Abstract
As opposed to the vast majority of prokaryotic repressors, the immunity repressor of temperate Escherichia coli phage P2 (C) recognizes non-palindromic direct repeats of DNA rather than inverted repeats. We have determined the crystal structure of P2 C at 1.8 Å. This constitutes the first structure solved from the family of C proteins from P2-like bacteriophages. The structure reveals that the P2 C protein forms a symmetric dimer oriented to bind the major groove of two consecutive turns of the DNA. Surprisingly, P2 C has great similarities to binders of palindromic sequences. Nevertheless, the two identical DNA-binding helixes of the symmetric P2 C dimer have to bind different DNA sequences. Helix 3 is identified as the DNA-recognition motif in P2 C by alanine scanning and the importance for the individual residues in DNA recognition is defined. A truncation mutant shows that the disordered C-terminus is dispensable for repressor function. The short distance between the DNA-binding helices together with a possible interaction between two P2 C dimers are proposed to be responsible for extensive bending of the DNA. The structure provides insight into the mechanisms behind the mutants of P2 C causing dimer disruption, temperature sensitivity and insensitivity to the P4 antirepressor.
INTRODUCTION
The binding of proteins to palindromic DNA sequences is widespread in many different biological systems. The majority of all proakryotic repressors bind palindromic DNA sequences. This is a structurally favored pattern because of the 2-fold symmetry of the systems that can be recognized by symmetric protein dimers. However, there are also a few examples of proteins binding directly repeated non-palindromic DNA sequences (1–3).
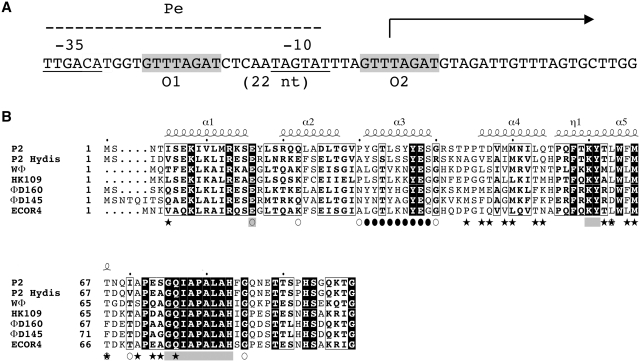
The P2 C protein is the immunity repressor of temperate E. coli phage P2. After infection, P2 can either enter the lytic cycle, leading to production of phage particles or form lysogeny. The outcome of the infection is dependent on two repressors, the immunity repressor P2 C and the Cox repressor, which control two converging promoters Pe and Pc. Formation of lysogeny requires expression of the P2 C repressor, which turns off the early Pe promoter (Figure 1A) and the integrase that promotes integration of the phage genome into the host chromosome. In the lysogenic stage, the cell survives and the phage genome will replicate as part of the host genome in each cell cycle. With a low frequency, however, the phage genome will excise from the host chromosome and enter the lytic cycle, a phenomenon-termed induction.
Figure 1.
(A) DNA sequence of the Pe operator, the direct repeats (O1 and O2). The –10 and –35 regions of the promoter are underlined and the transcriptional start site is indicated by an arrow. (B) Sequence alignment of a representative of the different immunity groups of P2-like coliphages. The location of the alpha helices in the structure of P2 C are indicated by numbered helices above the sequence. Helix 5 is initiated with a short 310 helix segment, this is indicated with η1.The amino acids mutated in the DNA-recognition helix is indicated by filled circles and the positions of the mutations previously studied are marked with open circles. Stars indicate the residues in the dimer interface. Residues in the conserved surface area are indicated by gray squares.
As opposed to most prokaryotic repressors, P2 C recognizes non-palindromic direct repeats of DNA, denoted half-sites, rather than inverted repeats. This seems to be a common property of most P2-like coliphages, and so far seven different immunity classes have been identified that all seem to recognize direct-repeat DNA sequences (4). In P2, the operator half-sites spans the −10 region of the early Pe promoter, and are separated by two helical turns (Figure 1A) (5). In the heteroimmune P2-like phage P2 Hy dis, the half-sites spans the −35 region of Pe and are separated by 2.5 helical turns (6). In the heteroimmune phage WΦ, the half-sites spans the −10 region, like in P2, but are separated by three helical turns (7). In fact, the WΦ C protein has been shown to repress the Pe promoter even if the distance between the half-sites is changed from 3 helical turns, to 2.5 or 2 helical turns (8). For several of the C repressors both half-sites have to be intact for the repressor to bind. This indicates cooperatively in the binding of the two dimers to the two half-sites. The binding of one P2 C dimer to one half-site strengthens the interaction between the other half-site and the second dimer. Both half-sites need to be intact for P2 C and P2 Hy dis C to bind while WΦ C will bind to only one half-site (6,8). P2 C as well as other members of this repressor family are DNA-bending proteins (8).
P2 C is a small slightly basic protein of 99 amino acids (5). In the absence of DNA, P2 C form dimers but not higher oligomeric forms (8–10). A recent NMR chemical shift assignment of P2 C indicates the presence of five helical segments and the single set of chemical shifts suggests a symmetric dimer (10).
The defective satellite phage P4 has the capacity to de-repress the unrelated prophage P2 after infection, thereby getting access to the late functions of the helper, which is required for P4 lytic growth. The de-repression of prophage P2 is mediated by the P4 E protein that functions as an anti-repressor by binding to P2 C. A P2 mutant, sos, that is insensitive to the action of the P4 E protein, has been isolated and the mutation mapped to residue 67 of P2 C (Thr67Ile) (9).
The immunity repressors of the P2-like phages can be divided into two types depending on sequence similarities and regulation, the P2 C-family of proteins and the 186 CI-family of proteins. The size of the 186 CI protein is twice the size of P2 C, and they share no sequence similarities (11). The structure of the 186 CI protein has been determined (12). The C-terminal domain forms an unusual heptamers of dimers, but the overall fold of the monomers of 186 CI and lambda CI is very similar. Here we present the crystal structure of the P2 C repressor, which is the first structure solved in the family of C proteins from P2-like bacteriophages (4) (Figure 2). P2 C is different from the two previously structurally characterized binders of direct DNA repeats; they utilize an asymmetric tetramer with large structural differences to P2 C or a completely different protein fold with β-sheets making the major groove interactions (1–3). We provide insight into the properties of the P2 C protein family. The C-terminus in the P2 C structure is disordered; we have shown that this region is dispensable for repressor function. Furthermore, we have identified the DNA-recognition motif in P2 C and studied the effects of mutating the individual amino acids in this helix (residues 31–39) to alanines.
Figure 2.
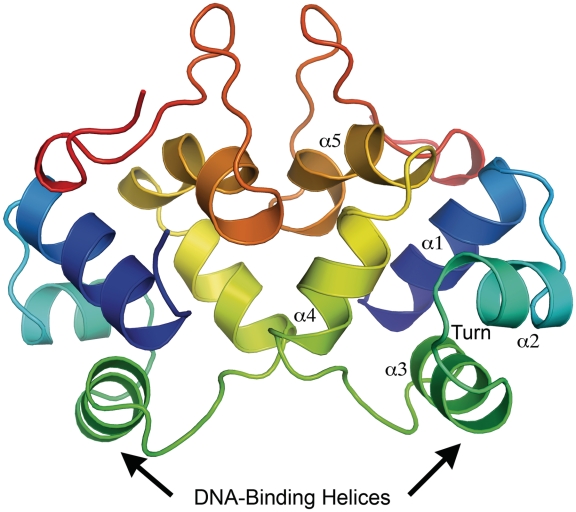
Overall ribbon representation of the P2 C dimer in rainbow color representation, from the N-terminus (blue) to the C-terminus (red). The positions of the DNA-binding helices are indicated. The helices are numbered α1–α5 and the position of the turn between α2 and α3 is indicated.
MATERIALS AND METHODS
Biological materials
Escherichia coli strains: C-1a, a prototrophic C strain (13); C-117, strain C-1a lysogenized with phage P2 (14); BL21(DE3), a B strain containing the T7 polymerase under the control of the lac promoter (15).
Plasmids: pEE675, a pKK232-8 derivative containing the P2 C-Pe-Pc region where Pe controls the cat gene (16), pEE679, a pET8c derivative containing P2 gene C under the control of the T7 promoter (8,9); pET8c, a pBR322 derivative containing the T7 promoter (15); pSS32-1, a pKK232-8 derivative containing the P2 Pe–Pc region where Pe controls the cat gene (17); pKK232-8, a pBR322 derivative containing a promoterless cat gene (18).
Site-directed mutagenesis
All plasmid constructions for the alanine scanning were performed by using QuickChange Site-Directed Mutagenesis Kit (Stratagene) and oligonucleotides were obtained from Thermo Electron, Corp, Germany and are available upon request, using plasmid pEE675 (16). All constructions were verified by DNA sequencing (Macrogen Inc., Korea).
Electrophoretic mobility shift analysis
Mutated C-genes were amplified from derivatives of pEE675 containing the respective mutation obtained by site directed mutagenesis. The amplified fragments were cloned into expression vector pET8c under the control of the T7 promoter. Crude extract from strain BL21(DE3) containing pertinent plasmids overexpressing the wild type or mutated P2 C upon induction were used. The respective crude extract was prepared by adding 10 µl of 1 M isopropyl-β-d-thiogalactoside to 20 ml culture at OD600 = 0.3 and after 2 h continued growth the culture was harvested and re-suspended in 2 ml 100 mM sodium phosphate, pH = 7.0. The cells were disrupted by sonication. After removing the cell debris, the supernatant was diluted 500- and 1000-fold before electrophoretic mobility shift assay. DNA fragment of P2 wt operator was amplified by using primers 7+28R (GCATTAAGACTATCTTCTC) and pKK240L (CCTTAGCTCCTGAAAATCTCG) with pEE675 plasmid as a template and 5′-end labeled using [γ-32P]-ATP (GE Healthcare) and T4 polynucleotide kinase (Fermentas). The labeled DNA was purified using MicroSpin G-25 columns (GE Healthcare) and incubated with different amounts of crude extract in a buffer containing 60 mM Hepes–NaOH pH 7.7, 60% glycerol, 20 mM Tris–HCl pH 7.9, 300 mM KCl, 5 mM EDTA, 0.05 µg/µl poly dI/dC, 0.3 µg/µl BSA and 5 mM DTT in a total volume of 25 µl. The samples were loaded onto a native 5% polyacryl-amide gel (PAA) after incubation 30 min at 37°C. The gel was vacuum dried before phosphor image analysis (Fuji Film FLA-3000).
Chloramphenicol acetyltransferase activity determination
At an OD600 of 0.8, 10 ml cultures were harvested. The cultures were washed in 100 mM Tris–HCl, pH 7.9 and lyzed by sonication in a total volume of 2 ml and the cell extracts were cleared by centrifugation. Total protein concentration was determined using bovine serum albumin as standard (19). The supernatants were diluted and an equal amount of protein was added for the chloramphenicol acetyltransferase (CAT) determinations with [14C]-chloramphenicol as described previously (20). The acetylated forms were separated by thin-layer chromatography, and the CAT activity was calculated after phosphor image analysis as the amount of acetylated chloramphenicol divided by the total amount of chloramphenicol.
Plating assay
A dilution of an overnight culture of each bacteria containing the mutagenized derivates of plasmid pEE675 was plated on the LA-plates containing 30 µg/ml chloramphenicol and 50 µg/ml ampicillin or LA-plates with only ampicillin. The plates were incubated over night at 37°C. The repression ability of P2 C was monitored as growth only on ampicillin plates and the loss of repression ability as growth on both chloramphenicol and ampicillin plates.
Plaque forming ability
To test the level of immunity, the plating efficiency of wild type P2 (21) was tested on LA plates supplemented with 2.5 mM CaCl2 with bacteria expressing the mutated or wild type P2 C as indicator.
P2 C expression and purification
Escherichia coli strain BL21 (DE3) containing plasmid pEE679 expressing P2 C was grown at 37°C in standard LB medium supplemented with ampicillin (100 µg/ml) for ∼4 h until the OD600 = 0.6 was reached. Protein expression was induced by addition of IPTG to a final concentration of 1 mM at 37°C for 4 h. The cells were harvested by centrifugation for 20 min at 9000g at 4°C and resuspended in 10 mM sodium phosphate buffer, pH 7.0. Cells were lysed by freeze/thawing together with sonication and thereafter centrifuged at 31 000g for 15 min at 4°C. The supernatant was collected and filtered with a 0.45-μm filter before starting the purification process. The protein was purified using an ÄKTATM FPLC-system in three steps. First, the filtered sample was adjusted to pH 8.0 with 5 M NaOH and loaded on a weak anion exchange column (DEAE, GE Healthcare) that had been equilibrated with 10 mM sodium phosphate buffer, pH 7.0 (running buffer). P2 C elutes with the flow through, as the pH of the running buffer is lower than the pI of P2 C. The second step was affinity chromatography using a HiTrap Heparin HP column equilibrated with running buffer. P2 C was eluted by a nine-column volume-gradient of 1 M NaCl. The eluted fractions containing P2 C were loaded on a Superdex 200 gel filtration column (GE Healthcare) for further purification using 25 mM Tris–HCl pH 7.5, 100 mM NaCl as running buffer. Finally, the sample was concentrated to 6 mg/ml using Amicon Ultra-15 centrifugal tubes (Millipore) with molecular weight cutoff 10 kDa. Glycerol was added to 10% final concentration before the protein was flash frozen in liquid nitrogen and stored at −80°C until further use.
Overexpression and purification of Selenomethionine-labeled P2 C
The E. coli strain BL21 (DE3) containing the P2 C expressing plasmid pEE679 was grown at 37°C in M9 minimal medium supplemented with ampicillin (100 µg/ml) for ∼8 h until OD600 = 0.6 was reached. A 100 ml of freshly prepared solution of 100 mg/l each of lysine, threonine, phenylalanine; 50 mg/l each of leucine, isoleucine, valine, l(+)-selenomethionine was added to each liter of the media 25 min before the induction. The induction and the purification processes were thereafter the same as for native P2 C.
Crystallization and data collection
Crystals of selenomethionine substituted P2 C were obtained in sitting drops at 20°C by mixing 300 nl protein solution (6 mg/ml) with 100 nl precipitant using a Mosquito nanodrop crystallization robot. The precipitant solution contained 3.9 M sodium formate and 0.1 M Tris pH 7.5. Crystals shaped as long rods grew in a few days and were frozen in liquid nitrogen directly before data collection. Crystals of native P2 C were obtained from hanging drops at 20°C by mixing 2.4 μl protein solution with 0.8 μl precipitant solution containing 3.9 M sodium formate and 0.1 M Tris pH 7.5. Crystals grew as long rods in a few days and were thereafter flash frozen in liquid nitrogen. Diffraction data was collected at 100 K from flash-frozen crystals in liquid nitrogen at beamline BL14.1 at the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY). A SAD dataset was collected for the selenomethionine crystal at a wavelength of 0.980 Å. The native data set was collected at a wavelength of 0.978 Å. Both datasets were processed using MOSFLM (22) and SCALA in the CCP4 program suite (23). Data collection and re?nement statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics
| Se-Met | Native | |
|---|---|---|
| Space group | P3221 | P3221 |
| Cell parameters | ||
| a (Å) | 91.1 | 91.6 |
| b (Å) | 91.1 | 91.6 |
| c (Å) | 80.75 | 81.3 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90 |
| Υ (°) | 120 | 120 |
| Resolution (Å) | 35−2.1 (2.21−2.1) | 35−1.8 (1.9−1.8) |
| Rmerge | 0.122 (0.589) | 0.071 (0.671) |
| Mn[I/σ(I)] | 14.9 (4.0) | 12.2 (2.0) |
| Completeness | 100% (100%) | 100% (100%) |
| Redundancy | 11.1 (11.1) | 4.9 (4.9) |
| Wavelength (Å) | 0.980 | 0.978 |
| Number of reflections work/test | 32 402/3125 | |
| Rwork/Rfree (%) | 14.9/17.3 | |
| RMSD | ||
| Bond lengths (Å) | 0.025 | |
| Bond angles (°) | 1.989 | |
| Ramachandran zone distribution (%) | 98.7/1.3/0/0 | |
Phasing, model building and refinement
The scaled selenomethionine data set was phased using Phenix Autosol (24,25), which also performed an automated initial model building. The structure given from Phenix Autosol was not further processed, but used to determine the phases for the native data set. Initial phases for the native data were obtained by molecular replacement in MOLREP (26) using the structure from the selenomethionine substituted P2 C. The model was automatically built using ARP/wARP (27) followed by cycles of manual building using coot (28) and refinement with refmac5 (29) in the CCP4 program suite (23) to complete the structure. TLS parameters were used during the last steps of refinement (30). The electron density for the N-terminal methionine and the last 14 C-terminal residues could however not be observed. These residues were therefore omitted from the model. Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 2xcj.
Modeling of the P2 C DNA complex
A linear double-stranded DNA molecule containing O1 and O2 half-sites with a center-to-center distance of 22 bp was created. Two homodimeric P2 C molecules were super-positioned on two P22 c2 repressor molecules bound to their DNA stretches (PDB code 2R1J). The RMSD of a super-positioning of P2 C and c2 is 1.9 Å over 66 residues (31). The major groove of the DNA stretch of each of the c2 repressors was aligned on each of the O1 and O2 half-sites, respectively. Additional nucleic acids were added in order to link both DNA strands to c2. Finally, the newly created DNA strand was bent by 40° and the linear DNA that contained both O1 and O2 was deleted.
RESULTS
Here we present the crystal structure of P2 C to 1.8 Å. P2 C is a symmetric homodimeric protein (Figure 2). The two subunits are related by a 2-fold rotational axis. Each protein monomer contains five alpha helices made up of residues Ile5-Glu16 (helix 1), Arg20-Thr27 (helix 2), Tyr31-Ser39 (helix 3), Thr46-Gln54 (helix 4), Gln57-Met66 (helix 5) and a β-sheet-like structure made up by residues Gln69-Gln76. The protein is 99 amino acids long and could be traced from residue 2–85 in the electron density.
Helices 2 and 3 of P2 C are separated by a 4-residue turn (Thr27-Gly28-Val29-Pro30). The sharp turn, of approximately 120°, formed by these four residues is one of the common properties of DNA-binding simple helix-turn-helix (HTH) motifs. There are only a few sequence elements widely observed among HTH motifs. The most common among these is the shs sequence in the turn between helices 2 and 3, where s is a small residue (usually glycine) and h is a hydrophobic residue (32). This sequence is also found in P2 C, which can be described as a HTH DNA-binding protein. In a HTH motif, the second helix constitutes the stabilizing helix and the third helix is known as the DNA-recognition helix, which forms the principal DNA–protein interaction by inserting itself into the major groove of the DNA duplex. Helix 3 of P2 C can, based on these properties, be assigned as the DNA-binding helix (Figures 2 and 3). This has also been confirmed by super-imposition of the P2 C structure on protein structures with known HTH domains the mutational analysis presented below.
Figure 3.
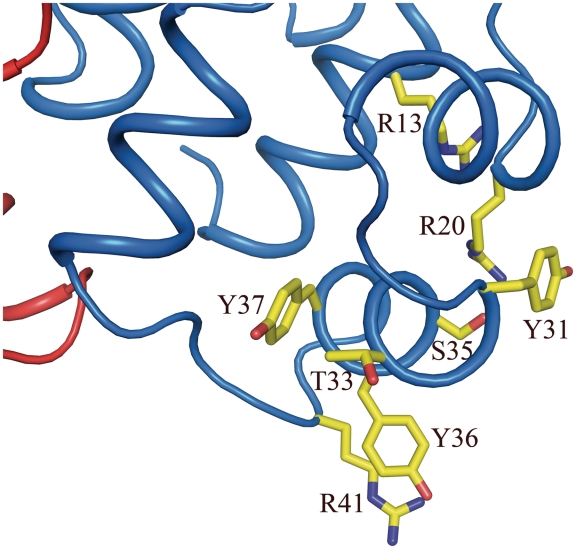
One of the two DNA-binding regions of the P2 C dimer. Residues important in DNA binding and recognition are shown as yellow sticks.
The structure of P2 C and comparisons with other HTH containing DNA-binding proteins allows us to identify residues expected to be important for interactions with DNA (31,33–35). Arg13 and Arg20 are highly likely to make unspecific interactions with the phosphate backbone of the DNA (Figure 3). The negative charge of Glu 38 is important for stabilizing and positioning Arg13 and Arg20 through several salt bridges. Both Arg13 and Glu38 are strictly conserved in the P2 C family of repressors (Figure 1B). Tyr 37 is also strictly conserved and is part of the hydrophobic foundation of helix three; it is also positioned to make unspecific interactions with the backbone of the DNA. Arg41 is located next to Tyr36 and likely has access both to sequence specific interactions in the major groove as well as proximity to the phosphate backbone. Since Tyr31, Thr33, Ser35 and Tyr36 are all positioned to make base specific interactions with the major groove of the DNA, we suggest these residues to be critical for the specificity of P2 C (Figure 3). Interestingly four of five residues positioned to make base specific interactions (Tyr31, Ser35, Tyr36 and Arg41) have a very low degree of conservation in the P2 family of repressors (Figure 1B). This is in agreement with the evolutionary pressure on the phage to have a unique immunity class. Each immunity repressor only recognizes the operators of members of the same immunity class. This prevents members of the same immunity group to grow lytically on each others lysogen. But, a superinfection of a phage with a different immunity than the resident prophage will allow lytic development of the super-infecting phage. Since P2-like prophages are common in E. coli, ∼30% of natural isolates contains a P2-like prophage (4), phages with a rare immunity will have an evolutionary advantage since it will be able to proliferate on most lysogens. The relative expression levels have been analyzed for wild-type P2 C and the biologically inactive mutants expected not to be important for the overall structure of P2 C (Y31A, T33A and Y36A). The expression levels of the mutants are as high as for wild type (Supplementary Figure S1).
Alanine scanning of the proposed DNA-recognition helix
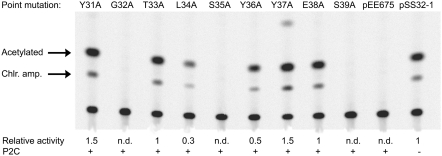
To determine the effects of alanine substitutions in the proposed DNA-recognition motif, helix 3, amino acids 31–39 were substituted one at a time by site-directed mutagenesis of plasmid pEE675 that contains the P2 C-Pe-Pc region upstream of the cat reporter gene where Pe controls expression of the cat gene. The mutations are listed in Table 2 and their positions in the P2 C structure are shown in Figure 4. The wild-type P2 C protein will repress the Pe promoter and the reporter gene in this construct. Chloramphenicol acetyltransferase assays clearly show that the Tyr31Ala, Thr33Ala, Leu34Ala, Tyr36Ala, Tyr37Ala and Glu38Ala substitutions, but not the Gly32Ala, Ser35Ala and Ser39Ala, abolished or reduced the repression capacity of the P2 C repressor in vivo (Figure 5). This result was confirmed by testing the capacity of the bacteria containing the respective mutated plasmid to grow on plates supplemented with chloramphenicol. All substitutions that abolished or reduced the capacity of P2 C to repress Pe were also able to grow on chloramphenicol plates.
Table 2.
P2 C mutants
| Name | Wild type | Mutant | Function | References |
|---|---|---|---|---|
| c5 | Glu 16 | Val | Temperature sensitive. | (5) |
| c8 | Gln 22 | Pro | Temperature sensitive. | (5,37) |
| vir 1 | Pro 30 | Gln | Unable to establish lysogeny, inactive. | (5) |
| Y31A | Tyr 31 | Ala | Inactive in vivo and in vitro | This work |
| G32A | Gly 32 | Ala | Active | This work |
| T33A | Thr 33 | Ala | Inactive in vivo and in vitro | This work |
| L34A | Leu 34 | Ala | Inactive in vivo | This work |
| S35A | Ser 35 | Ala | Active | This work |
| Y36A | Tyr 36 | Ala | Inactive in vivo and in vitro | This work |
| Y37A | Tyr 37 | Ala | Inactive in vivo and in vitro | This work |
| E38A | Glu 38 | Ala | Inactive in vivo and in vitro | This work |
| S39A | Ser 39 | Ala | Active | This work |
| c9 | Gly40 | Asp | Temperature sensitive. | (5) |
| c6 | Leu 63 | Phe | Dimer disrupted and temperature sensitive. | (5,37) |
| W64F | Trp 64 | Phe | Inactive in vivo | (37) |
| W64Y | Trp 64 | Tyr | Inactive in vivo | (37) |
| sos | Thr 67 | Ile | No binding to the P4 antirepressor. | (37) |
| c7 | Ile 70 | Asn | Less dimer, less interaction with P4 and temperature sensitive. | (5,37) |
| sly1 | Gly 85 | Glu | Lysogens are unable to liberate phage spontaneously. | (5) |
| S91ochre | Ser 91 | Stop codon | Active | This work |
Figure 4.
A ribbon representation of the P2 C dimer. Previously studied mutations are shown as yellow sticks in the blue monomer. The positions of the residues in the alanine scanning presented here are indicated by yellow sticks in the red monomer. The mutations are also listed in Table 2.
Figure 5.
The CAT assay showing effects of the alanine substitutions in helix 3. pSS32-1 is a control showing the strength of the Pe promoter in the absence of P2 C, and pEE675 is a vector expressing the wild-type P2 C. The relative CAT activity of the respective P2 C protein compared to the fully derepressed promoter (pSS32-1) is given below each lane. n.d., not detectable.
Immunity to super-infecting phages conferred by the mutated P2 C proteins
Since the chloramphenicol acetyl transferase assay showed that the repression capacity was abolished when six of the nine amino acids in helix 3 were substituted with alanines, the immunity level of the respective mutated P2 C protein was analyzed for its capacity to repress infecting P2 phages. Thus, the plaque forming capacity of phage P2 on strains C-1a containing the respective plasmid expressing the mutated P2 C protein was compared to strain C-1a without a plasmid and strain C-117, which is strain C-1a lysogenized with P2. P2 showed a high-plaque forming efficiency on strains expressing the P2 C protein with the following substitutions Tyr31Ala, Thr33Ala, Tyr36Ala, Tyr37Ala and Glu38Ala, but failed to form plaques on strains expressing all other substitutions (data not shown). Thus, all substitutions that failed or reduced the capacity to repress the reporter gene also failed to repress an infecting P2 phage except the Leu34Ala substitution. However, as can be seen in Figure 5, the Leu34Ala mutation shows a lower activity of the reporter gene, indicating some remaining DNA-binding capacity, which is enough to repress the super-infecting phage.
Alanine mutations affect the binding capacity of the P2 C repressor in vitro
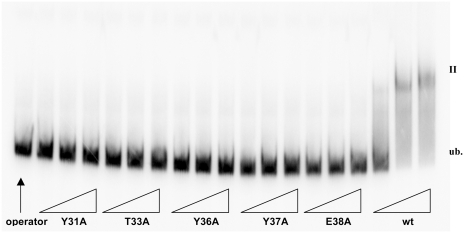
Since some alanine substitutions in helix 3 of P2 C abolished or reduced the repression capacity of P2 C in vivo, electromobility-shift analysis was carried out to analyze the capacities of the mutated P2 C to bind to the operator DNA in vitro. The mutated P2 C repressors were therefore cloned into the pET8c expression vector under the control of the inducible T7 promoter. The over-expression of the respective wild type and mutated P2 C proteins was analyzed by SDS-gel electrophoresis. As can be seen in Figure 6, crude extracts of cells expressing wild-type P2 C show one retarded band in the electromobility assay, and the substitutions Tyr31Ala, Thr33Ala, Tyr36Ala, Tyr37Ala, Glu38Ala abolish the operator recognition capacity of P2 C, since no retarded bands were detected. The Leu34Ala will under similar conditions not show any retarded band, but with increasing amounts of crude extract a specific shift, identical in migration to the wt P2 C protein is obtained (data not shown). Thus, the electromobility-shift analysis supports that helix 3 is the DNA-recognition helix.
Figure 6.
The electromobility-shift assay of alanine substitutions Tyr31Ala, Thr33Ala, Tyr36Ala, Tyr37Ala, Glu38Ala and wild-type P2C. Lane 1 is unbound probe without any P2 C protein. Crude extracts are added in increasing amounts, as indicated by the triangles.
A C-terminal truncation of 9 amino acids does not affect the biological activity of P2 C
The structure of P2 C shows a flexible C-terminal tail. A possible explanation is that this C-terminal part is folded upon DNA binding. It could be involved in strengthening the interaction to DNA and/or mediate contacts between dimers when bound to the DNA. To test this, a stop codon was inserted into the P2 C gene in plasmid pEE675 so that the last 9 amino acids are removed from P2 C and its capacity to block expression of the cat gene was investigated. All colonies transformed with the assay plasmid containing the truncated P2 C gene were unable to grow on chloramphenicol plates like the wild-type P2 C, indicating that the truncated P2 C protein is fully functional.
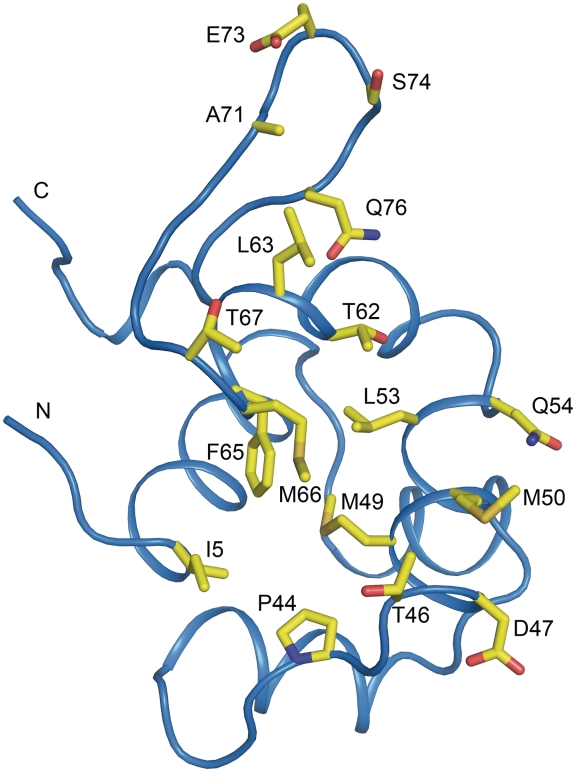
The dimer interface
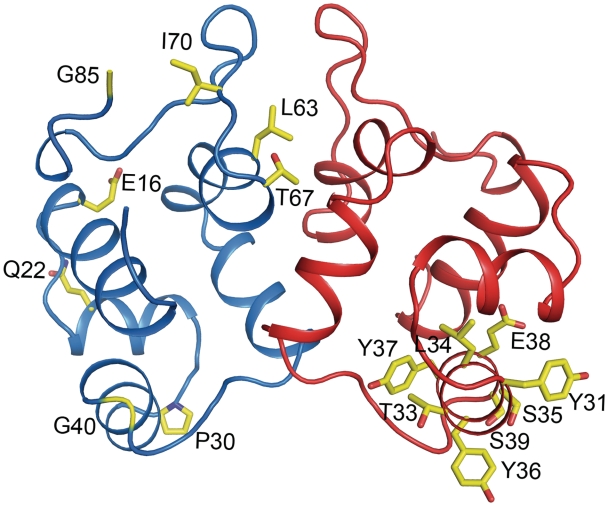
The majority of the contacts between the two monomers are made by helices 4 and 5. A pronounced area with hydrophobic interactions is present in the dimer interface; key residues in this interaction are Ile5, Leu53, Met49, Met50, Phe65 and Met66 (Figure 7). The residues Pro44, Thr46, Gln54, Met66, Ser74 and Glu73 make hydrogen bonds across the dimer interface, further stabilizing the interaction. All residues involved in the dimer interface are labeled in the alignment presented in Figure 1B and shown in Figure 7. P2 C has been shown to form dimers but not higher oligomers in solution in absence of DNA (9). This is in agreement with our gel-filtration profile of P2 C that shows one peak that corresponds to a dimeric form of P2 C with a size of 22 kDa (Supplementary Figure S2). The buried surface area of the homodimeric interface is 1700Å2, pointing to a strong interaction between the P2 C monomers (36).
Figure 7.
Representation of the dimer interface of the P2 C monomer. One monomer of P2 C is shown and the dimer interface is exposed. Residues involved in the dimer interface are shown as yellow sticks and are also indicated in Table 2. The positions of the N- and C-termini are indicated.
The amino acid sequence is less conserved in the N-terminal half compared to the C-terminal half of the protein. This is in agreement with our assignment of the DNA-recognition helix to the N-terminal part. The dimerization interface is however not, as previously suggested, conserved (Figures 1B and 7) (37). These residues thus appear to have a high-mutation rate. Still, the properties of the side chains are often retained. This is an interesting observation considering that there is an evolutionary pressure for preventing the formation of heterodimers with repressors from different immunities during super-infections. Heterodimers will hinder the super-infecting phage from forming lysogeny and the resident prophage will be induced.
The region from Gly75 to His83, not involved in the dimer interface, is however strictly conserved (Figure 1B), there must be a high-evolutionary pressure to retain the sequence and structure of this segment. The strictly conserved region is mainly located on the surface of the structure and facing away from the DNA-binding side of the P2 C dimer. This surface region is further expanded by four other strictly conserved residues, Glu16, Lys60, Tyr61 and Pro72 (Supplementary Figure S3). The temperature sensitive c5 mutation has a valine codon in the position corresponding to Glu16. The surface of a protein is normally least conserved and the high degree of conservation of this region indicates an important role. The position of this surface region is well situated to interact with other proteins simultaneously interacting with the DNA, for example RNA polymerase where the simultaneous presence P2 C has been shown to strengthen the interaction to the DNA (17). It should however also be noted that apart from being exposed on the surface of P2 C some residues (Glu16, Lys60, Tyr61, Pro79) in this conserved area are involved in salt bridges and interactions with the hydrophobic core and are likely to also be important for protein stability. This conserved region is involved in one of the largest crystal contacts that make up the continuous crystal lattice. Protein–protein interaction surfaces are often involved in crystal contacts. For steric reasons, it is highly unlikely that this interaction between dimers, present in the crystal lattice, mediates the direct interaction between the two dimers when they are bound to the O1 and O2 regions of the DNA. The DNA-binding helices of P2 C would in this case be oriented so that they could not simultaneously bind to the DNA.
DISCUSSION
The structure suggests that the P2 C dimer has the ability to bind two consecutive DNA major grooves. Unexpectedly, the P2 C dimer has great similarities to binders of palindromic sequences. However, the two identical DNA-binding helixes of the symmetric P2 C dimer have to bind different DNA sequences because the 8-bp repeats do not provide a palindromic DNA sequence. The repeats are not long enough to interact simultaneously with both monomers in the P2 C dimer and a center-to-center distance of 22 bp separates the repeats. We suggest a model where one of the two DNA-binding helices of the P2 C dimer makes sequence specific interactions with the 8-bp repeat while the other makes less specific interactions outside the repeats.
Binding to direct repeats
The P2 C structure reveals large differences to previously structurally studied repressors that binds directly repeated sequences as the cII protein of bacteriophage λ (λ cII) (2). The tetrameric cII, as opposed to many other HTH DNA-binding proteins, recognizes direct repeats of DNA, like P2 C (2). The tetramerization of cII occurs by the dimerization of dimers that form an asymmetric tetramer also in the absence of DNA. Each dimer binds to one of the two half-sites of the DNA, but with only one monomer of each dimer making sequence-specific interactions with the DNA. Even though both P2 C and λ cII contain the HTH motif there are large structural differences between the proteins.
There have been suggestions of P2 C using a similar structural solution to the binding of direct repeats as the λ cII, but there are no indications of the formation of a non-symmetric tetramer in the P2 C structure. This is supported by the fact that some of the members of the C family have the direct repeats separated by 2.5 helical turns of the DNA, placing the recognition sequences on opposite sides of the DNA (6). This would preclude a non-symmetric tetramer, similar to the one produced by λ cII, to reach both sites (35).
Indirect read-out
It is possible that P2 C like the P22 c2 repressor induces a B’ state in the DNA upon binding of the P2 C dimer, and that a similar mechanism of indirect readout is used. Indirect readout is defined as contributions to affinity other than the direct interaction between the protein and the DNA bases, for example the possibility to physically distort a particular sequence outside the protein interaction area (35). This mechanism allows the recognition of a sequence that is not in direct contact with the protein. One possibility for the cooperative binding of the two P2 C dimers is that the change in DNA conformation by binding of the first dimer leads to an altered DNA conformation also at the second binding site. This would explain the transfer of effect also when the repressor binding sites are located on opposing sides of the DNA helix. A similar mechanism has previously been observed for the cooperative DNA binding of two QacR dimers to DNA (38). There is a high-AT content in the region connecting the O1 and O2 half sites of P2 C, which is favorable for the B’ state and flexibility of the DNA (35,39). It is possible that mutations introducing GC base pairs in the linker region lead to a lowered affinity for the P2 C, as seen for P22 c2 (35).
The DNA-binding mode of P2 C by comparison to structurally related proteins
DNA–protein contacts of a specific complex cannot be reliably predicted. With new protein–DNA complex structures emerging, some general principles on base-amino acid interactions can however be found (40). Predictions made from these general rules together with comparisons with homologous proteins can give a notion of the state in which the 2-fold symmetric P2 C homodimer binds its directly repeated non-palindromic recognition sequence.
Today, no structure is known for any of the C-family of repressor proteins in the P2-like bacteriophages. However, a number of structurally related proteins available in the Protein Data Bank (PDB) were identified using a secondary structure matching server (31). The structural homologues represent different protein families, but are all prokaryotic DNA-binding proteins containing a HTH domain. Superimposing these structures on P2 C, combined with mutational analysis of P2 C, reveals some general features of the DNA-binding mechanism of P2 C that are described below.
The P22 c2 repressor
The P22 c2 repressor directs the temperate lambdoid bacteriophage P22 to the lysogenic developmental pathway (35). P22 c2 is monomeric in solution but forms dimers upon interaction with operators (41). The c2 repressor recognizes two symmetric DNA half-sites, i.e. a palindromic sequence, as opposed to the non-palindromic sequence recognized by P2 C. Superimposition of the P2 C structure on the structure of the N-terminal DNA-binding domain of P22 c2 in complex with DNA (35) shows some common features between the two repressors (RMSD 1.9Å over 66 amino acids) (31). The distance between the DNA-binding helices in the dimers are similar (26Å) and P22 c2 also introduces a bend in the target DNA. DNA–protein contacts in the P22 c2–DNA complex are mainly constituted by five residues in or near the DNA-recognition helix in the HTH domain. The DNA-binding mode will most likely be similar between the repressor proteins, i.e. helix 3 of P2 C inserted into the major groove. The sequence identity between the two DNA-recognition helices is very low, but the amino acid side chain properties are conserved. As often observed in HTH domains, the major part of the amino acids possess polar side chains, enabling hydrogen bonding to the nucleotides. The hydrophobic valine 33 of P22 c2 fits perfectly into a cleft in the DNA, caused by methyl groups in the TTAA sequence element in the major groove. The P2 C target DNA does not include a TTAA sequence, and therefore no valine cleft. This could explain the glycine at the corresponding position in P2 C (Gly32).
The repressor of the temperate bacteriophage 434
Another protein with a homologous structure to P2 C is the DNA-binding domain of the cI repressor of the temperate bacteriophage 434 (R1-69). R1-69 is a monomer in solution and dimerizes with a 2-fold symmetry upon binding to its palindromic DNA-recognition sequence (33). The DNA–protein interactions of R1-69 are typical for a HTH protein; as also seen in the P22 c2 structure, polar interactions are important in the major groove. Except from the dimerization, there are no major conformational changes in the protein upon binding to DNA, as can be seen by comparison of the crystal structures of the apo-protein and the R1-69–DNA complex (42). Both R1-69 and P22 c2 cause a major distortion in the DNA upon binding of the repressor (8,33). Interactions with the DNA backbone are likely to be important for structural changes observed in the DNA, thus positioning the recognition helices correctly in the major groove. The ability of a DNA strand to be deformed is also thought to depend on the base pair sequence (43). To only look at amino acid–base interactions when discussing specificity of a DNA-binding protein is therefore an oversimplification.
The controller protein of the Esp13961 system
Although not a bacteriophage repressor protein, the controller protein of the restriction-modification genes, the Esp13961 system (Esp13961 C) (34) is structurally similar to P2 C. Esp13961 C cooperatively binds to direct repeated palindromic sequences with one dimer to each repeated half-site. The tetrameric complex formed by the two dimers is stabilized by interactions between Arg35 and Glu25 of one subunit from each dimer. The center-to-center distance between the half-sites of the Esp13961 C is only 15 bp.
Interactions between P2 C and DNA
As often seen in DNA–protein complexes, arginines contribute to a major part of the DNA backbone–protein interactions in the homologous structures studied above. This is also most likely the case for P2 C, as judged by the superimpositions discussed above. Arg14 of P22 c2 forms a salt bridge with the backbone phosphates and superimposes perfectly with Arg13 of P2 C (Figure 3). This arginine in P2 C has a very similar position as Arg10 of 434 R1-69 (33,42). Based on the P2 C structure and model of the DNA complex, we further suggest that also Arg20 and Arg41 of P2 C are involved in non-specific interactions with the phosphate backbone of the DNA (Figure 3).
In the alanine scanning experiment, residues Tyr31–Ser39 were replaced by alanines one at a time by site-directed mutagenesis. The results strongly support our assignment of helix 3 as the DNA-recognition helix. Mutating residues Tyr31, Thr33, Leu34, Tyr36, Tyr37 and Glu38 to alanine remove or reduce the repression of the Pe promoter, i.e. decrease the DNA-binding capacity of P2 C. Furthermore, the mutants Tyr31Ala, Thr33Ala, Tyr36Ala, Tyr37Ala and Glu38Ala abolish the capacity of P2 C to bind operator DNA in vitro. The structure shows that Tyr31, Thr33, Ser35, Tyr36 and Tyr37 are positioned to make direct interaction with the DNA (Figure 3). Leu34 is forming the hydrophobic base for helix 3, replacing this residue is likely to disrupt the stability of this region of the protein. Glu38 positions and provides stability to Arg13 and Arg20 that both are likely to provide unspecific interactions with the backbone of the DNA, disruption of this salt bridge network could explain the loss of the interaction with DNA. The Gly32Ala, Ser35Ala and Ser39Ala mutations had no effect on the DNA-binding capacity of P2 C. The structure of P2 C suggests Gly32 and Ser35 to play a role for specificity; they are located on the same face of helix 3 and can make DNA interactions. It is possible that mutations introducing bulkier side chains at these positions will disrupt DNA binding.
P2 C binding bends the operator DNA
The DNA-binding helix of P2 C contains three tyrosines, all in positions allowing interaction with the DNA. The ring-shaped amino acid side chains of phenylalanine, histidine and proline have been seen forming ring-stacking with bases in DNA, thereby stabilizing and causing distortion of the double helix (40). Tyrosines are thought to have the ability to make the same kind of interactions, but this is more rarely observed in DNA–protein complexes (40). The tyrosines of P2 C could be one of the reasons for the severe distortion of the DNA observed in electrophoretic mobility shift assays (8).
The distance between adjacent major grooves in a B-DNA duplex is 34 Å. This is utilized in systems like the Trp repressor to change the affinity of the repressor. The binding of tryptophan induces a structural change moving the two DNA-binding helixes to an ideal spacing for interaction with DNA (44). The spacing between the DNA-binding helixes in the P2 C dimer is considerably smaller than the ideal 34 Å as the distance is only 26 Å between the C-alphas of Tyr36 in the two monomers.
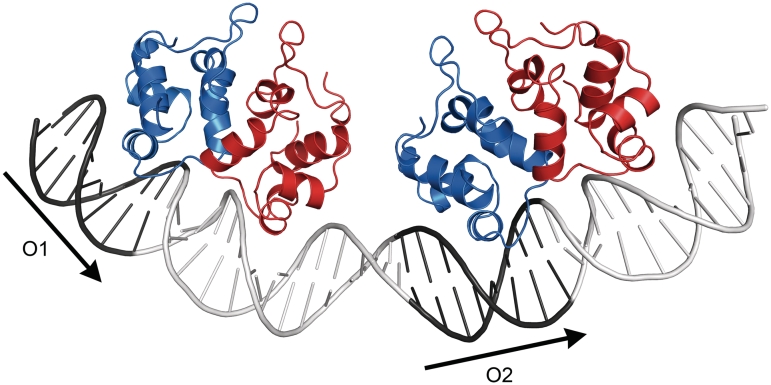
The distance between the DNA-binding helices (Gln37-Gln37) in the P22 c2 DNA complex is also 26 Å, this protein bind palindromic sequences but has considerable structural similarity to the P2 C dimer (31). The P22 c2 dimer induces a 16° curvature in the DNA. We have modeled the approximate interaction between P2 C and DNA based on this structure (35). Previous studies have also shown that binding of P2 C induces an ∼90° bend in DNA containing both recognition repeats (8). The two DNA-recognition helixes of the P2 C dimer cannot fit into the major groove of two consecutive turns of the DNA unless a considerable bend in the DNA is induced (Figure 8). Bending of the DNA makes major groove interactions possible with both recognition helices simultaneously. We suggest that each P2 C dimer induces a bend in the DNA and that an additional bending of the DNA occurs between the two dimers (Figure 8). The additional bending could be the result of a favorable interaction between both dimers when bound to the DNA, the model suggests that the P2 C dimers could be close enough for a direct interaction; this interaction could also contribute to cooperativety in repressor binding. We suggest that the total bend of ∼90° is the sum of these three contributions, the bending at each dimer as well as the bending between dimers. The 14-bp sequence connecting the two DNA repeats consists of 11 AT and 3 GC base pairs. The high AT contents of the linker region between the O1 and O2 repeats increases the DNA flexibility in this region. Further studies are necessary to validate our suggested binding model and elucidate the details of the DNA interactions.
Figure 8.
A hypothetical model of two P2 C dimers bound to two direct repeats, O1 and O2, of the operator. The dimers are presented in a ribbon representation with one monomer in blue and the other in red. The binding of the individual dimers is based on a superposition of P2 C with the complex of the P22 c2 and its palindromic DNA-operator region (PDB code 2R1J). It should be noted that this is a model and the exact position of P2 C on the repeat is not known, only the distance between the dimers based on the distance between the DNA repeats. The P2 C monomer making interactions outside the DNA repeats may face either side of the repeats; there are currently no clear experimental data available to distinguish these alternatives. However, previous footprint analysis of the binding is in agreement with the model presented here (17). The positions of the repeats are indicated with arrows.
The disordered C-terminal region of P2 C
In the structure presented here, the C-terminal 14 residues are disordered. It is tempting to speculate that the disordered C-terminal makes interactions between the dimers when they are bound to the DNA simultaneously and/or are critical for the direct DNA binding. However, the deletion of the C-terminal 9 amino acids does not lead to inactivation of the protein, showing that this part of the disordered C-terminal is not critical for interactions with the DNA. This is also observed for λ cII where the disordered C-terminal is dispensable for DNA binding (2). The C-terminal of P2 C is thus not critical for DNA binding, but could be involved in the interaction with other proteins like P4 E (16) or RNA polymerase (17), especially considering the close proximity to the highly conserved surface region described above.
Previously isolated P2 C mutations
Several naturally occurring and in vitro-introduced mutations of P2 C have been studied (Table 2). A naturally occurring P2 sos (from support of satellite phage) variant with Thr67 replaced by isoleucine has been shown to prevent the P4 E protein, described in the ‘Introduction’ section, from turning the transcriptional switch from lysogenic to lytic mode. The P4 E protein is an anti-repressor with no DNA-binding capacity on its own, but relies on the interaction with C to derepress the P2 prophage. The formation of an E–C multi-subunit complex, unable to bind DNA, turns the switch to lytic growth (9). The association of the E protein with P2 C could lead to a steric block of the interaction with the DNA. Even though Thr67 is located at the rim of the dimer interface the sos variant of P2 C forms functional dimers. The sos variant is unable to interact with the E protein and therefore retains its DNA-binding capacity (37). Threonine 67 is situated in the fifth helix of P2 C, on the opposite side of the dimer compared to the DNA-binding helix (helix 3), (Figure 4). However, Thr67 is located at the surface of the protein allowing possible interactions with the P4 E-protein. This can explain the reduced affinity between the E protein and P2 C in the presence of an isoleucine instead of a threonine in position 67. Furthermore, it is possible that the sos (Thr67Ile) mutation induces structural changes in the anti-repressor interaction area of P2 C.
Among the other mutations studied in P2 C are the Trp64Phe and Trp64Tyr variants, which were introduced by in vitro mutagenesis (37). The results from these mutation studies indicated that the substitutions at position 64 make the repressor biologically inactive at both low and high temperatures. In the presented P2 C structure, Trp64 is positioned in the fifth helix pointing into the hydrophobic core of the protein and is not directly involved in the dimeric contacts as previously suggested (37). Replacement of tryptophan 64 with a smaller amino acid, like tyrosine or phenylalanine, will destabilize the hydrophobic contacts in the core of the protein, thereby affecting the overall structure. The likely instability and miss-folding of the Trp64 mutations explain their inactivity and their inability to form dimers.
Two additional mutations of P2 C have been studied; the c6 and c7 mutations (5). These mutations have mainly been investigated for the ability to interact with the P4 E anti-repressor, but also for their dimerization capabilities (37). The c6 mutation generates a repressor where a phenylalanine replaces leucine 63 and the c7 mutation generates a repressor where an asparagine replaces isoleucine 70. Both the c6 and c7 mutations make the repressor temperature-sensitive, i.e. the repressor is active at 30°C but not at 42°C. The C6 repressor seems to interact with the E protein to the same extent as the wild-type repressor, but does not dimerize in vivo, whereas the C7 repressor seems to have less affinity for the P4 E anti-repressor and reduced dimerization (37). The c7 mutation (Ile70Asp) is not positioned in the dimer interface, but on the surface of the protein. However, the residue is likely to stabilize the loop region in which it is situated, and thereby also stabilize the P2 C dimer indirectly. The decrease in affinity for the P4 E protein could be explained by the surface position of this side chain, positioning it close to the area indicated by the sos mutation to be important for the anti-repressor interaction (Figure 4). Leu63, the site of the c6 mutation, makes extensive hydrophobic interactions in the dimer interface (Figure 7). Exchanging leucine with the larger phenylalanine will severely affect the hydrophobic packing of the residues in the dimer interface. This explains the diminished dimerization of the c6 (Leu63Phe) mutation of P2 C.
Three additional mutations causing temperature sensitive variants of P2 C have been described earlier, c5 (Glu16Val), c8 (Gln21Pro) and c9 (Gly40Asp) (5,37). Glu16, mutated in C5, interacts with Tyr61 via a hydrogen bond and forms a salt bridge with Lys60, these interactions stabilize the structure in this part of the protein and are all part of the strictly conserved surface area of P2 C. The replacement of glutamic acid by valine will result in loss of these interactions and decreased stability due to the exposure of the hydrophobic valine to solvent. The c8 (Gln21Pro) and c9 (Gly40Asp) mutations that cause temperature sensitivity as well as the vir1 (Pro30Gln) mutation that has been shown to inactivate the repressor are all located in turns and loops of the structure. The backbone properties of these residues play an important role. All of these three mutations above (c8, c9, vir1) remove or introduce prolines or glycines. These residues have unique backbone properties, with glycines being highly flexible and prolines having restricted flexibility.
Interestingly, two of the mutations described above, c9 (Gly40Asp) and vir1 (Pro30Gln), are located precisely at the ends of the DNA-binding helix. Both mutations are likely to affect the position of the helix and the stability of this region. The vir1 mutation inactivates P2 C and has since its isolation in 1960 been extensively used as a research tool because of its inability to establish lysogeny (45).
ACCESSION NUMBER
PDB: 2xcj.
FUNDING
Wenner-Gren Foundations; Magn. Bergvall Foundation; Swedish Foundation for Strategic Research and the Center for Biomembrane Research (to P.S.); Swedish Research Council; Center for Biomembrane Research and the Knut and Alice Wallenberg foundation (to M.H.); Estonian Science Foundation (to P.D.); Swedish Research Council (to E.H.L.). Funding for open access charge: Magn. Bergvall Foundation and the center for Biomembrane Research.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Uwe Müller at BESSY (Berlin, Germany) for support at beamline BL14.1.
REFERENCES
- 1.Datta AB, Panjikar S, Weiss MS, Chakrabarti P, Parrack P. Structure of lambda CII: implications for recognition of direct-repeat DNA by an unusual tetrameric organization. Proc. Natl Acad. Sci. USA. 2005;102:11242–11247. doi: 10.1073/pnas.0504535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain D, Kim Y, Maxwell KL, Beasley S, Zhang R, Gussin GN, Edwards AM, Darst SA. Crystal Structure of Bacteriophage [lambda]cII and Its DNA Complex. Mol. Cell. 2005;19:259–269. doi: 10.1016/j.molcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson JL, Cardoso-Palacios C, Nilsson AS, Haggard-Ljungquist E. Evolution of immunity and host chromosome integration site of P2-like coliphages. J. Bacteriol. 2006;188:3923–3935. doi: 10.1128/JB.01953-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljungquist E, Kockum K, Bertani LE. DNA sequences of the repressor gene and operator region of bacteriophage P2. Proc. Natl Acad. Sci. USA. 1984;81:3988–3992. doi: 10.1073/pnas.81.13.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renberg-Eriksson SK, Ahlgren-Berg A, DeGrooth J, Haggard-Ljungquist E. Characterization of the developmental switch region of bacteriophage P2 Hy dis. Virology. 2001;290:199–210. doi: 10.1006/viro.2001.1162. [DOI] [PubMed] [Google Scholar]
- 7.Liu T, Haggard-Ljungquist E. The transcriptional switch of bacteriophage WPhi, a P2-related but heteroimmune coliphage. J. Virol. 1999;73:9816–9826. doi: 10.1128/jvi.73.12.9816-9826.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlgren-Berg A, Henriksson-Peltola P, Sehlen W, Haggard-Ljungquist E. A comparison of the DNA binding and bending capacities and the oligomeric states of the immunity repressors of heteroimmune coliphages P2 and WPhi. Nucleic Acids Res. 2007;35:3167–3180. doi: 10.1093/nar/gkm171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, Renberg SK, Haggard-Ljungquist E. The E protein of satellite phage P4 acts as an anti-repressor by binding to the C protein of helper phage P2. Mol. Microbiol. 1998;30:1041–1050. doi: 10.1046/j.1365-2958.1998.01132.x. [DOI] [PubMed] [Google Scholar]
- 10.Massad T, Papadopoulos E, Henriksson-Peltola P, Haggard-Ljungquist E, Damberg P. Assignment of 1H, 13C, and 15N chemical shift resonances of P2 C-repressor protein. Biomol. NMR Assign. 2008;2:215–217. doi: 10.1007/s12104-008-9124-6. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson AS, Haggård-Ljungquist E. In: The Bacteriophages. Calendar R, editor. New York: Oxford Press; 2006. pp. 365–390. [Google Scholar]
- 12.Pinkett HW, Shearwin KE, Stayrook S, Dodd IB, Burr T, Hochschild A, Egan JB, Lewis M. The structural basis of cooperative regulation at an alternate genetic switch. Mol. Cell. 2006;21:605–615. doi: 10.1016/j.molcel.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki I, Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. Virology. 1965;40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- 14.Bertani LE. Abortive induction of bacteriophage P2. Virology. 1968;36:87–103. doi: 10.1016/0042-6822(68)90119-0. [DOI] [PubMed] [Google Scholar]
- 15.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Renberg SK, Haggard-Ljungquist E. Derepression of prophage P2 by satellite phage P4: cloning of the P4 epsilon gene and identification of its product. J. Virol. 1997;71:4502–4508. doi: 10.1128/jvi.71.6.4502-4508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha S, Lundqvist B, Haggard-Ljungquist E. Autoregulation of bacteriophage P2 repressor. EMBO J. 1987;6:809–814. doi: 10.1002/j.1460-2075.1987.tb04823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Gorman CM, Moffat LF, Howard BH. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertani G. Studies on lysogenesis I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. Warrington, U.K: Daresbury Laboratories; 1992. [Google Scholar]
- 23.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta. Crystallogr. D. Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 27.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D-Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D-Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta. Crystallogr. D. Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta. Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 32.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 34.McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36:4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins D, Hsiao C, Woods KK, Koudelka GB, Williams LD. P22 c2 Repressor-Operator Complex: Mechanisms of Direct and Indirect Readout. Biochemistry. 2008;47:2325–2338. doi: 10.1021/bi701826f. [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Eriksson SK, Liu T, Haggard-Ljungquist E. Interacting interfaces of the P4 antirepressor E and the P2 immunity repressor C. Mol. Microbiol. 2000;36:1148–1155. doi: 10.1046/j.1365-2958.2000.01937.x. [DOI] [PubMed] [Google Scholar]
- 38.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002;21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hud NV, Plavec J. A unified model for the origin of DNA sequence-directed curvature. Biopolymers. 2003;69:144–158. doi: 10.1002/bip.10364. [DOI] [PubMed] [Google Scholar]
- 40.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Anda J, Poteete AR, Sauer RT. P22 c2 repressor. Domain structure and function. J. Biol. Chem. 1983;258:10536–10542. [PubMed] [Google Scholar]
- 42.Rodgers DW, Harrison SC. The complex between phage 434 repressor DNA-binding domain and operator site OR3: structural differences between consensus and non-consensus half-sites. Structure. 1993;1:227–240. doi: 10.1016/0969-2126(93)90012-6. [DOI] [PubMed] [Google Scholar]
- 43.Albright RA, Matthews BW. How Cro and lambda-repressor distinguish between operators: the structural basis underlying a genetic switch. Proc. Natl Acad. Sci. USA. 1998;95:3431–3436. doi: 10.1073/pnas.95.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang RG, Joachimiak A, Lawson CL, Schevitz RW, Otwinowski Z, Sigler PB. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987;327:591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- 45.Bertani LE. Host-dependent induction of phage mutants and lysogenization. Virology. 1960;12:553–569. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.