Abstract
CPEB3 is a sequence-specific RNA-binding protein and represses translation of its target mRNAs in neurons. Here, we have identified a novel function of CPEB3 as to interact with Stat5b and inhibit its transcription activity in the nucleus without disrupting dimerization, DNA binding and nuclear localization of Stat5b. Moreover, CPEB3 is a nucleocytoplasm-shuttling protein with predominant residence in the cytoplasm; whereas activation of NMDA receptors accumulates CPEB3 in the nucleus. Using the knockdown approach, we have found the receptor tyrosine kinase, EGFR, is a target gene transcriptionally activated by Stat5b and downregulated by CPEB3 in neurons. The increased EGFR expression in CPEB3 knockdown neurons, when stimulated with EGF, alters the kinetics of downstream signaling. Taken together, CPEB3 has a novel function in the nucleus as to suppress Stat5b-dependent EGFR gene transcription. Consequently, EGFR signaling is negatively regulated by CPEB3 in neurons.
INTRODUCTION
Long-term memory requires synthesis of plasticity-related proteins (PRPs) to strengthen synaptic efficacy and consequently consolidate memory. RNA-binding proteins play indispensable roles to control spatial-temporal PRP production by regulating transport, localization, translation and/or degradation of PRP RNAs (1–4). Cytoplasmic polyadenylation element binding protein (CPEB)-like proteins, CPEB2, CPEB3 and CPEB4, in vertebrates likely influence PRP synthesis for the following reasons. CPEB3 and CPEB4 are expressed predominantly in neurons and CPEB3-repressed translation of a reporter RNA is abrogated by the activation of N-methyl-d-aspartic acid receptor (NMDAR) (5). CPEB3 and CPEB4 mRNAs are elevated in the hippocampus after kainate-induced seizure, indicating they are immediate early gene products upon synaptic activation and likely modulate neuronal function (6). Importantly, Orb2 in Drosophila is required for long-term conditioning of male courtship behavior (7), implicating that its mammalian homologs, CPEBs2–4, may also have roles in memory. A recent study has shown that a single nucleotide polymorphism in the CPEB3 gene is associated with human episodic memory (8).
CPEBs2–4 were first identified based on sequence similarity with CPEB (or CPEB1) in the carboxyl terminal RNA-binding domain (9). However, CPEBs2–4 could interact with RNA sequences identified from a SELEX (systematic evolution of ligands by exponential enrichment) screen that are different from the conventional CPEB1-binding site (UUUUA1-2U) (5). Despite CPEB1-controlled translation is characterized at the molecular details and plays important roles in development, cell cycle, neuronal plasticity and cellular senesce (10), much less is known about the functional entities of CPEBs2–4 once they bind to RNAs. A previous study has shown that CPEB3 repressed translation of a reporter RNA and Glu2 RNA (5). Interestingly, a prion-like property has been observed in Orb2 as well as Aplysia CPEB in yeasts (11) and a recent study has shown that multimeric state of CPEB is required for maintaining long-term facilitation in Aplysia (12). Nonetheless, whether any mammalian CPEB possesses prion-like change to modulate its target RNA translation is still in question. To understand how CPEB3 regulates translation, we used a yeast two-hybrid screen to identify its binding partners. Unexpectedly, the screen identified a transcription factor, signal transducer activated transcription (Stat) 5b, interacted with CPEB3. Stat5b is one of the seven Stat family members of which transcriptional activity are modulated by Janus tyrosine kinases (JAKs), that are activated by cytokines and hormones (13,14). Translocation of dimerized Stat to the nucleus activates target gene transcription (15). Using promoter assays, CPEB3 inhibits Stat5b-dependent transcription without affecting DNA binding, nuclear translocation and dimerization of Stat5b. Moreover, CPEB3 shuttles between the nucleus and cytoplasm and activation of NMDARs increases nuclear level of CPEB3, suggesting that neuronal activity regulates CPEB3′s roles in transcription and translation.
One target gene transcriptionally regulated by Stat5b and CPEB3 interaction identified from this study is the receptor tyrosine kinase, epidermal growth factor receptor (EGFR). Upon ligand binding, the receptors become phosphorylated on tyrosine residues within their cytoplasmic kinase domain and activated which then initiate several downstream signaling pathways, such as JAK-Stat, mitogen-associated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)-Akt. The elevated EGFR level in CPEB3 knockdown neurons, when stimulated with EGF, results in extended and amplified downstream signaling measured by phosphorylation of Stat5b and Akt. Although EGFR has been studied extensively in cell proliferation (including neurogenesis), anti-apoptosis and cancer progression (16–18), its function in post-mitotic neurons is less characterized. In the EGFR null mice, abnormal astrocyte development and neuronal death impede the study of EGFR function in the adult brain (19,20), but it has been demonstrated that EGF enhances long-term potentiation in the hippocampal slices and dentate gyrus of anesthetized rats after tetanic stimulation (21,22), suggesting its corresponding receptor, EGFR, may function as a neuronal modulator. Using pharmacological approach, activation or deprivation of EGFR’s kinase activity by infusing EGF or gefitinib (23), respectively, in the brain, affects spatial learning and memory performance in mice. Together, this study first identifies a novel transcriptional function for the CPEB family members besides their characterized roles in translation (5,10,24,25). By interacting with Stat5b, CPEB3 downregulates the expression of EGFR of which kinase activity modulates learning and memory.
MATERIALS AND METHODS
Antibodies
Antibodies used for the study are, Akt (cat #4691), pT308-Akt (cat #2965), pY1068-EGFR (cat #2236S) and pY699-Stat5 (cat #9359) from Cell Signaling; EGFR (cat #SC-03) and Stat5b (cat #SC1656) from Santa Cruz Biotechnology; synaptophysin (cat #MAB5258) and MAP2 (cat #MAB378) from Chemicon and flag epitope (cat #F1804) from Sigma-Aldrich.
Yeast two-hybrid screen
Matchmaker Two-Hybrid Library Construction and Screening Kit (Clontech) was used to construct a random-primed adult mouse brain cDNA library. Briefly, mouse brain poly(A) RNA (Ambion) and the CDS III/6 primer were used to generate double-stranded cDNA that was co-transformed with linearized pGAD-Rec vector into the yeast strain, AH109. Approximately 5.5 × 105 transformants were harvested, mixed and stored at −80°C. The N-terminus of hCPEB3 (amino acids 1–427) was cloned into pGBKT7 and transformed into the yeast strain, Y187 (screening bait). Both yeast strains were mated for 24 h at 30°C with gentle swirling at 40 rpm then plated in nutrient-selective plates (SD/X-α-gal/-Leu/-Trp/-Ade/-His) to screen for positive interaction clones.
Plasmid construction
The shRNA sequences, ACAAACCTGTTCAAATCC and CAATACTGGGAATAAATC targeted against rat CPEB2 and CPEB4 mRNAs respectively, were cloned into lentiviral vector pLL3.7-Syn (26). The shRNA clone, TRCN0000012554 against rat Stat5b (CGGCCAAAGGATGAAGTATAT), was obtained from the RNAi Core Facility (Academia Sinica). The various truncated Stat5b mutants were PCR amplified and cloned into pGADT7 for two-hybrid interaction.
Cell culture and lentivirus infection
HEK-293T and COS7 cells were cultured in DMEM with 10% FBS. Cultures of rat hippocampal neurons (27) were grown at a cell density of 8000/cm2 on coverslips or 30 000/cm2 on dishes. Hippocampal neurons of DIV 6 were infected with lentivirus overnight and harvested on DIV 11 for RNA or protein extraction. On the day of harvest, the concentrations of reagents used to treat neurons are: 50 µM NMDA, 15 µM AMPA, 50 µM DHPG, 30 mM KCl, 30 µM APV, 20 µM NBQX, 5 ng/ml leptomycin B, 100 ng/ml EGF, 30 µg/ml cycloheximide and 2 µg/ml actinomycin D.
Co-immunoprecipitation (Co-IP)
For co-IP, 4 µg of DNA mixtures containing equal amounts of two or three plasmids were co-transfected in a 6-cm dish of 293T cells using lipofectamine 2000. The overnight transfected cells were lysed in 120 µl IP buffer (20 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 0.1% NP40, 10% glycerol, 0.5 mM DTT, 1× protease inhibitor cocktail and 100 µg/ml RNaseA) and centrifuged at 10 000 × g for 5 min at 4°C. The supernatant was incubated with protein G beads bound with myc or flag antibody (Ab) for 2 h at 4°C. The beads were washed five times with 300 µl IP buffer and the precipitated proteins were used for western blotting. For reciprocal IP of endogenous CPEB3 and Stat5b, ∼4 × 107 cortical neurons were lysed in 1.6 ml IP buffer. Equal volumes of lysate were incubated with Stat5b, CPEB3 or control IgG and the immunoprecipitates were probed with CPEB3 and Stat5b antibodies.
RNA extraction, cDNA synthesis and quantatitive PCR (Q–PCR)
Total RNA was extracted with Trizol (Invitrogen). The cDNA was synthesized using oligo-dT primer and ImPromII Reverse Transcriptase (Promega). Quantitative PCR was conducted using the Universal Probe Library and Lightcycler 480 system (Roche). Data analysis was done using the comparative Ct (threshold cycle value) method with the non CPEB3-targeted RNA, NF-M mRNA as the reference. The PCR primers are: EGFR, 5′-TGCACCATCGACGTCTACAT-3′ and 5′-AACTTTGGGCGGCTATCAG-3′; CPEB3, 5′-TGGAGAACTCCTTAATGGATATGA-3′ and 5′-AAGTTTATTCCCATGCGTCCT-3′; Stat5b, 5′-GGAGAGCCTACGGATCCAA-3′ and 5′-AGGGACACTTGCTTCTGCTG-3′; NF-M, 5′-CGTCATTTGCGAGAATACCA-3′ and 5′-TCTTCACCCTCCAGTAGTTTCC-3′.
Chromatin immunoprecipitation (ChIP)
Approximately 3 × 107 cortical neurons of DIV11 were crosslinked with 1.42% formaldehyde for 15 min and quenched with 125 mM glycine for 5 min at room temperature. The fixed cells were washed three times with ice-cold PBS, lysed in 3 ml of ChIP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP40 and 1% Triton) and sonicated for 10 min with 30 s on/off cycles. The sonicated sample was centrifuged for 5 min at 12 000 × g at 4°C and equal volumes of supernatant were incubated overnight with the protein G beads with Stat5b or mouse IgG. The beads were washed three times with ChIP buffer containing 500 mM NaCl followed by another three washes with ChIP buffer. One-third of beads were eluted with Laemmli sample buffer for western blot analysis and the remaining beads along with the saved input sample were incubated at 95°C for 15 min and digested with Proteinase K (1 mg/ml in 50 mM Tris, pH 7.4, 10 mM EDTA, 50 mM NaCl and 1% Triton X-100) at 55°C for 30 min. The supernatants were phenol/chloroform extracted, ethanol precipitated in the presence of 10 µg yeast tRNA as a carrier and used for PCR analysis. Primers used to detect binding to the −3000 to +300 promoter region and a negative control spanning intron 24 and exon 25 of the EGFR gene are: −2790 to −2581: 5′-CGACTGACCTCTGAATGTCC-3′ and 5′-TGCTTGGTTGGTAGTCCAGT-3′; −2299 to −1981: 5′-GACCTAAGTTCTTATTCGGAAC-3′ and 5′-TACCACATGGTAGCCTGGCA-3′; −2001 to −1641: 5′-ATGCCAGGCTACCATGTGGTA-3′ and 5′-AAGCTGTGGAAATGTAGAGCA-3′; −1475 to −1061: 5′-CCACTTTATATAAAACCCCTG-3′ and 5′-AAGACCAACTAATGTCACTGC-3′; −510 to −101: 5′-GGTAGGACAGTAACCGAGG-3′ and 5′-CTAGCTAGCCCTGGTCGG-3′; Intron24 and exon25: 5′-TGGCAGACCTTGTAGACAGT-3′ and 5′-CGTCTTCCATGTCCTCCTC-3′.
Luciferase reporter assay
A mixture of ∼0.5 µg of plasmid DNA containing 0.2 µg Stat5b, 0.2 µg CPEB3, 0.1 µg firefly luciferase reporter and 5 ng Renilla luciferase were simultaneously transfected into 293T cells with Fugene HD (Roche). Cells were harvested 20–24 h after transfection and analyzed using Dual-Luciferase Reporter Assay System (Promega).
Animal surgery and Morris water maze
All experimental protocols were carried out following guidelines of the Institutional Animal Care & Utilization Committee. For intracerebroventricular drug administration, the 8-week-old male C57BL/6J mouse was anesthetized with ketamine before implanting a cannula at the right lateral ventricle (1.0 mm lateral and 0.5 mm caudal to bregma to a depth of 3.0 mm from the skull) that was attached to an Alzet osmotic Y pump (Model 1004) to infuse drug at 0.11 µl/h for 14 days. Recombinant EGF (Sigma-Aldrich) and gefitinib (first constituted to 5 mM in DMSO, Astra Zeneca) were diluted at the concentration of 50 µg/ml and 50 µM, respectively, in the vehicle (artificial cerebrospinal fluid containing 1 mg/ml BSA). One week after the surgery, mice with complete wound-healing were subject to Morris water maze task (28). Training for the hidden platform version consisted of four trials each day for four consecutive days. The probe trial was administered on the day after training completed. The visible platform task, which consisted of four trials for 1 day with the escape platform marked by a visible flag, was conducted to ensure the intact vision of mice. For all the trials, the maximal swimming duration was 60 s and the inter-trial interval was 60 min. The trajectories of mice were recorded and analyzed with a video tracking system, TrackMot (Singa Technology, Taiwan). Data were represented with mean ± S.E.M. and one-way and two-way ANOVA and post hoc LSD tests were used for statistics.
RESULTS
CPEB3 interacts with Stat5b and inhibits Stat5b-dependent transcription
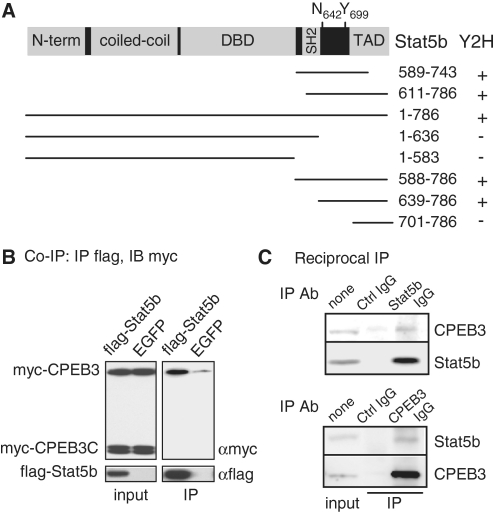
A yeast two-hybrid screen was performed using the N-terminal 427 amino acids of human CPEB3 as the bait to probe a mouse brain cDNA library. Two positive clones contained the C-terminal regions of Stat5b were identified (Figure 1A). Further analysis showed that the amino acids 639–700 between Src-homology (SH2) and transactivation domain (TAD) of Stat5b were crucial for its association with the N-terminus of CPEB3 (Figure 1A). In this region, tyrosine 699 (Y699) is phosphorylated upon cytokine and growth hormone stimulation that enhances Stat5b’s transactivation ability (13,14). When myc-tagged full length (myc-CPEB3) and C-terminal RNA-binding domain (myc-CPEB3C) of CPEB3 along with flag-tagged Stat5b were expressed in 293T cells, myc-CPEB3 but not myc-CPEB3C was co-precipitated with flag-Stat5b in the presence of RNase A treatment, indicating this interaction is mediated through the CPEB3 N-terminus in an RNA-independent manner (Figure 1B). Furthermore, using cortical neuronal lysate to perform reciprocal immunoprecipitation, endogenous CPEB3 was pulled down in the Stat5b immunoprecipitate and vice versa (Figure 1C).
Figure 1.
CPEB3 interacts with Stat5b. (A) Salient features of Stat5b showing the N-terminal region, coiled-coil domain, DNA-binding domain (DBD), Src-homology (SH2) and transactivation domain (TAD). The transcriptional activity of Stat5b is enhanced when the denoted tyrosine 699 (Y699) is phosphorylated upon growth hormone stimulation. When asparagine 642 mutated to histidine (N642H), it improves Stat5b’s activity by hindering dephosphorylation of phosphorylated Y699. Using the N-terminus (amino acids 1–427) of CPEB3 as the bait, a yeast two hybrid (Y2H) screen identified two clones corresponding to the amino acids 589–743 and 611–786 of Stat5b. The wild-type and truncated Stat5b mutants were tested for positive (+) or negative (–) interaction with the N-terminus of CPEB3. (B) Co-immunoprecipitation (Co-IP) assay. The plasmid expressing flag-Stat5b or a control EGFP along with plasmids encoding myc-tagged full-length (myc-CPEB3) and the C-terminal RNA-binding domain of CPEB3 (myc-CPEB3C) were cotransfected into 293T cells. The cell lysates were precipitated with flag antibody (Ab) and immunoblotted with myc Ab. IB: immunoblotting, IP: immunoprecipitation. (C) Reciprocal immunoprecipitation assay. The lysate from cortical neurons was precipitated with control, Stat5b or CPEB3 Ab and immunoblotted with CPEB3 and Stat5b Abs.
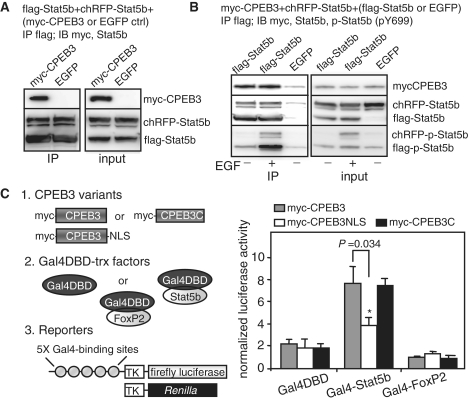
Since CPEB3 binds to a region of Stat5b containing the critical residue Y699, we questioned whether CPEB3 could interfere with Stat5b-mediated transcription and whether Y699 phosphorylation affects Stat5b and CPEB3 interaction. To monitor Stat5b-dependent transcription, we used the IGF1 promoter containing two Stat5b-binding sites or as a control, the both sites were mutated (mut) (29). The nuclear localization sequence (NLS) of SV40 large T antigen, PKKKRKVG, was appended to the C-terminus of CPEB3 to facilitate its nuclear localization since CPEB family proteins are predominantly cytoplasmic (Figure 2B, immunostained pictures). The wild-type (WT) and constitutively active (CA) forms of Stat5b, carrying the N642H mutation that hindered dephosphorylation of Y699 (30), were employed for the assay. The Renilla luciferase plasmid was included to normalize variation in transfection efficiencies. The ratio of normalized luciferase activity (i.e. firefly/Renilla) obtained in the presence of CA versus WT Stat5b was considered as fold of transcriptional activation caused by phosphorylated Y699 (p-Y699) (Figure 2A). This approach mimics cytokine-induced Stat5b phosphorylation-activated transcription while avoids simultaneously activating other signaling molecules to compromise the promoter assay (31). In the presence of enhanced green fluorescent protein (EGFP, a control), a 3-fold transactivation was observed in the wild-type but not the mutant promoter. Such activation was decreased to 2.2-fold with myc-CPEB3 and further reduced to 1.5-fold with nucleus-localized form of CPEB3 (myc-CPEB3NLS). In contrast, myc-CPEB3C, which was predominantly nuclear, did not affect Stat5b-induced transcription since it did not bind Stat5b (Figure 1B). Because Stat5bWT and Stat5bCA interact similarly with myc-CPEB3 (Figure 2C), the N642H replacement and Y699 phosphorylation have no effect on Stat5b’s association with CPEB3.
Figure 2.
CPEB3 inhibits Stat5b-dependent transcription. (A) The 293T cells were transfected with plasmids encoding (i) EGFP, myc-CPEB3, myc-CPEB3 fused with the nuclear localization sequence (myc-CPEB3NLS) or myc-CPEB3C, and (ii) wild-type (WT) or constitutively active (CA) Stat5b with N642H mutation, and (iii) reporters expressing firefly luciferase driven by the IGF1 promoter containing two wild-type or mutated Stat5b-binding sites along with Renilla luciferase. TK: thymidine kinase promoter. Stat5b-mediated transactivation is expressed as fold of induction of normalized luciferase activity (firefly/Renilla) obtained in the presence of CA verse WT Stat5b. Three independent experiments were analyzed and expressed as mean ± s.e.m. and the asterisks denote significant difference (Student’s t-test). (B) Subcellular distribution of myc-CPEB3, myc-CPEB3NLS and myc-CPEB3C in 293T cells. (C) CPEB3 interacts similarly with Stat5b and Stat5bCA. The extracts containing myc-CPEB3 and WT or CA flag-Stat5b, were precipitated with myc Ab and probed with flag, p-Stat5b and myc Abs.
CPEB3 suppresses Stat5b’s transcription activity without affecting dimerization and DNA binding of Stat5b
To examine whether CPEB3 could disrupt dimerization of Stat5b and hence inhibit Stat5b’s activity, flag-Stat5b and red fluorescence protein cherry (chRFP)-fused Stat5b along with myc-CPEB3 or EGFP were co-expressed in 293T cells. Figure 3A shows the amount of chRFP-Stat5b dimerized with flag-Stat5b is not influenced by myc-CPEB3. Moreover, growth factor-induced Y699 phosphorylation does not change Stat5b’s self-dimerization and association with CPEB3 (Figure 3B). Because p-Y699 is evidently detected in Stat5bCA (Figure 2B), Stat5b is constantly phosphorylated and dephosphorylated in 293T cells even without growth factor stimulation. To test whether CPEB3 could inhibit the transcription mediated by non-phosphorylated Stat5b, the Y699F mutant was used. The nuclear form of CPEB3 further diminished Stat5b-mediated transcription regardless of the phosphorylation status of Y699 (Supplementary Figure S1). To confirm that CPEB3-mediated suppression does not require interference with Stat5b’s DNA binding, Stat5b and a control, FoxP2, were fused to Gal4 DNA-binding domain (Gal4DBD) that bound to the Gal4-binding promoter sequences. In the presence of myc-CPEB3, myc-CPEB3NLS or myc-CPEB3C expression, the transactivation ability of Gal4DBD-Stat5b but not that of the controls was perceptibly inhibited by nucleus-localized CPEB3 (Figure 3C). Because CPEB3-NLS represses Stat5b’s activity greater than CPEB3, it is unlikely such inhibition is caused by sequestering Stat5b in the cytoplasm. Thus, CPEB3 suppresses Stat5b-dependent transcription without disrupting DNA binding, dimerization and nuclear localization of Stat5b and such a negative regulation is not dependent on Y699 phosphorylation of Stat5b.
Figure 3.
CPEB3 inhibits Stat5b-dependent transcription without affecting its dimerization and DNA-binding ability. (A) CPEB3 does not affect Stat5b dimerization. The 293T cells expressing flag-Stat5b and red fluorescence protein cherry (chRFP)-tagged Stat5b along with myc-CPEB3 or EGFP, were precipitated with flag Ab and probed with myc and Stat5b Abs. (B) Y699 phosphorylation in Stat5b has no effect on its dimerization as well as association with CPEB3. The 293T cells expressing myc-CPEB3 and red fluorescence protein cherry (chRFP)-tagged Stat5b along with flag-Stat5b or EGFP, were treated with or without EGF for an hour. The extracts were precipitated with flag antibody and probed with myc, Stat5b and pY699-Stat5b antibodies. (C) CPEB3 represses Stat5b-activated transcription without disrupting Stat5b’s DNA binding. The 293T cells were transfected with plasmids encoding (i) myc-CPEB3, myc-CPEB3NLS or myc-CPEB3C, and (ii) Gal4 DNA-binding domain (Gal4DBD) or Gal4DBD fused to Stat5b or FoxP2, and (iii) reporters containing firefly luciferase driven by the promoter containing five Gal4-binding sites and Renilla luciferase. The data from three sets of experiments is expressed as mean ± s.e.m. and the asterisks denote significant difference (Student’s t-test).
Activation of NMDARs influences nucleocytoplasmic distribution of CPEB3
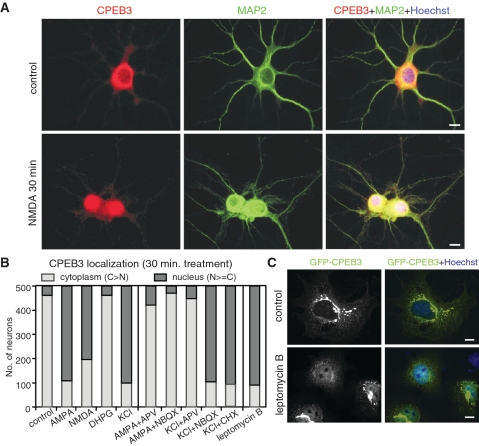
Because CPEB3-repressed Stat5b-dependent transcription was manifest even when CPEB3 was not appended with NLS (Figure 2A), we questioned if CPEB3 was a nucleocytoplasmic shuttling protein with longer residency in the cytoplasm, and whether neuronal activity could modulate CPEB3 distribution. Hippocampal neurons of day in vitro (DIV) 12 were stimulated with various glutamate receptor agonists, α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA), NMDA and 3,5-dihydroxyphenylglycine (DHPG), for 30 min and immunostained with affinity-purified CPEB3 antibody. Five hundred neurons were scored with the staining signal greater in the cytoplasm than in the nucleus categorized as cytoplasm-localized; while the rest was considered as nucleus-localized. The majority of unstimulated neurons showed a stronger CPEB3 signal in the cytoplasm with <10% of cells displaying nuclear distribution. AMPA and NMDA induced >60% of cells exhibited stronger or similar CPEB3 signals in the nucleus (examples in Figure 4A and quantified results in Figure 4B). To confirm the above observation was not caused by antibody’s cross-reactivity or changes in nuclear permeability, neurons expressing myc-CPEB3 and a control flag-FKBP8 were stimulated. Both AMPA and NMDA stimulations shifted the distribution of myc-CPEB3 but not flag-FKBP8 from cytoplasmic to nuclear dominance (Supplementary Figure S2A). KCl depolarization which triggered presynaptic release of glutamate and subsequently activated postsynaptic glutamate receptors, also resulted in nuclear CPEB3 accumulation in a NMDAR-dependent manner since the addition of NMDAR blocker, (2R)-amino-5-phosphonovaleric acid (APV), but not the AMPAR antagonist, 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzoquinoxaline-2, 3-dione (NBQX), was sufficient to prevent this event (Figure 4B). Interestingly, the AMPA effect on CPEB3 distribution was mediated through NMDAR signaling because it was blocked not only by NBQX but also by APV. Therefore, it appears that AMPAR activation, similar to KCl treatment, results in depolarization of neurons that potentiates synaptic release of glutamate and expels Mg2+ clogs from NMDARs, hence facilitates NMDAR activation. A translation inhibitor, cycloheximide (CHX), did not affect KCl-induced CPEB3 redistribution, so the nuclear accumulated CPEB3 is not newly synthesized (Figure 4B). Despite the NLS and nuclear export sequence (NES) in CPEB3 have not been identified, nuclear export was mediated by chromosome region maintenance 1 (CRM1) since blocking CRM1 by leptomycin B (LMB) induced nuclear accumulation of CPEB3. Moreover, application of LMB in COS cells expressing GFP-tagged CPEB3 also caused GFP-CPEB3 retained in the nucleus (Figure 4C), suggesting CPEB3 constantly shuttles between nucleo-cytoplasmic compartments and activation of NMDARs modulates its distribution equilibrium. In contrast, Stat5b was present in both nucleus and cytoplasm and the stimulation with AMPA, NMDA and DHPG did not change its distribution (data not shown). Although NMDAR signaling increased nuclear CPEB3, it did not affect Stat5b distribution (see immunostaining in Supplementary Figure S2B and biochemical fractionation in Supplementary Figure S3), indicating the two factors were unlikely co-transported in response to neuronal activity. Moreover, the co-immunoprecipitation assay demonstrated that CPEB3 and Stat5b remained associated in both cytoplasmic and nuclear extracts isolated from neurons treated with NMDA (Supplementary Figure S3).
Figure 4.
NMDAR signaling increases nuclear distribution of CPEB3. (A) Immunostaining of CPEB3 in cultured hippocampal neurons stimulated with or without NMDA for 30 min. MAP2 staining was used to identify neurons. (B) The summary result of CPEB3 localization from analyzing 500 neurons treated with various reagents for 30 min. The staining signal in the cytoplasm greater than that in the nucleus is considered as cytoplasm-localized (light gray bar) and the rest is grouped as nucleus-localized (dark gray bar). (C) The COS7 cells expressing GFP-CPEB3 were treated for 30 min with leptomycin B, fixed and stained for nuclei with Hoechst 33342. Scale: 10 µm.
Stat5b-activated EGFR transcription is downregulated by CPEB3
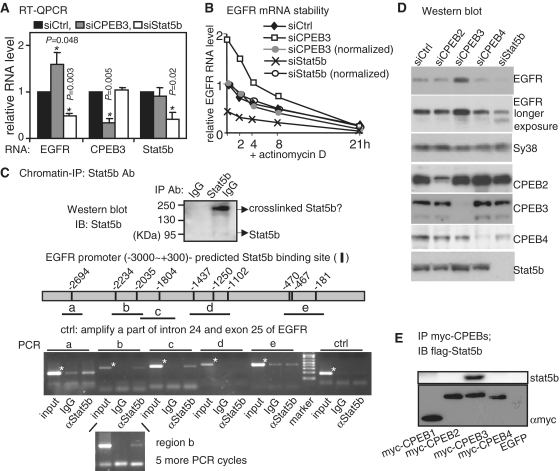
To identify a target gene regulated by Stat5b and CPEB3 interaction, we focused on EGFR because hepatic EGFR RNA was reduced in Stat5b knockout mice in the pervious microarray study (32) and the isotope-coded affinity tag (ICAT) proteomic analysis displayed a two-fold increase of EGFR protein in CPEB3 knockdown (KD) neurons (unpublished data). Hippocampal neurons of DIV6 were infected overnight with lentivirus containing or lacking a short hairpin sequence for rat CPEB3 or Stat5b. The infected neurons were harvested on DIV11 for RNA and protein analysis. The RNA levels of EGFR, CPEB3 and Stat5b were measured by quantitative PCR following reverse transcription (RT–QPCR). If EGFR transcription is activated by Stat5b whose transactivation ability is offset by CPEB3, a decrease and an increase in EGFR RNA level is expected, respectively, in Stat5bKD and CPEB3KD neurons as seen in Figure 5A. These changes are results of transcription since EGFR RNA stability is similar in the control and knockdown neurons (Figure 5B, normalized curves). To ascertain that Stat5b binds to the EGFR promoter, we performed chromatin immunoprecipitation (ChIP). Stat5b-binding sites in the proximal 3 kb EGFR promoter (Supplementary Figure S4) were predicted by MATCH analysis (33). Formaldehyde-fixed cortical neurons were lysed and precipitated with Stat5b antibody. In Figure 5C, an extra immunostained band of higher molecular weight likely represents crosslinked dimeric Stat5b as it was not observed when using unfixed neurons (Figure 1C). The promoter regions spanning the predicted Stat5b-binding sites at −2694, −2234, −2035 and −1804 were preferentially PCR-amplified from the Stat5b IgG-precipitated substance. In contrast, the areas around −1437, −1250, −1102, −470, −467, −181 as well as a control region spanning intron 24 and exon 25 of EGFR gene were not. The same Stat5b-binding regions in the EGFR promoter were also identified in another ChIP assay (Supplementary Figure S5). Finally, the EGFR protein level was examined using CPEB2, CPEB3, CPEB4 and Stat5b knockdown neurons. As expected, a reduction in CPEB3 leads to an increase in EGFR synthesis whereas a deficiency in Stat5b results in a decrease in EGFR expression (Figure 5D). The elevated EGFR expression in CPEB3 KD neurons could be rescued with the expression of full length human CPEB3 (hCPEB3), to a lesser extent with the N-terminus but not the C-terminus of hCPEB3 (Supplementary Figure S6). Intriguingly, the knockdowns of CPEB2 and CPEB4 have no obvious effect on EGFR expression (5). Since the nuclear history of mRNA could influence its expression pattern later in the cytoplasm (34), we determined if CPEB2 and CPEB4 did not regulate EGFR synthesis because of lacking interaction with Stat5b. In 293T cells that expressing flag-Stat5b along with various myc-tagged CPEBs, only myc-CPEB3 was co-immunoprecipitated with flag-Stat5b (Figure 5E). The inability of CPEB2 and CPEB4 to bind Statb5b is not due to their failure to translocate to the nucleus, as when CRM1-mediated export was blocked, GFP-CPEB2 and GFP-CPEB4 were accumulated in the nucleus (Supplementary Figure S7).
Figure 5.
EGFR transcription is regulated by Stat5b and CPEB3. (A) The RNA levels of EGFR, CPEB3 and Stat5b in CPEB3 and Stat5b knockdown (siCPEB3 and siStat5b) hippocampal neurons were measured by quantitative RT–PCR and expressed as a relative ratio to the control knockdown (siCtrl) which was arbitrarily set to 1. The results are expressed as mean ± s.e.m. from three independent experiments and the asterisks denote the difference of statistical significance (Student’s t-test). (B) The stability of EGFR RNA in siCtrl, siCPEB3 and siStat5b neurons was measured in the presence of actinomycin D. The amount of EGFR RNA expressed as a relative ratio to the time zero siCtrl which was arbitrarily set to 1. To normalize the differential EGFR transcript level caused by CPEB3 or Stat5b knockdown, the amounts of EGFR RNA at time zero of siCPEB3 and siStat5b were arbitrarily set to 1 (normalized, siCPEB3: gray circle, siStat5b: open circle). (C) Chromatin immunoprecipitation of cortical neuronal lysate with Stat5b or control IgG. The pulled-down substances were analyzed for proteins and DNA. The Western blot shows precipitated Stat5b and the band of 190 kDa molecular weight is likely to be crosslinked dimeric Stat5b. The predicted Stat5b-binding sites within the –3000 to +300 EGFR promoter and the PCR-amplified regions (a–e) are illustrated. The specific amplified bands are indicated with asterisks. The regions, a, b and c, were amplified more evidently in the Stat5b immunoprecipitate. To increase signal intensity of the region b detected in the Stat5b immunoprecipitate, five more PCR cycles were applied. (D) Immunoblots using extracts from hippocampal neurons knocked down with CPEB2, CPEB3, CPEB4 or Stat5b. (E) Stat5b interacts with CPEB3 but not other CPEBs. The lysates containing flag-Stat5b and one of myc-tagged CPEBs1–4 were precipitated with myc Ab and probed with myc and flag Abs.
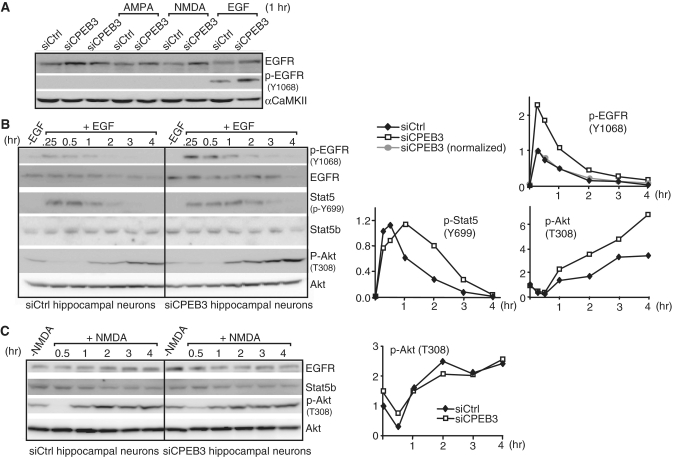
The kinetics of EGFR downstream signaling is altered in CPEB3 knockdown neurons
In cultured hippocampal neurons, the kinase activity of EGFR as assessed by autophosphorylation of Y1068 is relatively inert. Stimulating neurons with DHPG (data not shown), AMPA and NMDA apparently did not release, if any, sufficient amount of endogenous EGFR ligands to yield detectable p-Y1068 (Figure 6A). To address whether aberrant EGFR expression in CPEB3KD neurons changed downstream signaling, the neurons were stimulated with EGF and monitored for the activation of EGFR (p-Y1068), Stat5b (p-Y699) and Akt (p-T308) (Figure 6B). The increased p-Y1068 signal in KD neurons was caused by elevated EGFR expression; once normalized; the kinetics of EGFR activation (p-Y1068) was similar in control and KD neurons (Figure 6B, top plot). Since ligand-activated EGFR was endocytosed and degraded in proteasome and lysosome-dependent manner (35,36), a decline of EGFR level was observed in EGF-treated samples. In addition, EGF-stimulated CPEB3KD neurons displayed prolonged and augmented activation of Stat5b (p-Y699) and Akt (p-T308), respectively (Figure 6B, bottom plots). As a control, Akt activation showed similar kinetics in NMDA-stimulated control and CPEB3KD neurons, suggesting CPEB3KD effect on Akt signaling is distinctively tailored to specific stimulus like EGF (Figure 6C). In addition, the Y699 phosphorylation of Stat5b is not detectable upon NMDA stimulation, indicating NMDAR signaling does not affect Stat5b transcriptional activity via Y699 phosphorylation.
Figure 6.
The kinetics of EGFR downstream signaling is altered in CPEB3-deficient hippocampal neurons. (A) The tyrosine kinase activity of EGFR as measured by autophosphorylation of Y1068 (p-Y1068) was assessed using siCtrl and siCPEB3 neurons treated with AMPA, NMDA and EGF. (B) Western blots of p-EGFR, EGFR, p-Stat5, Stat5b, p-Akt and Akt proteins in neurons treated with EGF at the indicated time. The signals were quantified and plotted against the time of treatment. The p-EGFR signal (top plot) in siCPEB3 neurons was also normalized with the fold of increase in EGFR protein level that was determined from non-EGF treated samples (normalized, gray circle). The signals of p-Stat5b at Y699 and p-Akt at T308 are displayed in the bottom plots. The signals of p-EGFR and p-Stat5b at 0.25 h and p-Akt at 0 h (i.e. no EGF treatment) from the siCtrl samples were arbitrarily set to 1. (C) Same as the above except neurons were treated with NMDA. The p-EGFR and p-Stat5b were undetectable (data not shown). The plot shows the kinetics of Akt T308 phosphorylation in siCtrl and siCPEB3 neurons.
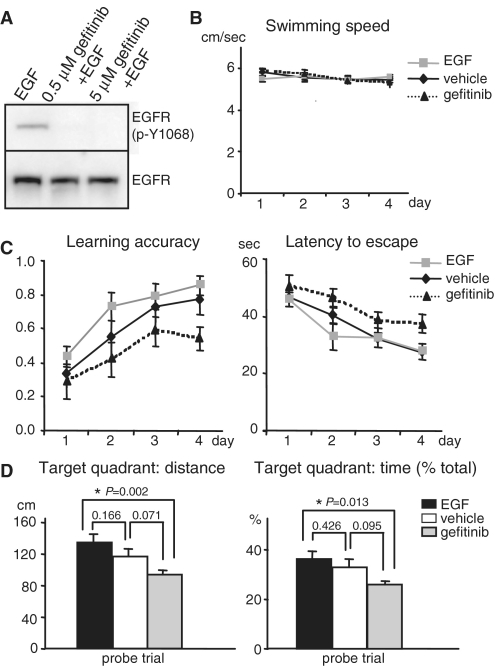
EGFR is involved in modulating learning and memory
Because activation of EGFR initiates several signaling pathways, such as PI3K-Akt and MAPK, in parallel that have been implicated in plasticity and memory, perturbation of this receptor tyrosine kinase activity locally in the brain may influence memory performance. To assess whether EGFR is a PRP that functions in learning and memory, pharmacological treatment of mice with EGF ligand and the inhibitor gefitinib was used. EGF-induced EGFR activation in the cultured neurons was effectively suppressed by gefitinib (Figure 7A). Male C57/BL6J mice with indwelling cannulae in the lateral ventricle infused with vehicle, EGF or gefitinib were tested for spatial learning and memory using Morris water maze. The mice received four spatial trials per day with a 60 min inter-trial interval and learned to locate a hidden platform using extra maze visual cues during 4-day spatial acquisition trials. No difference in swimming speed of all drug-treated animals was observed (Figure 7B). Animals infused with EGF trended towards enhanced performance in terms of accuracy and latency of locating the platform (Figure 7C). Such a mild change could be due to the reduced surface level of EGFR since persistent activation by EGF resulted in continuous endocytosis and degradation of the receptors (35,36). During the probe trial, the hidden platform was removed and the amount of time and distance mice spent in the original platform quadrant was recorded to test their consolidated memory after 4-day trials (Figure 7D). Deprivation of EGFR activity with gefitinib clearly retarded learning and memory performance as demonstrated in decreased accuracy and increased latency in each trial (Figure 7C) as well as reduced retention in the target quadrant in the probe trial (Figure 7D).
Figure 7.
EGFR influences spatial memory. (A) To test gefitinib potency, the hippocampal neurons were stimulated with EGF in the presence or absence of gefitinib. The EGFR kinase activity was indicated by p-Y1068. (B) No difference in the swimming speed in the water maze task among vehicle, EGF and gefitinib-treated mice. (C) Intervention of the kinase activity of EGFR in the brain with EGF (activation) and gefitinib (inhibition) affects the learning accuracy and latency to escape in Morris water maze tasks. The accuracy was calculated per day as the trial number that the mice successfully climb onto the invisible platform divided by total four trials. The gefitinib group (n = 10) showed slower and impaired learning in Morris water maze when compared to the control vehicle group (n = 11) as judged by reduced learning accuracy (left plot, P = 0.021) and increased latency (right plot, P = 0.016) to locate the invisible platform for the four consecutive training days. In contrast, the EGF-treated group (n = 12) showed a trend of enhanced learning accuracy (left plot, P = 0.074) and decreased latency (right plot, P = 0.452). The P-value was calculated using the two-way ANOVA least square difference (LSD) analysis. (D) For the probe trial when the platform was removed after the 4-day training, the distance (left plot) and the percentage of time (right plot) that EGF- and gefitinib-treated mice swam in the target quadrant where the platform used to be were increased and reduced, respectively, when compared to the vehicle-infused mice. The P-value was calculated using the one-way ANOVA LSD test to compare the statistical difference between groups.
DISCUSSION
We have identified a novel role for the RNA-binding protein, CPEB3 in transcription. All CPEBs, originally thought to be localized in the cytoplasm, are now proteins shuttling between nucleocytoplasmic compartments. Two recent studies have shown that CPEB1 shuttles to the nucleus where it possibly plays a role in ribosomal biogenesis (37) and alternative splicing (38). One nuclear function of CPEB3 is to downregulate Stat5b’s transcriptional activity; whereas the roles of nuclear CPEB2 and CPEB4 remain to be determined. Interestingly, NMDAR signaling reorganizes CPEB3 distribution from cytoplasmic to nuclear prevalence, suggesting neuronal activity could regulate and partition CPEB3′s functions between the two compartments to control gene expression. One target gene transcriptionally regulated by CPEB3 is EGFR of which function in modulating learning and memory has been pharmacologically demonstrated in the present study.
The classical cytokine-Stat-signaling draws a paradigm in which cytokine-induced phosphorylation on a critical tyrosine residue of cytoplasmic Stats is the prerequisite to initiate dimerization and subsequent nuclear translocation of Stats. Thus, this phosphorylation is essential to transduce the extracellular signal to the nucleus and to activate Stat-dependent gene transcription (15,39). However, the finding that non-phosphorylated Stat1 (Y701F mutant) could bind to and activate the low molecular weight polypeptide-2 promoter (40) and accumulating evidences have subsequently demonstrated this phosphorylation event is not necessarily required for Stat’s dimer formation, constitutive transit in and out of nucleus and transcriptional activation (41) that are in accordance with our findings in this study. Silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT), which binds to the N-terminal coiled-coil domain of STAT5 and suppresses the induction of STAT5 target genes (42) is the only identified transcriptional co-repressor of Stat5. Here, we have uncovered CPEB3 as another negative regulator for Stat5b without affecting Stat5b’s dimerization, nuclear localization and DNA binding. The activation domains of transcription factors are known to activate gene transcription via association with a diverse array of coactivators, some of which promote chromatin remodeling, whereas others, such as TFIID and mediator complexes, direct activator recruitment to the transcriptional machinery (43–45). Thus, we hypothesize that the binding of CPEB3 to a region next to the Stat5b’s activation domain likely interferes the association of Stat5b with other coactivators to repress Stat5b-activated transcription. Thus, identification of which coactivator complexes are involved in integrating Stat5b signal and convey it to activate the basal transcription apparatus needs to be first investigated prior to testing our hypothesis.
Although several RNA-binding proteins, such as Pbx-regulating protein 1 (46,47) and FUSE-binding proteins (48), are also able to bind DNAs and function as transcription factors, to our knowledge, nothing is known for a translational repressor in complex with a transcription factor to regulate gene expression. A recent study has identified ribosomal protein S3 (RPS3), a KH domain RNA-binding protein, was in complex with NF-κB and enhanced NF-κB-mediated transcription (49). Although RPS3 is a subunit of 40S ribosome and has a role in general translation, no specific mRNA has been identified to associate with RPS3. Despite CPEB2 and CPEB4 share identical RNA-binding specificity with CPEB3 and constantly shuttle between nucleocytoplasmic compartments, they have no obvious influence on the level of EGFR once knocked down in neurons. Since CPEB2 and CPEB4 do not associate with Stat5b, it appears that the interaction between a translation regulator and a transcription factor may help CPEB3 has regulatory roles in a spectrum of genes different from CPEB2 and CPEB4. The nuclear import of Stat5b is not influenced by CPEB3 and NMDA. Because the fact that Stat5b activates transcription of EGFR gene and CPEB3 offsets its activity, it seems illogical for CPEB3 and Stat5b to import simultaneously to the nucleus. Moreover, CPEB3 and Stat5b interaction is not disrupted by NMDA treatment and a nuclear form of CPEB3, CPEB3NLS, has stronger suppression effect on Stat5b-dependent transcription than CPEB3 in the promoter assays, so it is expected that NMDA-induced nuclear accumulation of the repressor CPEB3 is likely to further downregulate Stat5b-dependent EGFR gene transcription in neurons. Nonetheless, due to the long half-life of EGFR mRNA (>4 h, Figure 5B) and prolonged NMDAR activation-induced neuronal toxicity, we can not directly demonstrate that EGFR transcription is downregulated by NMDA stimulation in a CPEB3-dependent manner. It remains to be identified the specific importins and non-canonical nuclear localization sequences in CPEB3 responsible for its import to further address the physiological significance of NMDA-induced nuclear accumulation of CPEB3.
Because pharmacological intervention of the kinase activity of EGFR in the brain affects spatial memory, EGFR is a PRP that modulates learning and memory. Hence, there should be caution regarding potential cognitive side-effects when using EGFR-targeted drugs to treat brain tumors. Since EGFR signaling leads to parallel activation of several processes, such as MAPK and PI3K-Akt signaling pathways that have been demonstrated to play important roles in plasticity and memory (50–53), even a subtle change in EGFR expression likely has significant impact on plasticity. In CPEB3-deficient neurons, the EGFR level is elevated. Interestingly, EGFR expression was found upregulated in forebrain regions of schizophrenics (54) and EGF administration in neonatal rats induced various behavioral hallmarks of schizophrenia (55). Generation of conditional CPEB3 knockout mice is in process to address whether aberrant EGFR expression will cause abnormal signaling and aberrant behaviors in mice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council and National Health Research Institute in Taiwan; National Research Program for Genomic Medicine Grants for the RNAi Core. Funding for open access charge: National Science Council in Taiwan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Peter Rotwein for Stat5b and IGF1 plasmids, Young-Sun Lin for the Gal4 system, Roger Tsien for cherry RFP plasmid, Pang-Hsien Tu for TDP43 antibody and Joel Richter for CPEB3 antibody. The authors appreciate Chun- Kuei Su for sharing stereotaxic instrument, Pan-Chyr Yang for advice on Iressa, Yu-Chia Lin and Ching-Pang Chang for assistance on behavior study. The authors thank Ya-Ping Lin in the institutional core for conducting ICAT analysis and the RNAi Core Facility in Academia Sinica for the Stat5b shRNA clone.
REFERENCES
- 1.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 2.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 3.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl Acad. Sci. USA. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 8.Vogler C, Spalek K, Aerni A, Demougin P, Muller A, Huynh KD, Papassotiropoulos A, de Quervain DJ. CPEB3 is Associated with Human Episodic Memory. Front Behav. Neurosci. 2009;3:4. doi: 10.3389/neuro.08.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 10.Richter JD. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 12.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Johnston JA, Bacon CM, Finbloom DS, Rees RC, Kaplan D, Shibuya K, Ortaldo JR, Gupta S, Chen YQ, Giri JD, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc. Natl Acad. Sci. USA. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertens C, Darnell JE., Jr SnapShot: JAK-STAT signaling. Cell. 2007;131:612. doi: 10.1016/j.cell.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 16.Ninomiya M, Yamashita T, Araki N, Okano H, Sawamoto K. Enhanced neurogenesis in the ischemic striatum following EGF-induced expansion of transit-amplifying cells in the subventricular zone. Neurosci. Lett. 2006;403:63–67. doi: 10.1016/j.neulet.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 17.Torroglosa A, Murillo-Carretero M, Romero-Grimaldi C, Matarredona ER, Campos-Caro A, Estrada C. Nitric oxide decreases subventricular zone stem cell proliferation by inhibition of epidermal growth factor receptor and phosphoinositide-3-kinase/Akt pathway. Stem Cells. 2007;25:88–97. doi: 10.1634/stemcells.2006-0131. [DOI] [PubMed] [Google Scholar]
- 18.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Sibilia M, Steinbach JP, Stingl L, Aguzzi A, Wagner EF. A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. EMBO J. 1998;17:719–731. doi: 10.1093/emboj/17.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornblum HI, Hussain R, Wiesen J, Miettinen P, Zurcher SD, Chow K, Derynck R, Werb Z. Abnormal astrocyte development and neuronal death in mice lacking the epidermal growth factor receptor. J. Neurosci. Res. 1998;53:697–717. doi: 10.1002/(SICI)1097-4547(19980915)53:6<697::AID-JNR8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 21.Terlau H, Seifert W. Influence of epidermal growth factor on long-term potentiation in the hippocampal slice. Brain Res. 1989;484:352–356. doi: 10.1016/0006-8993(89)90380-6. [DOI] [PubMed] [Google Scholar]
- 22.Ishiyama J, Saito H, Abe K. Epidermal growth factor and basic fibroblast growth factor promote the generation of long-term potentiation in the dentate gyrus of anaesthetized rats. Neurosci. Res. 1991;12:403–411. doi: 10.1016/0168-0102(91)90071-6. [DOI] [PubMed] [Google Scholar]
- 23.Arteaga CL, Johnson DH. Tyrosine kinase inhibitors-ZD1839 (Iressa) Curr. Opin. Oncol. 2001;13:491–498. doi: 10.1097/00001622-200111000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell. Biol. 12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 25.Hagele S, Kuhn U, Boning M, Katschinski DM. Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1alpha mRNA 3′-UTR and modulate HIF-1alpha protein expression. Biochem. J. 2009;417:235–246. doi: 10.1042/BJ20081353. [DOI] [PubMed] [Google Scholar]
- 26.Huang YS, Richter JD. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol. 2007;431:143–162. doi: 10.1016/S0076-6879(07)31008-2. [DOI] [PubMed] [Google Scholar]
- 27.Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- 28.Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 29.Woelfle J, Chia DJ, Rotwein P. Mechanisms of growth hormone (GH) action. Identification of conserved Stat5 binding sites that mediate GH-induced insulin-like growth factor-I gene activation. J. Biol. Chem. 2003;278:51261–51266. doi: 10.1074/jbc.M309486200. [DOI] [PubMed] [Google Scholar]
- 30.Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, Kitamura T. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J. Biol. Chem. 2000;275:24407–24413. doi: 10.1074/jbc.M909771199. [DOI] [PubMed] [Google Scholar]
- 31.Toda T, Wada H, Ogawa S, Tani-ichi S, Ikuta K. A reporter assay for Stat5-dependent promoters without cytokine stimulation. Anal. Biochem. 2008;372:250–252. doi: 10.1016/j.ab.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol. Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 33.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell. Dev. Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J. Biol. Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 36.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol. Biol. Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CL, Evans V, Shen S, Xing Y, Richter JD. The nuclear experience of CPEB: Implications for RNA processing and translational control. RNA. 2009;16:338–348. doi: 10.1261/rna.1779810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin. Cell Dev. Biol. 2008;19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 2001;20:6836–6844. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis BA, Reinberg D. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell. Sci. 2003;116:3667–3675. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 44.Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 46.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villaescusa JC, Buratti C, Penkov D, Mathiasen L, Planaguma J, Ferretti E, Blasi F. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One. 2009;4:e5213. doi: 10.1371/journal.pone.0005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 49.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 50.Opazo P, Watabe AM, Grant SG, O'Dell TJ. Phosphatidylinositol 3-kinase regulates the induction of long-term potentiation through extracellular signal-related kinase-independent mechanisms. J. Neurosci. 2003;23:3679–3688. doi: 10.1523/JNEUROSCI.23-09-03679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin CH, Yeh SH, Lin CH, Lu KT, Leu TH, Chang WC, Gean PW. A role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron. 2001;31:841–851. doi: 10.1016/s0896-6273(01)00433-0. [DOI] [PubMed] [Google Scholar]
- 52.English JD, Sweatt JD. Activation of p42 mitogen-activated protein kinase in hippocampal long term potentiation. J. Biol. Chem. 1996;271:24329–24332. doi: 10.1074/jbc.271.40.24329. [DOI] [PubMed] [Google Scholar]
- 53.Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat. Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 54.Futamura T, Toyooka K, Iritani S, Niizato K, Nakamura R, Tsuchiya K, Someya T, Kakita A, Takahashi H, Nawa H. Abnormal expression of epidermal growth factor and its receptor in the forebrain and serum of schizophrenic patients. Mol. Psychiatry. 2002;7:673–682. doi: 10.1038/sj.mp.4001081. [DOI] [PubMed] [Google Scholar]
- 55.Futamura T, Kakita A, Tohmi M, Sotoyama H, Takahashi H, Nawa H. Neonatal perturbation of neurotrophic signaling results in abnormal sensorimotor gating and social interaction in adults: implication for epidermal growth factor in cognitive development. Mol. Psychiatry. 2003;8:19–29. doi: 10.1038/sj.mp.4001138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.