense that if a particular protein belongs to one category it does not belong to any other category. There is now growing evidence that protein structure space is in fact continuous and that classification of the fold space into discrete categories is necessarily reductionist (e.g. 40–43). In the absence of a standard definition of structural similarity, manual curation is clearly necessary. Despite these difficulties, alignments of distantly related proteins, with divergent primary sequences and conserved tertiary structures, represent the difficult test cases that are crucial for a useful benchmark.
Figure 1.
(A) 3D structure superposition of protein domains, 1tvxA and 1prtF, using the DaliLite server (RMSD = 2.5, %id = 16). (B) Sequence alignment inferred from the 3D structure superposition. Secondary structure elements are shown above and below the alignment (red = helix; green = beta-strand). (C) Classification of the two domains in the CATH and SCOP databases.
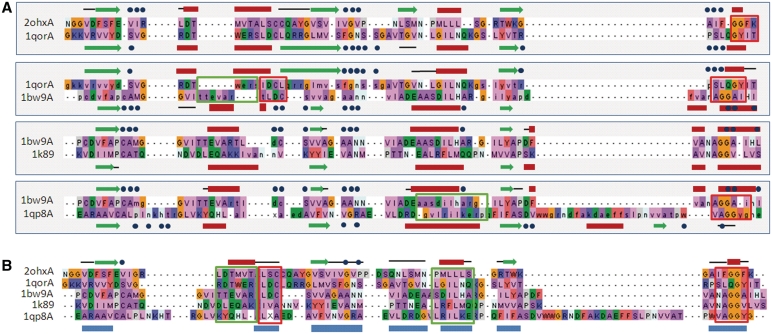
3D structure superposition can also lead to misleading sequence alignment at the local similarity level. As an example, Figure 2A shows a number of pairwise sequence alignments from the Prefab benchmark. The sequences all belong to the same homologous superfamily [NAD(P)-binding Rossmann-like Domain] according to the CATH classification. The pairwise alignments are inferred from 3D superpositions where two different superposition algorithms agreed on 50 or more positions. Nevertheless, a number of local regions can be identified where the sequence alignment could be improved, by including information about sequence conservation in the superfamily or known functional residues, as shown in Figure 2B.
Figure 2.
(A) Pairwise alignments from Prefab benchmark, based on automatic 3D superpositions (only part of the full length alignments are shown for the sake of clarity). Residues in upper case represent the ‘consensus’ regions that are superposed consistently by two different superposition methods, while lower case characters represent residues that are superposed inconsistently and are excluded from the alignment test. Secondary structure elements are shown above and below the alignment (red = helix; green = beta strand). Black lines above and below the alignment indicate consensus regions that do not have the same secondary structure. Blue dots indicate known functional residues. (B) Multiple alignment of the same set of sequences based on 3D structure superposition and sequence conservation. Blue boxes below the alignment indicate ‘core blocks’ according to the definition used in the BAliBASE benchmark. Secondary structure elements conserved in all sequences are shown above and below the alignment (red = helix; green = beta strand). Black lines above the alignment indicate core blocks that do not have a conserved secondary structure. Outlined boxes indicate sequence segments (red = consensus; green = non-consensus) that are aligned differently in (A) and (B).
Another criterion that is often used to judge the quality of a sequence alignment is the conservation of secondary structure elements. It is assumed that residues that are assigned to different secondary structure classes cannot be considered to be homologous. This ignores the fact that small changes in sequence can lead to large changes in structure in naturally occurring proteins (44). The alignments in Figure 2 show examples (highlighted by black lines above or below the sequences) where well aligned sequence segments adopt different local conformations. At the extreme are proteins that adopt multiple functional states with different 3D folds, such as the lymphotactin which adopts two distinct structures in equilibrium, one corresponding to the canonical chemokine fold and one with an all-beta-sheet arrangement having no structural similarity to any other known protein (45).
To overcome the problems associated with the structural superposition of divergent proteins, the benchmarks listed above have incorporated one or more solutions: (i) a combination of superposition algorithms is used and the regions where a consensus is observed are assumed to be reliable, (ii) the structure-based alignments are verified manually and corrected to ensure that annotated functional residues are aligned correctly, (iii) reliable regions (also known as core blocks) are identified based on a combination of sequence and/or secondary structure conservation. This ensures that the sequence alignment tasks defined by the benchmark are solvable and have a true biological significance.
Test case selection: when perfect is the enemy of good
It is impossible for a multiple alignment benchmark to be exhaustive. Given the size of the current protein structure databases (the wwPDB database, www.wwpdb.org, contains over 50 000 structures), including all possible alignments would clearly pose problems of scalability. Furthermore, exhaustivity is not a requirement for a good benchmark. It is sufficient to provide enough representative tests to perform statistical tests and to differentiate between alignment methods. A benchmark should however include as many different types of proteins as possible. In the case of a multiple sequence alignment benchmark based on 3D structure comparisons, the largest source of protein structures is the PDB database (26), although this set contains a certain amount of bias due to over-represented structures (46), as well as under-represented categories, such as transmembrane proteins. Protein structure databases, such as SCOP or CATH, thus provide useful resources by classifying proteins at various levels of structural similarity. Therefore, most benchmarks select representative protein families from a protein structure database in order to include as many different structural fold types as possible.
However, the complexity of a multiple sequence alignment does not depend only on the structural class of the proteins, but also on the nature of the sequences themselves. One of the main features affecting alignment quality is the degree of similarity between the sequences to be aligned. It has been shown that sequences sharing more than ∼30% residue identity are generally well aligned (36,47), while errors are often observed for more divergent sequences. Other determinant factors include the number and lengths of the sequences, the presence of large insertions or deletions, or the presence of low complexity sequences, transmembrane helices or disordered regions. Other problems arise from the availability of the sequences in the public databases, from bias in the phylogenetic distributions to the huge volume of sequences from genome projects and the associated prediction errors. By explicitly representing these diverse alignment problems, a multiple alignment benchmark can be used to identify and improve the specific weak points of the alignment algorithms.
Quality assessment
Apart from the alignment test sets, a good benchmark should also include a means of comparing an alignment produced by an automatic method with the ‘gold standard’ alignment. Two of the most widely used alignment scores are the sum-of-pairs and the column score (47). The sum-of-pairs score is defined as the percentage of correctly aligned pairs of residues in the alignment produced by the program and is used to determine the extent to which the programs succeed in aligning some, if not all, of the sequences in an alignment. The column score corresponds to the percentage of correctly aligned columns in the alignment, which tests the ability of the programs to align all of the sequences correctly. However, these measures only consider correctly aligned residues. An alternative approach was included in the Oxbench benchmark, where the Position Shift Error score is used to measure the average magnitude of error, so that misalignments that cause a small shift between two sequences are penalized less than large shifts. All these scores are generally calculated only in the regions of the alignment that are identified as being reliable, i.e. the core block regions. Including the unreliable or badly aligned regions can lead to a significant bias in the benchmark results (47), which are then pernicious for both the developer and user communities.
The most appropriate score will depend on the set of sequences to be aligned and the requirements of the user. For very divergent sequences, it might be more useful to use the sum-of-pairs score, since many programs will not be able to align all the sequences correctly. The Position Shift Error score can also be useful in this case, since small misalignments are scored higher than large ones. There are other situations where the column score is more meaningful. This is the case, for example, for alignments containing many closely related sequences with only a small number of more divergent, or ‘orphan’ sequences. Here, most alignment programs will align the closely related sequences correctly and will obtain a high sum-of-pairs score even if the more divergent sequences are misaligned. In this case, the column score will better differentiate the programs that successfully align the orphan sequences.
For sequences that are only partially related, it can be useful to distinguish the regions that are homologous from the unrelated regions. The SABmark benchmark proposes two metrics to address this problem. The ratio fD of correctly aligned residues divided by the length of the ‘gold standard’ alignment (equivalent to the sum-of-pairs score) is used to measure the sensitivity, while the ratio fM of correctly aligned residues divided by the length of the automatic alignment, measures the specificity of the program.
Statistical tests
As we have seen above, most of the multiple alignment benchmarks define some sort of score that measures the quality of an alignment compared to the ‘gold standard’. Many other measures have been defined in other fields that are more or less specific to the particular field. For example, in binary classification tasks (48), where a program predicts something to be either ‘true’ or ‘false’, the accuracy can be represented by four numbers: TP (true positives) = number of true cases in the benchmark predicted to be true, (ii) TN (true negatives) = number of false cases in the benchmark predicted to be false, (iii) FP (false positives) = number of false cases in the benchmark predicted to be true and (iv) FN (false negatives)=number of true cases in the benchmark predicted to be false. These are often summarized by sensitivity [TP/(TP+FN)] and specificity [TP/(TP+FP)]. In the case where the output of a program is not simply ‘true’ or ‘false’, but is a continuous number, a threshold must be selected to differentiate between true and false predictions and there is normally a trade-off between the number of false positives and false negatives. Here, a ROC (receiver operating curve) can be used, showing the FPR (false positive rate) on the x-axis and the TPR (true positive rate, also known as recall) on the y-axis. The FPR measures the proportion of false cases predicted to be true, while the TPR measures the proportion of true cases that are correctly predicted. An alternative is the precision-recall curve, with precision (the proportion of true predictions that are actually true) on the x-axis and recall on the y-axis.
Regardless of the score used to measure program performance, it is crucial to determine the statistical significance of the differences observed, using for example, hypothesis testing and the associated P-value (49). The P-value reflects the measure of evidence against the null hypothesis, for example that no difference exists between the performances of two programs. The smaller the P-value, the less plausible is the null hypothesis. Unfortunately, in many studies, a low P-value is often misinterpreted to imply that the result is of practical significance, or that there is a large difference in performance. An alternative statistic is the confidence interval (51), which indicates the reliability of an estimated value. For example, a confidence level of 95% means that the confidence interval covers the true value in 95 of 100 studies performed.
Evolution and progress
The benchmarks have been used in the past to compare different multiple alignment programs and have led to significant progress. From their beginnings in 1975, until 1994 when McClure (6) first compared different methods systematically, the main innovation was the introduction of the heuristic progressive method that allowed the multiple alignment of larger sets of sequences within a practical time limit, e.g. MultAlign (50), MultAl (51) or Clustal (52). Soon after this initial comparison (6), various new methods were introduced that exploited novel iterative algorithms, such as genetic algorithms in SAGA (53), iterative refinement in PRRP (54) or segment-to-segment alignment in Dialign (35). A comparison study of these new algorithms based on BAliBASE (47) showed that no single method was capable of producing high quality alignments for all the test cases studied. For the first time, the study revealed a number of specificities in the different algorithms. For example, while most of the programs successfully aligned sequences sharing >40% residue identity, an important loss of accuracy was observed for more divergent sequences with <20% identity. Another important discovery was the fact that while global alignment methods in general performed better for sets of sequences that were of similar length, local algorithms were generally more successful at identifying the most conserved motifs in sequences containing large extensions and insertions. As a consequence, the first methods were introduced that combined both global and local information in a single alignment program, such as DbClustal (55), T-Coffee (56), MAFFT (57), Muscle (33), Probcons (58) or PROMALS (59). Other authors introduced different kinds of information in the sequence alignment, such as 3D structure in 3DCoffee (60) and MUMMALS (61) or domain organization in REFINER (62). A number of methods were also developed to address specific problems, such as the accurate alignment of closely related sequences in PRANK (63) or the alignment of sequences with different domain organizations in POA (64).
In the post-genomic era, the ever-increasing amount of sequence information available in the public databases means that the size and complexity of the data sets that need to be routinely analysed are increasing (65). Alignments of several thousands of sequences are now commonplace, for instance the largest alignments in the Pfam database (27) have over 100 000 sequences. Clearly, the CPU and memory requirements of the alignment methods will soon become a critical factor. Furthermore, the sequencing of many eukaryotic genomes is providing full length sequences for numerous large, multi-domain proteins. Novel approaches will be required to decipher the complex evolutionary relationships between such proteins (domain insertions/deletions, repeats, permutations). Another growing problem is the quality of the sequences produced by the high throughput sequencing technologies, with a high proportion of partial and/or badly predicted sequences, which cause particular problems for the traditional alignment algorithms. If the sequence alignment methods are to evolve to cope with these new challenges, the alignment benchmarks must also evolve in order to provide new test cases that are representative of the new alignment requirements.
It may also be useful in the future to address other alignment criteria. For example, most of the current benchmarks evaluate the ability of the programs to correctly align the most conserved segments of the sequences. Nevertheless, the accurate alignment of the regions between these ‘core blocks’ is often essential for subsequent applications, such as accurate phylogenetic reconstruction (66), 3D structure modeling (67) or the identification of important functional sites (68).
CONCLUSIONS AND PERSPECTIVES
The issues raised by multiple alignment benchmarking can be applied more generally. Although good benchmark data is useful by itself, it is more constructive when accompanied by a thoughtful analysis. Good benchmarking leads to a better understanding of the problems underlying poor performance, by highlighting specific bottlenecks. Understanding why a program performs as it does on specific benchmarks is often as important as the actual benchmark results. Thus, benchmarking can help the developer improve the performance of his software. In turn, this implies that the benchmarks must continually evolve to represent the current problems and challenges in the domain. The design of a benchmark is therefore closely related to the scientific paradigm for an area: deciding what to include and exclude is a statement of values.
Many benchmarks focus entirely on one particular aspect of performance, such as computational speed or a specific accuracy score, neglecting other important features, including software reliability, accessibility, portability, compatibility and stability. These aspects of software usability (69) can be quantified using measures of effectiveness e.g. counting the number of times a particular task is completed successfully by a group of users, and of efficiency e.g. measuring the time it takes to perform the task. There are often real trade-offs between these different qualities, and all are important. For example, the multiple alignment program, ClustalW (70,71), is one of the most highly cited programs in bioinformatics, but it is not the most accurate in many situations. Nevertheless, it remains in widespread use today because the software is easily available, it can be quickly installed on most computer systems and it is easy to use. Such usability issues will take on an ever-increasing role as the software systems for analysing biological data become more and more complex.
Although intelligent benchmarking requires detailed analysis of current problems and is clearly time-consuming, the benefits are far-reaching. Benchmark studies not only have a strong positive effect in advancing a single research effort, they also encourage transparency of research results and collaboration between laboratories. They thus set a baseline for research and promote a stronger consensus on the research goals within the bioinformatics community. Ultimately, more systematic benchmarking will benefit the biologist, by providing clear guidance about the capabilities and limitations of the available algorithms and enabling him to identify the most appropriate methods for a particular project.
FUNDING
Centre National de la Recherche Scientifique; Institut National de la Santé et de la Recherche Médicale; Université de Strasbourg. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Laboratory of Integrative Bioinformatics and Genomics (LBGI) for many fruitful discussions and the Strasbourg Bioinformatics Platform (BIPS) for their support.
REFERENCES
- 1.Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142–149. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy R. To dream the possible dream – Turing award lecture. Commun. ACM. 1996;39:105–112. [Google Scholar]
- 3.Tichy WF. Should computer scientists experiment more? IEEE Computer. 1998;31:32–40. [Google Scholar]
- 4.Sim SE, Easterbrook S, Holt RC. In Proceedings of the 25th International Conference on Software Engineering. Washington DC, USA: IEEE Computer Society; 2003. Using benchmarking to advance research: a challenge to software engineering; pp. 74–83. [Google Scholar]
- 5.McClure MA, Vasi TK, Fitch WM. Comparative analysis of multiple protein-sequence alignment methods. Mol. Biol. Evol. 1994;11:571–592. doi: 10.1093/oxfordjournals.molbev.a040138. [DOI] [PubMed] [Google Scholar]
- 6.Pearson WR. Comparison of methods for searching protein sequence databases. Protein Sci. 1995;4:1145–1160. doi: 10.1002/pro.5560040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burset M, Guigó R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 8.Thompson JD, Plewniak F, Poch O. BAliBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics. 1999;15:87–88. doi: 10.1093/bioinformatics/15.1.87. [DOI] [PubMed] [Google Scholar]
- 9.Gardner PP, Wilm A, Washietl S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005;33:2433–2439. doi: 10.1093/nar/gki541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilm A, Mainz I, Steger G. An enhanced RNA alignment benchmark for sequence alignment programs. Algorithms Mol. Biol. 2006;1:19. doi: 10.1186/1748-7188-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll H, Beckstead W, O'Connor T, Ebbert M, Clement M, Snell Q, McClellan D. DNA Reference Alignment Benchmarks Based on Teritary Structure of Encoded Proteins. Bioinformatics. 2007;23:2648–2649. doi: 10.1093/bioinformatics/btm389. [DOI] [PubMed] [Google Scholar]
- 12.Pollard DA, Bergman CM, Stoye J, Celniker SE, Eisen MB. Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics. 2004;5:6. doi: 10.1186/1471-2105-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Sinha S. Towards realistic benchmarks for multiple alignments of non-coding sequences. BMC Bioinformatics. 2010;11:54. doi: 10.1186/1471-2105-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kryshtafovych A, Fidelis K, Moult J. CASP8 results in context of previous experiments. Proteins. 2009;9:217–228. doi: 10.1002/prot.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuff AL, Sillitoe I, Lewis T, Redfer OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, Mintseris J, Janin J, Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 18.Sonego P, Pacurar M, Dhir S, Kertész-Farkas A, Kocsor A, Gáspári Z, Leunissen JA, Pongor S. A Protein Classification Benchmark collection for machine learning. Nucleic Acids Res. 2007;35:D232–D236. doi: 10.1093/nar/gkl812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huttenhower C, Hibbs MA, Myers CL, Caudy AA, Hess DC, Troyanskaya OG. The impact of incomplete knowledge on evaluation: an experimental benchmark for protein function prediction. Bioinformatics. 2009;25:2404–2410. doi: 10.1093/bioinformatics/btp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cope LM, Irizarry RA, Jaffee HA, Wu Z, Speed TP. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 21.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toronen P, Pehkonen P, Holm L. Generation of Gene Ontology benchmark datasets with various types of positive signal. BMC Binformatics. 2009;10:319. doi: 10.1186/1471-2105-10-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drelie Gelasca E, Obara B, Fedorov D, Kvilekval K, Manjunath B. A biosegmentation benchmark for evaluation of bioimage analysis methods. BMC Bioinformatics. 2009;10:368. doi: 10.1186/1471-2105-10-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Poch O. Multiple sequence alignment as a workbench for molecular systems biology. Current Bioinformatics. 2006;1:95–104. [Google Scholar]
- 25.Mizuguchi K, Deane CM, Blundell TL, Overington JP. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 27.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahr A, Thompson JD, Thierry JC, Poch O. BAliBASE (Benchmark Alignment dataBASE): enhancements for repeats, transmembrane sequences and circular permutations. Nucleic Acids Res. 2001;29:323–326. doi: 10.1093/nar/29.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 31.Perrodou E, Chica C, Poch O, Gibson TJ, Thompson JD. A new protein linear motif benchmark for multiple sequence alignment software. BMC Bioinformatics. 2008;9:213. doi: 10.1186/1471-2105-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghava GP, Searle SM, Audley PC, Barber JD, Barton GJ. OXBench: a benchmark for evaluation of protein multiple sequence alignment accuracy. BMC Bioinformatics. 2003;4:47. doi: 10.1186/1471-2105-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Walle I, Lasters I, Wyns L. SABmark–a benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian AR, Weyer-Menkhoff J, Kaufmann M, Morgenstern B. DIALIGN-T: an improved algorithm for segment-based multiple sequence alignment. BMC Bioinformatics. 2005;6:66. doi: 10.1186/1471-2105-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackshields G, Wallace IM, Larkin M, Higgins DG. Analysis and comparison of benchmarks for multiple sequence alignment. In Silico Biol. 2006;6:321–339. [PubMed] [Google Scholar]
- 37.Edgar RC. Quality measures for protein alignment benchmarks. Nucleic Acids Res. 2010;38:2145–2153. doi: 10.1093/nar/gkp1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoye J, Evers D, Meyer F. Rose: generating sequence families. Bioinformatics. 1998;14:157–163. doi: 10.1093/bioinformatics/14.2.157. [DOI] [PubMed] [Google Scholar]
- 39.Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shakhnovich BE, Max Harvey J. Quantifying structure-function uncertainty: a graph theoretical exploration into the origins and limitations of protein annotation. J. Mol. Biol. 2004;337:933–949. doi: 10.1016/j.jmb.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Goldstein RA. The structure of protein evolution and the evolution of protein structure. Curr. Opin. Struct. Biol. 2008;18:170–177. doi: 10.1016/j.sbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Petrey D, Honig B. Is protein classification necessary? Toward alternative approaches to function annotation. Curr. Opin. Struct. Biol. 2009;19:363–368. doi: 10.1016/j.sbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valas RE, Yang S, Bourne PE. Nothing about protein structure classification makes sense except in the light of evolution. Curr. Opin. Struct. Biol. 2009;19:329–334. doi: 10.1016/j.sbi.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier S, Jensen PR, David CN, Chapman J, Holstein TW, Grzesiek S, Ozbek S. Continuous molecular evolution of protein-domain structures by single amino acid changes. Curr. Biol. 2007;17:173–178. doi: 10.1016/j.cub.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 45.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl Acad. Sci. USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenner SE, Chothia C, Hubbard TJ. Population statistics of protein structures: lessons from structural classifications. Curr. Opin. Struct. Biol. 1997;7:369–376. doi: 10.1016/s0959-440x(97)80054-1. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 49.du Prel JB, Hommel G, Röhrig B, Blettner M. Confidence interval or p-value?: part 4 of a series on evaluation of scientific publications. Dtsch Arztebl Int. 2009;106:335–339. doi: 10.3238/arztebl.2009.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barton GJ, Sternberg JE. A strategy for the rapid multiple alignment of protein sequences. Confidence levels from tertiary structure comparisons. J. Mol. Biol. 1987;198:327–337. doi: 10.1016/0022-2836(87)90316-0. [DOI] [PubMed] [Google Scholar]
- 51.Taylor WR. Multiple sequence alignment by a pairwise algorithm. J. Mol. Evol. 1988;28:161–169. [Google Scholar]
- 52.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 53.Notredame C, Higgins DG. SAGA: sequence alignment by genetic algorithm. Nucleic Acids Res. 1996;24:1515–1524. doi: 10.1093/nar/24.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gotoh O. Significant improvement in accuracy of multiple protein sequence alignments by iterative refinement as assessed by reference to structural alignments. J. Mol. Biol. 1996;264:823–838. doi: 10.1006/jmbi.1996.0679. [DOI] [PubMed] [Google Scholar]
- 55.Thompson JD, Plewniak F, Thierry JC, Poch O. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 2000;28:2919–2926. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 57.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 60.O'Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 61.Pei J, Grishin NV. MUMMALS: multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006;34:4364–4374. doi: 10.1093/nar/gkl514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakrabarti S, Lanczycki CJ, Panchenko AR, Przytycka TM, Thiessen PA, Bryant SH. Refining multiple sequence alignments with conserved core regions. Nucleic Acids Res. 2006;34:2598–2606. doi: 10.1093/nar/gkl274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 65.Kemena C, Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25:2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dessimoz C, Gil M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010;11:R37. doi: 10.1186/gb-2010-11-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cozzetto D, Tramontano A. Relationship between multiple sequence alignments and quality of protein comparative models. Proteins. 2005;58:151–157. doi: 10.1002/prot.20284. [DOI] [PubMed] [Google Scholar]
- 68.Sankararaman S, Sha F, Kirsch JF, Jordan MI, Sjölander K. Active site prediction using evolutionary and structural information. Bioinformatics. 2010;26:617–624. doi: 10.1093/bioinformatics/btq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolchini D, Finkelstein A, Perrone V, Nagl S. Better bioinformatics through usability analysis. Bioinformatics. 2009;25:406–412. doi: 10.1093/bioinformatics/btn633. [DOI] [PubMed] [Google Scholar]
- 70.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]