Abstract
The human Ena/Vasp-like (EVL) protein is considered to be a bifunctional protein, involved in both actin remodeling and homologous recombination. In the present study, we found that human EVL forms heat-stable multimers of circular single-stranded DNA (ssDNA) molecules in the presence of a type I topoisomerase in vitro. An electron microscopic analysis revealed that the heat-stable ssDNA multimers formed by EVL and topoisomerase were ssDNA catemers. The ssDNA catenation did not occur when either EVL or topoisomerase was omitted from the reaction mixture. A deletion analysis revealed that the ssDNA catenation completely depended on the annealing activity of EVL. Human EVL was captured from a human cell extract by TOPO IIIα-conjugated beads, and the interaction between EVL and TOPO IIIα was confirmed by a surface plasmon resonance analysis. Purified TOPO IIIα catalyzed the ssDNA catenation with EVL as efficiently as the Escherichia coli topoisomerase I. Since the ssDNA cutting and rejoining reactions, which are the sub-steps of ssDNA catenation, may be an essential process in homologous recombination, EVL and TOPO IIIα may function in the processing of DNA intermediates formed during homologous recombination.
INTRODUCTION
Human Ena/Vasp-like (EVL) is a member of the ENA/VASP family, which is involved in actin-remodeling processes (1). We previously reported (2) that EVL may also function in homologous recombination, because it directly binds to RAD51 and RAD51B, which are essential proteins for meiotic homologous recombination and mitotic recombinational repair of DNA double-strand breaks (3–6). Biochemical studies revealed that EVL forms tetramer-based multimers, and actually stimulates the RAD51-mediated recombinase reactions, such as homologous pairing and strand exchange, in vitro (2,7). In addition to the RAD51-stimulating activity, EVL also possesses ssDNA annealing activity (2), which is considered to be important for the homologous-recombination processes. Therefore, EVL may have dual functions in cytoplasmic actin remodeling and nuclear homologous recombination.
Topoisomerases promote DNA strand cutting and rejoining, and are known to be important in homologous recombination. Escherichia coli RecA, a bacterial homolog of RAD51, forms homologous joint molecules between circular ssDNA and closed circular dsDNA by its recombinase activity (8). Escherichia coli topoisomerase I (Topo I) reportedly converts the homologous joint molecules formed by RecA into hemicatemers (8). Escherichia coli topoisomerase III efficiently catenates closed circular dsDNAs in the presence of RecQ helicase (9,10), which is suggested to function in homologous recombination. In humans, TOPO IIIα forms a complex with BLM and BLAP75 (11,12), and the complex is reportedly involved in the dissolution of the Holliday junction (13–18), which is a DNA intermediate formed in the late stage of homologous recombination. These facts suggest that the DNA cutting and rejoining activities of topoisomerases play important roles in homologous recombination.
In the present study, we unexpectedly found that EVL, with either E. coli Topo I or human TOPO IIIα, catalyzed ssDNA catenation. The omission of either EVL or topoisomerase quenched the ssDNA-catenating reaction, indicating that both proteins are essential for this reaction. A deletion analysis revealed that the EVL C-terminal domain, which possesses the annealing activity, is responsible for the ssDNA catenation. We also found that EVL physically interacted with human TOPO IIIα in a human cell extract and in vitro. These new findings suggest that EVL may function with a type I topoisomerase, such as TOPO IIIα, by catalyzing ssDNA cutting and re-joining reactions in the conversion of DNA intermediates during homologous recombination.
MATERIALS AND METHODS
Protein preparation
Human EVL, EVL(1–221) and EVL(222–418) were prepared by the methods described earlier (2,7). In these methods, human EVL, EVL(1–221) and EVL(222–418) were expressed as His6-tagged proteins, and the His6 tag was removed by a thrombin treatment during the purification procedure. Human RPA was produced in E. coli cells and was prepared according to the published protocol (19).
The DNA fragment encoding human TOPO IIIα was isolated from a human cDNA pool (purchased from Clontech Laboratories, Mountain View, CA, USA) by the polymerase chain reaction. The TOPO IIIα DNA fragment was cloned in the NdeI site of the pET15b vector (Novagen, Darmstadt, Germany). In this construct, the His6 tag sequence was fused to the N terminus of the protein. Human TOPO IIIα was produced in the E. coli BL21(DE3) codon plus-RP strain (Stratagene, La Jolla, CA, USA) and was purified by the following procedure. The cells producing His6-tagged TOPO IIIα were resuspended in a 50 mM Tris–HCl buffer (pH 7.5), containing 1M NaCl, 5 mM 2-mercaptoethanol and 10% glycerol, and were disrupted by sonication. The cell debris was removed by centrifugation for 20 min at 30 000g, and the lysate was mixed gently by the batch method with Ni-NTA agarose beads (3 ml, Qiagen, Hilden, Germany) at 4°C for 1 h. The His6-tagged TOPO IIIα-bound beads were washed with 40 ml of 20 mM potassium phosphate buffer (pH 7.4), containing 500 mM NaCl, 5 mM 2-mercaptoethanol, 40 mM imidazole and 10% glycerol, and then were washed again with 40 ml of 20 mM potassium phosphate buffer (pH 7.4), containing 500 mM NaCl, 5 mM 2-mercaptoethanol, 30 mM imidazole and 10% glycerol. The beads were then packed into an Econo-column (Bio-Rad Laboratories, Hercules, CA, USA) and were washed with 90 ml of 20 mM potassium phosphate buffer (pH 7.4), containing 500 mM NaCl, 5 mM 2-mercaptoethanol, 30 mM imidazole and 10% glycerol. His6-tagged TOPO IIIα was eluted with a linear gradient of 30–300 mM imidazole (13-column volumes), in 20 mM potassium phosphate (pH 7.4), 100 mM NaCl, 5 mM 2-mercaptoethanol and 10% glycerol. Peak fractions containing His6-tagged TOPO IIIα were diluted 5-fold with a 20 mM potassium phosphate buffer (pH 7.4), containing 5 mM 2-mercaptoethanol and 10% glycerol and were mixed gently by the batch method with Hydroxyapatite resin (5 ml, Bio-Rad Laboratories) at 4°C for 1 h. The unbound fraction was then dialyzed against a 20 mM potassium phosphate buffer (pH 7.4), containing 100 mM NaCl, 5 mM 2-mercaptoethanol and 10% glycerol. After the dialysis, the sample was loaded on an SP Sepharose column (1 ml, GE Healthcare Biosciences, Uppsala, Sweden), which was equilibrated with 20 ml of 20 mM potassium phosphate buffer (pH 7.4), containing 100 mM NaCl, 5 mM 2-mercaptoethanol and 10% glycerol. The resin was washed with 20 ml of 20 mM potassium phosphate buffer (pH 7.4), containing 225 mM NaCl, 5 mM 2-mercaptoethanol and 10% glycerol, and the His6-tagged TOPO IIIα was then eluted with a linear gradient of 225–600 mM NaCl (24-column volumes). The purified His6-tagged TOPO IIIα was dialyzed against 20 mM HEPES–NaOH buffer (pH 7.5), containing 100 mM NaCl, 5 mM 2-mercaptoethanol and 20% glycerol, and was stored at −80°C. The concentration of the purified His6-tagged TOPO IIIα was determined by the Bradford method (20), using bovine serum albumin as the standard.
Assays for DNA binding
The ϕX174 circular ssDNA (20 µM) was mixed with EVL in 10 µl of a standard reaction solution, containing 36 mM HEPES–NaOH (pH 7.5), 1 mM dithiothreitol, 4 mM 2-mercaptoethanol, 80 mM NaCl, 1 mM MgCl2, 24% glycerol and 0.1 mg/ml bovine serum albumin. The reaction mixtures were incubated at 37°C for 15 min, and were then analyzed by 0.8% agarose gel electrophoresis in 1× TAE buffer (40 mM Tris–acetate and 1mM EDTA) at 3.3 V/cm for 2 h. The bands were visualized by ethidium bromide staining.
ssDNA-catenating assay
The ϕX174 circular ssDNA (20 µM) was incubated with EVL and Topo I (New England Biolabs, Ipswich, MA, USA), in a reaction buffer containing 24 mM HEPES–NaOH (pH 7.5), 1 mM MgCl2, 1 mM Tris–HCl (pH 7.5), 1.1 mM dithiothreitol, 1 mM 2-mercaptoethanol, 20 mM NaCl, 5 mM KCl, 3.5 mM ammonium sulfate, 0.01 mM EDTA, 11% glycerol and 0.1 mg/ml bovine serum albumin, at 37°C for 1 h. For the experiments with TOPO IIIα, the reactions were conducted in a buffer containing 30 mM HEPES–NaOH (pH 7.5), 1 mM MgCl2, 1 mM dithiothreitol, 2.5 mM 2-mercaptoethanol, 70 mM NaCl, 14% glycerol and 0.1 mg/ml bovine serum albumin. The samples were then deproteinized by a treatment with 0.2% SDS and 1.3 mg/ml proteinase K at 37°C for 15 min. The products were then incubated at 100°C for 5 min and were chilled on ice for 5 min. The products were separated by 1% agarose gel electrophoresis, and the bands were visualized by SYBR Gold (Invitrogen, Carlsbad, CA, USA) staining.
ssDNA-annealing assay
The ssDNA-annealing assay was performed as described earlier (2). Briefly, the ssDNA oligonucleotide 49-mer (0.2 µM) was incubated with the indicated amounts of EVL or the EVL fragments at 30°C for 5 min, in 9 µl of reaction buffer, containing 28 mM HEPES–NaOH (pH 7.5), 50 mM NaCl, 2 mM 2-mercaptoethanol, 12% glycerol, 0.1 mM MgCl2, 1 mM DTT and 0.1 mg/ml bovine serum albumin. The reactions were initiated by the addition of 0.2 µM antisense 32P-labeled 49-mer oligonucleotide. At the times indicated, the reactions were quenched with an excess of the unlabeled 49-mer oligonucleotide. The DNA substrates and products were deproteinized by a treatment with 0.2% SDS and 1.5 mg/ml proteinase K at 30°C for 10 min. The products were fractionated by 10% PAGE in 0.5× TBE. The gels were dried, exposed to an imaging plate and visualized using an FLA-7000 imaging analyzer (Fujifilm, Tokyo, Japan).
Electron microscopic analysis
The ssDNA catemers formed by EVL and Topo I were extracted by phenol/chloroform and precipitated with ethanol. The ssDNA catemers were then coated with RecA in the absence of ATP and were stained on a copper-plated carbon grid with 2% uranyl acetate. The samples were visualized by tungsten rotary shadowing, and were observed with a JEOL JEM 2000FX electron microscope (JEOL, Akishima, Tokyo, Japan).
Pull-down assays with TOPO IIIα-conjugated beads
Purified human TOPO IIIα was covalently conjugated to Affi-Gel 10 beads (100 µl, Bio-Rad), according to the manufacturer's instructions. To block the remaining active ester sites, ethanolamine (pH 8.0) was added to a final concentration of 100 mM, and the resin was incubated at 4°C overnight. The unbound proteins were removed by washing the Affi-Gel 10-TOPO IIIα beads three times with 500 µl of binding buffer, which contained 20 mM HEPES–NaOH (pH 7.5), 300 mM NaCl, 5 mM 2-mercaptoethanol, 20% glycerol and 0.05% Triton X-100. After washing the resin, the Affi-Gel 10-protein matrices were adjusted to 50% slurries, and were stored at 4°C. The control beads were made by the same method, except that the TOPO IIIα was replaced by the TOPO IIIα storage buffer. For the EVL-binding assay, the TOPO IIIα-beads were incubated with an MCF7 whole-cell extract (1.8 mg of protein), and the beads were washed three times with 200 µl of washing buffer, containing 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). The proteins that copelleted with the TOPO IIIα beads were fractionated by 7.5% SDS–PAGE. The EVL protein was detected with the EVL-specific rabbit polyclonal antibodies.
Surface plasmon resonance analysis
The surface plasmon resonance (SPR) signals were measured with a Biacore X100 instrument (GE Healthcare Biosciences, Uppsala, Sweden). Flow cells were maintained at 25°C during the measurement, and the instrument was operated at the mid-flow rate (∼30 µl/min). Purified EVL was conjugated to the activated surface of the CM5 sensor chip (GE Healthcare Biosciences, Uppsala, Sweden), using the standard amine coupling conditions recommended by the manufacturer. The level of the conjugated EVL protein was 5600 resonance units. The SPR signals of the flow cell containing a sensor chip without the proteins were subtracted from those of the SPR signals of the flow cell containing the EVL-conjugated sensor chip. The running buffer was 20 mM HEPES–NaOH (pH 7.5), 200 mM NaCl, 2.5% glycerol, 1 mM dithiothreitol and 0.1% Tween-20. For the binding assay, 0.1 µM TOPO IIIα, RPA, Topo I or bovine serum albumin was injected for 2 min.
RESULTS
EVL promotes the formation of circular ssDNA multimers
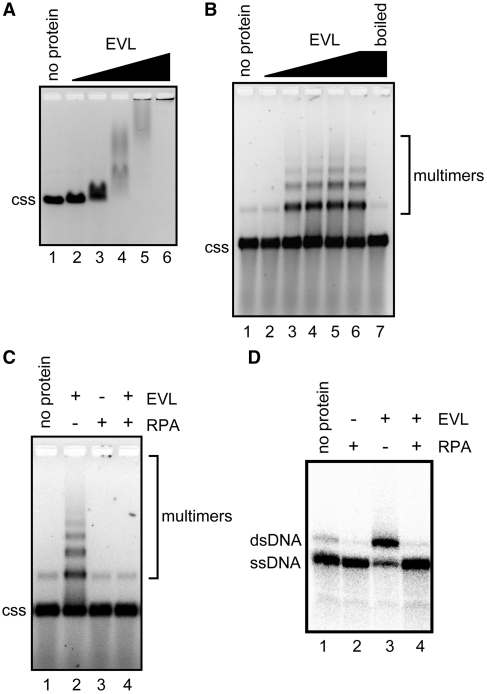
As described earlier (2), EVL bound to ϕX174 circular ssDNA (5386 bases) and formed large complexes (Figure 1A). We found that ssDNA multimers were formed within this EVL–ssDNA complex, after the EVL protein was removed from the complex by an SDS and proteinase K treatment (Figure 1B). Since we previously reported that EVL promotes the annealing of complementary ssDNAs (2), we suspected that the ssDNA multimers may be annealed products between short, complementary sequences within the ϕX174 circular ssDNA. As anticipated, the ssDNA multimers were dissociated into monomers, when the samples were incubated at 100°C for 5 min (Figure 1B, lane 7). In addition, the formation of the ssDNA multimers by EVL was completely suppressed by an ssDNA-binding protein, RPA (Figure 1C), which significantly inhibited the ssDNA annealing by EVL (Figure 1D). Therefore, we concluded that EVL promotes annealing between short, complementary sequences within the ϕX174 circular ssDNA and forms ssDNA multimers.
Figure 1.
EVL promotes the formation of circular ssDNA multimers. (A) The ssDNA-binding assay. ϕX174 ssDNA (20 µM) was incubated with the EVL protein at 37°C for 15 min. The samples were then separated by 0.8% agarose gel electrophoresis in TAE buffer and were visualized by ethidium bromide staining. Lane 1 indicates a negative control experiment without the protein. Lanes 2–6 indicate the experiments with EVL. The concentrations of EVL were 0.1 µM (lane 2), 0.2 µM (lane 3), 0.4 µM (lane 4), 0.8 µM (lane 5) and 1.6 µM (lane 6). The ϕX174 circular ssDNA molecule is indicated by css. (B) Multimer formation. ϕX174 circular ssDNA (20 µM) was incubated with EVL. The samples were then separated by 1% agarose gel electrophoresis and were visualized by SYBR Gold staining. The concentrations of EVL were 0.125 µM (lane 2), 0.25 µM (lane 3), 0.5 µM (lane 4), 1 µM (lane 5) and 2 µM (lanes 6 and 7). Lane 7 was incubated at 100°C for 5 min. Lane 1 indicates a negative control experiment without the protein. (C) Inhibition of ssDNA multimer formation by RPA. ϕX174 circular ssDNA (20 µM) was incubated with EVL (1 µM) in the presence or absence of RPA (2 µM). (D) Inhibition of the EVL-mediated ssDNA annealing by RPA. EVL (1 µM) was first incubated with ssDNA (0.2 µM) in the presence or absence of RPA (80 nM), followed by the addition of a complementary ssDNA. The reactions were conducted at 30°C for 8 min.
EVL promotes ssDNA catenation in the presence of Topo I
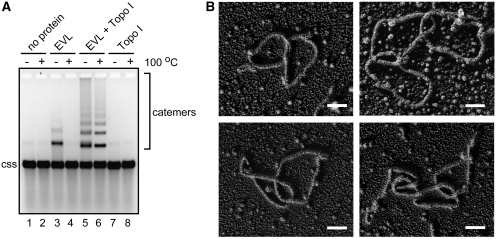
The ssDNA multimers formed by EVL may be annealed products, because the multimers were dissociated by heating (Figure 1B). Interestingly, we found that the ssDNA multimers formed by EVL in the presence of a type I topoisomerase, E. coli Topo I, were not resolved after incubation at 100°C for 5 min (Figure 2A, lane 6). These heat-stable ssDNA multimers may be ssDNA catemers. To assess whether ssDNA catemers were formed, we visualized the heat-stable ssDNA multimers by electron microscopy. To do so, the ssDNA multimers were purified, and were then coated with RecA to visualize the ssDNA. As anticipated, circular ssDNA catemers, containing two or three ssDNA molecules, were observed (Figure 2B). We counted a total of 102 molecules from the DNA samples and found that about 72.5, 18.6, 6.9 and 2% of ssDNA molecules were single circles, two ssDNA catemers, three ssDNA catemers and four or five ssDNA catemers, respectively. Therefore, we concluded that the heat-stable ssDNA multimers are ssDNA catemers.
Figure 2.
EVL promotes the ssDNA-catenating activity in the presence of Topo I. (A) Catemer formation. ϕX174 circular ssDNA (20 µM) was incubated with EVL (1 µM) and Topo I (0.4 nM). Lanes 1 and 2 indicate negative control experiments without the protein. Lanes 3 and 4 indicate the experiments in the presence of EVL. Lanes 5 and 6 indicate the experiments in the presence of EVL and Topo I. Lanes 7 and 8 indicate the experiments in the presence of Topo I. In the experiments shown in lanes 2, 4, 6 and 8, the samples were incubated at 100°C for 5 min before loading on the gel. Lanes 1, 3, 5 and 7 indicate control experiments without the 100°C incubation step. The ϕX174 circular ssDNA molecule is indicated by css. (B) Electron microscopic visualization of the ssDNA catemers. Scale bars denote 100 nm.
The ssDNA-catenating reaction did not occur when either EVL or Topo I was omitted from the reaction mixture (Figure 3A, lanes 2 and 7), indicating that both EVL and Topo I are essential for the ssDNA-catenating reaction. In addition, the formation of EVL-dependent DNA catemers was not detected when circular dsDNA was used as a substrate (Figure 3B). Protein titration experiments revealed that a very small amount of Topo I (0.04 nM) was sufficient to form the ssDNA catemers (Figure 3A, lane 3). This Topo I concentration (0.04 nM) was far below the amount required for inducing a topological change in supercoiled DNA, because 0.4 nM of Topo I is not sufficient to relax supercoiled DNA (Figure 3B, lanes 3 and 5). Therefore, these results suggested that Topo I catalytically functions in the EVL-dependent ssDNA-catenating reaction.
Figure 3.
Catalytic function of Topo I in the EVL-mediated ssDNA-catenating reaction. (A) ϕX174 circular ssDNA (20 µM) was incubated with Topo I in the presence of EVL. To eliminate the annealed products, the reaction products were treated at 100°C for 5 min before loading on the gel. Lane 1 is a control experiment without the proteins. Lane 2 indicates a negative control experiment without Topo I in the presence of EVL (1 µM). Lanes 3–6 indicate the experiments with various amounts of Topo I in the presence of EVL (1 µM). The concentrations of Topo I are 0.04 nM (lane 3), 0.2 nM (lane 4), 0.4 nM (lane 5) and 0.8 nM (lane 6). Lane 7 indicates a negative control experiment with Topo I (0.8 nM) in the absence of EVL. The ϕX174 circular ssDNA molecule is indicated by css. (B) ϕX174 superhelical dsDNA (20 µM) was incubated with EVL (1 µM) and Topo I (0.4 nM or 40 nM). Lane 1 indicates the negative control experiment without the protein. Lanes 2, 5 and 6 indicate the experiments in the presence of EVL (1 µM). Lanes 3 and 5 indicate the experiments in the presence of a low amount of Topo I (0.4 nM). Lanes 4 and 6 indicate the experiments in the presence of a high amount of Topo I (40 nM). ϕX174 superhelical and nicked or open circular dsDNA molecules are indicated by sc and nc/oc, respectively.
EVL-mediated ssDNA annealing plays an essential role in the ssDNA-catenating reaction
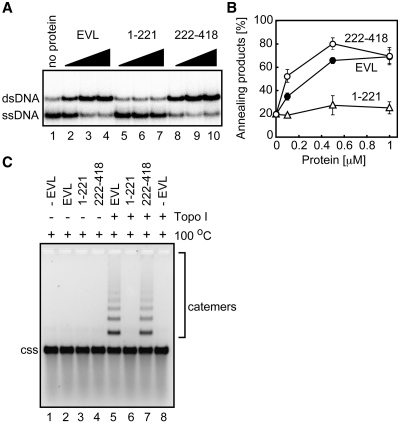
We next tested whether the ssDNA annealing by EVL plays an essential role in the ssDNA-catenating reaction with Topo I. To do so, we prepared two EVL fragments, EVL(1–221) and EVL(222–418), which contained amino acid residues 1–221 and 222–418, respectively (7). As shown in Figure 4A and B, EVL(222–418) promoted annealing to a similar extent as full-length EVL, but EVL(1–221) did not. We then tested whether EVL(1–221) and EVL(222–418) support the ssDNA-catenating reaction. As anticipated, EVL(222–418), which possesses the annealing activity, promoted the ssDNA-catenating reaction, with indistinguishable efficiency from the full-length EVL (Figure 4C, lane 7). In contrast, EVL(1–221) did not promote the ssDNA-catenating reaction (Figure 4C, lane 6). These results indicated that the EVL annealing activity is essential for the ssDNA-catenating reaction with Topo I.
Figure 4.
The EVH2 domain is responsible for the ssDNA-annealing activity and the ssDNA-catenation. (A) ssDNA annealing activities of EVL(222–418). EVL, EVL(1–221) or EVL(222–418) was first incubated with ssDNA (0.2 µM), followed by the addition of a complementary ssDNA. The reactions were conducted at 30°C for 8 min. Lane 1 indicates a control experiment without protein, and lanes 2–4, lanes 5–7 and lanes 8–10 indicate the experiments with EVL, EVL(1–221) and EVL(222–418), respectively. The EVL, EVL(1–221) and EVL(222–418) concentrations were 0.1 µM (lanes 2, 5 and 8), 0.5 µM (lanes 3, 6 and 9) and 1 µM (lanes 4, 7 and 10). (B) Graphical representation of the experiments shown in panel A. The band intensities of the annealed products were quantified, and the average values of three independent experiments are shown with the standard deviation values. Closed circles, open circles and open triangles indicate experiments with EVL, EVL(222–418) and EVL(1–221), respectively. (C) Catemer formation with the EVL(222–418) fragment. Lane 1 indicates a control experiment without protein. Lanes 2 and 5 indicate the experiments with EVL. Lanes 3 and 6 indicate the experiments with EVL(1–221). Lanes 4 and 7 indicate the experiments with EVL(222–418). Lanes 5–8 indicate the experiments in the presence of Topo I (0.4 nM). The ϕX174 circular ssDNA molecule is indicated by css.
Human TOPO IIIα catalyzes the ssDNA-catenating reaction in the presence of EVL
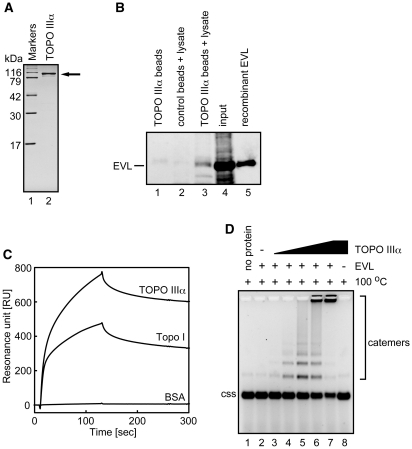
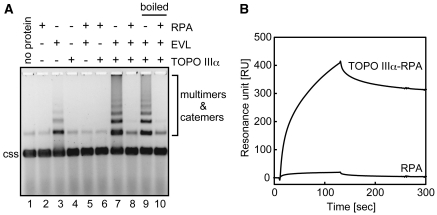
We then determined whether human topoisomerase can also promote the EVL-dependent ssDNA-catenating reaction. A eukaryotic type I topoisomerase, human TOPO IIIα, reportedly functions in homologous recombination (11–18). Therefore, we purified human TOPO IIIα as a recombinant protein (Figure 5A). The purified TOPO IIIα was chemically conjugated to Affi-Gel 10, and pull-down assays were performed with MCF7 cell extracts. As shown in Figure 5B (lane 3), the endogenous EVL protein in the MCF7 cells was clearly detected in the TOPO IIIα-bound fraction. A SPR analysis also revealed that purified TOPO IIIα efficiently bound to EVL (Figure 5C). We next performed the EVL-dependent ssDNA-catenating assay with TOPO IIIα. As shown in Figure 5D, TOPO IIIα significantly stimulated the ssDNA-catenating reaction in the presence of EVL. Like E. coli Topo I (Figure 3A), the ssDNA catemers were formed with a very small amount of TOPO IIIα (0.01 nM, Figure 5D, lane 4), suggesting its catalytic function in the reaction. The ssDNA catemers were not formed when either EVL or TOPO IIIα was omitted from the reaction mixture (Figure 5D, lanes 2 and 8). In addition, we found that the ssDNA-catenating reaction mediated by EVL and TOPO IIIα was significantly inhibited by RPA (Figure 6A), which suppresses the EVL-dependent ssDNA annealing (Figure 1C and D). The EVL-TOPO IIIα interaction was still observed in the presence of RPA (Figure 6B). These results suggested that RPA inhibits the ssDNA-catenating reaction by inhibiting the EVL-mediated ssDNA annealing.
Figure 5.
Human TOPO IIIα catalyzes the ssDNA-catenating reaction in the presence of EVL. (A) Human TOPO IIIα was purified and analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. (B) Pull down assay. Affi-Gel 10 beads chemically conjugated with human TOPO IIIα were incubated with MCF7 whole cell extract. Proteins bound to the TOPO IIIα beads were separated by SDS–PAGE and were analyzed by western blotting. Endogenous EVL was probed with an anti-EVL polyclonal antibody. Lanes 2 and 3 indicate experiments with the control Affi-Gel 10 beads and the TOPO IIIα beads, respectively, in the presence of the MCF7 cell lysate. Lane 1 indicates a control experiment with the TOPO IIIα beads in the absence of the MCF7 cell lysate. The input MCF7 cell lysate (20 µg of protein) and the purified EVL (2 ng) protein were applied in lanes 4 and 5, respectively. (C) SPR analysis of the TOPO IIIα-EVL interaction. Sensorgrams for TOPO IIIα, Topo I and bovine serum albumin (BSA) binding to the immobilized EVL protein are presented. (D) ϕX174 circular ssDNA (20 µM) was incubated with TOPO IIIα in the presence of EVL. To eliminate the annealed products, the reaction products were treated at 100°C for 5 min before loading on the gel. Lane 1 is a control experiment without the proteins. Lane 2 indicates a negative control experiment without TOPO IIIα in the presence of EVL (1 µM). Lanes 3–7 indicate the experiments with various amounts of TOPO IIIα in the presence of EVL (1 µM). The concentrations of TOPO IIIα are 0.001 nM (lane 3), 0.01 nM (lane 4), 0.05 nM (lane 5), 0.1 nM (lane 6) and 0.5 nM (lane 7). Lane 8 indicates a negative control experiment with TOPO IIIα (0.5 nM) in the absence of EVL. The ϕX174 circular ssDNA molecule is indicated by css.
Figure 6.
RPA inhibits the ssDNA-catenating reaction mediated by EVL and human TOPO IIIα. (A) ϕX174 circular ssDNA (20 µM) was incubated with TOPO IIIα and EVL in the presence or absence of RPA. Lanes 1–8 indicate control experiments without boiling. Lanes 9 and 10 indicate the ssDNA-catenating experiments, in which the reaction products were treated at 100°C for 5 min before loading on the gel. Proteins added to the reactions are indicated on the top of panel A. Protein concentrations were RPA (2 µM), EVL (1 µM) and TOPO IIIα (0.1 nM). (B) SPR analysis of the TOPO IIIα–EVL interaction in the presence of RPA. Sensorgrams for TOPO IIIα/RPA and RPA binding to the immobilized EVL protein are presented.
Intriguingly, we also found that EVL bound to E. coli Topo I, with reduced affinity as compared to human TOPO IIIα in vitro (Figure 5C). Although this EVL-Topo I combination does not exist in the natural context, it suggested that EVL binding to the Topo I region, which may be evolutionarily conserved with TOPO IIIα, may enhance the ssDNA-catenating reaction in vitro.
DISCUSSION
In the present study, we found that human EVL and a type I topoisomerase promote the catenation of ssDNA molecules. It has been reported that RecA, a bacterial recombinase, promotes hemicatenation between ssDNA and dsDNA in the presence of Topo I (8); however, no report for the ssDNA-catenating reaction has been published thus far. We also confirmed that both prokaryotic and eukaryotic type I topoisomerases are functional for the ssDNA catenation with EVL.
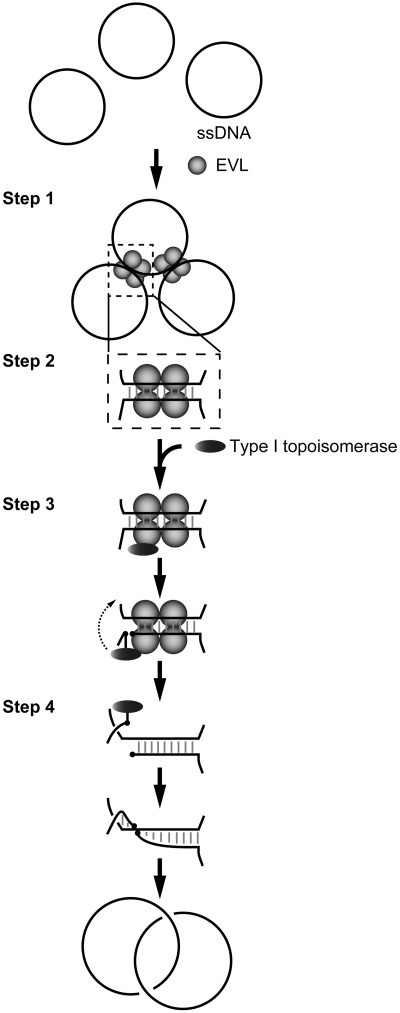
Type I topoisomerase promotes cutting and rejoining reactions on a strand within dsDNA. In the present study, we showed that the annealing activity of EVL plays an essential role in the ssDNA-catenating reaction with a type I topoisomerase. Therefore, we have proposed an annealing-mediated model for the ssDNA-catenating reaction (Figure 7). In this model, EVL first binds to ssDNA and forms large complexes, each containing multiple ssDNA molecules (Step 1). Since human EVL contains a tetramerization domain at its C-terminus (21), the formation of multiple ssDNA complexes may be mediated through its tetramerization activity. The short complementary sequences of the ssDNA may be annealed within the EVL-multiple ssDNA complexes (Step 2). A type I topoisomerase, which may be recruited on the ssDNA annealed sites through binding to EVL, then cuts an ssDNA strand within the annealed region (Step 3). This nicked site rotates and rejoins (Step 4).
Figure 7.
Model for the ssDNA-catenating reaction by EVL and a type I topoisomerase.
The functional significance of the ssDNA catenation by EVL and topoisomerases remains to be elucidated; however, it may be involved in homologous recombination because EVL directly interacts with the eukaryotic RecA homologs, RAD51 and RAD51B (2). Our previous analyses suggested that EVL may function as a RAD51 mediator, which stimulates the RAD51-mediated homologous-pairing and strand-exchange reactions (2,7). The ssDNA-catenating activity, comprising the ssDNA cutting and rejoining reactions, may be utilized to process the DNA intermediates formed by the homologous-pairing and strand-exchange reactions by RAD51. In the homologous-recombination process, it has been proposed that a DNA intermediate containing a D-loop, in which the ssDNA has invaded a homologous region of the intact dsDNA, is first formed by the RAD51-mediated homologous pairing. After this homologous-pairing step, the invading strand primes repair synthesis of the DNA strands, and the D-loop structure is converted to a four-way junction, the Holliday-junction intermediate, which moves along the DNA to expand the newly paired heteroduplex region. The ssDNA cutting and rejoining activities of EVL and topoisomerase may be involved in the conversion process from the D-loop to Holliday junction intermediates.
Alternatively, the ssDNA cutting and rejoining activities may function in the resolution of the Holliday junction. The Holliday-junction intermediate must be resolved into two parental DNA molecules by the ssDNA-nicking activity in the late stage of the homologous-recombination pathway. Interestingly, human TOPO IIIα, which was found to promote the ssDNA-catenating reaction with EVL, is reportedly involved in Holliday junction dissolution (13–16). In addition, EVL is known to bind RAD51B, which preferentially binds to the Holliday junction (2,22). Therefore, EVL may also be involved in the late stage of the HRR pathway, together with TOPO IIIα. Further analyses are required to clarify these issues.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Japanese Society for the Promotion of Science (JSPS) of Japan. Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
H. K. is a research fellow in the Waseda Research Institute for Science and Engineering.
REFERENCES
- 1.Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell. Biol. 2003;13:386–392. doi: 10.1016/s0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 2.Takaku M, Machida S, Hosoya N, Nakayama S, Takizawa Y, Sakane I, Shibata T, Miyagawa K, Kurumizaka H. Recombination activator function of the novel RAD51- and RAD51B-binding protein, human EVL. J. Biol. Chem. 2009;284:14326–14336. doi: 10.1074/jbc.M807715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 5.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.Takaku M, Machida S, Nakayama S, Takahashi D, Kurumizaka H. Biochemical analysis of the human EVL domains in homologous recombination. FEBS J. 2009;276:5841–5848. doi: 10.1111/j.1742-4658.2009.07265.x. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham RP, Wu AM, Shibata T, DasGupta C, Radding CM. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981;24:213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- 9.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 10.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J. Biol. Chem. 2003;278:42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 12.Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 16.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J. Biol. Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 17.Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J. Biol. Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 21.Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama H, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]