Abstract
MicroRNAs (miRNAs) are small regulatory RNAs targeting multiple effectors of cell homeostasis and development, whose malfunctions are associated with major pathologies such as cancer. Herein we show that GAM/ZFp/ZNF512B works within an intricate gene regulatory network involving cell-cycle regulators, TGFβ effectors and oncogenic miRNAs of the miR-17-92 cluster. Thus, GAM impairs the transcriptional activation of the miR-17-92 promoter by c-Myc, downregulates miR-17-92 miRNAs differentially, and limits the activation of genes responsive to TGFβ canonical pathway. In contrast, TGFβ decreases GAM transcripts levels while differentially upregulating miR-17-92 miRNAs. In turn, miR-17, miR-20a and miR-92a-1 target GAM transcripts, thus establishing a feedback autoregulatory loop. GAM transcripts are also targeted by miRNAs of the let-7 family. GAM downregulates Drosha, the main effector of miRNA maturation in the nucleus, and interacts with it in a RNA-dependent manner. Finally, GAM modulates the levels of E2F1 and Ras, and increases apoptosis while reducing cell proliferation. We propose that GAM represents a new kind of vertebrate regulator aimed at balancing the opposite effects of regulators of cell homeostasis by increasing the robustness of gene circuitries controlling cell proliferation, differentiation and development.
INTRODUCTION
MicroRNAs (miRNAs) have been implicated in the regulation of a number of fundamental processes, including muscle, cardiac, neural and lymphocyte development, or the regulation of both the innate and adaptive immune responses (1,2). MiRNAs originate from primary transcripts (pri-miRNAs) converted in the nucleus into precursor miRNAs (pre-miRNAs) by the RNase III Drosha, associated with DGCR8 to form the small microprocessor complex (3). Pre-miRNAs are then exported in the cytoplasm where the miRNA hairpin is cleaved by the RNase III Dicer within the RISC loading complex. The guide strand, which corresponds to the mature miRNA, is then incorporated into the RISC complex (3). MiRNAs and their transcriptional regulators usually form autoregulatory loops aimed at controlling their respective levels (4).
MiRNAs participate in gene regulatory networks whose molecular malfunctions are associated with major pathologies such as cancer (5–7). Thus, the miR-17-92 cluster on chromosome 13, which appeared with the vertebrates, contains six miRNAs (miR-17, -18a, -19a, -20a, -19b-1 and -92a-1). The miR-17-92 cluster is amplified and overexpressed in B-cell lymphomas and solid tumors such as breast or small-cell lung cancers (5), where it may enhance oncogenesis by potentially targeting E2F1, p21/CDKN1A and BCL2L11/BIM (8). However, there is also some evidence suggesting that loss-of-function of miR-17-92 miRNAs might be advantageous for cancer cells in certain settings. Indeed, loss-of-heterozygosity at the 13q31.3 locus has been observed in multiple tumor types, and a genome-wide analysis of copy number alterations in cancer revealed that the miR-17-92 cluster was deleted in 16.5% of ovarian cancers, 21.9% of breast cancers and 20% of melanomas (9). The transcription of the miR-17-92 cluster is controlled by c-Myc and E2F factors (10–13). Interestingly, miR-17 and miR-20a target E2F1, while miR-20a also targets E2F2 and E2F3 (10,12). Two ortholog clusters are found on chromosomes 7 (miR-106b-93-25) and X (miR-106a-18b-20b-19b-2-92a-2-363), respectively. In contrast to miR-17-92 miRNAs that appeared late in evolution, let-7 miRNAs belong to a family of early developmental regulators well conserved during evolution (14). The let-7 gene family expanded from one member in nematods to more than 10 in mammals. In C. elegans, let-7 works as a master regulator of temporal patterning by controlling the transition from undifferentiated, proliferating stem cells to differentiated, quiescent cells (14). In human, let-7 miRNAs are frequently downregulated in cancers like lung, colon or other solid tumors (5), and are therefore considered as tumor suppressor miRNAs in these types of cancers. In particular, let-7 miRNAs target oncogenes of the Ras family (15) and c-Myc, and their expression in colon tumors results in reduced levels of both Ras and c-Myc (16).
GAM/ZFp/ZNF512B, a vertebrate-specific zinc-finger factor, was recently identified as a potential developmental regulator whose isoform shifts correlated with the main stages of skin and heart morphogenesis in chicken (17). In human, GAM transcripts are present in all tissues and cell types tested so far. Although GAM may possibly bind TC- and/or AG-rich DNA sequences (17), none of its target genes has been identified to date, and its functions remain elusive. Interestingly, GAM 3′-UTR (untranslated region) contains target sequences for ∼150 miRNAs, several of whose, including miR-17-92 and let-7 miRNAs, have been implicated in the regulation of development and/or have been described either as oncomirs or tumor suppressor miRNAs. Given that miRNAs can potentially target tens to hundreds of different transcripts, we hypothesized that GAM and at least some of the miRNAs targeting its 3′-UTR may form regulatory gene circuitries aimed at regulating cell homeostasis through their combined effects on key regulators of cell proliferation. Here, we present the first evidence that GAM indeed may work as a molecular sensor potentially aimed at maintaining cell homeostasis by affecting the levels of regulators of cell homeostasis such as E2F1 and Ras, by modulating the levels of miR-17-92 miRNAs, by regulating the levels of and interacting with Drosha, and by interfering with TGFβ signaling.
MATERIALS AND METHODS
Cell culture and transfection
HEK-293, HepG2 and MCF7 were transfected using Lipofectamine 2000 (Invitrogen). K562 cells were electroporated using the AMAXA kit. Whenever needed, cells were mock-treated or stimulated 34 h after transfection with TGFβ (100 ng/ml, from R&D) for 14 h. Stealth oligonucleotides targeting GAM (siGAM), Drosha (siDrosha) and c-Myc (siMyc) were from Invitrogen.
Preparation of constructs
The human GAM cDNA was amplified by PCR from a mix of human cDNAs purchased from Clontech. The miR-17-92 cluster promoter, the whole miR-17-92 cluster sequence and the 3′-UTR of GAM were cloned by PCR from genomic DNA extracted from HEK-293 cells. GAM 3′-UTR was cloned downstream of the luciferase gene in the XbaI site of the pGL3-Control vector (Promega) or downstream of GAM encoding sequence in the pCMV-HA vector. The whole sequence of the miR-17-92 cluster was inserted into the pCMV vector. The c-Myc expression vector was from Invitrogen. The sequences of the oligonucleotides used for cloning are available upon request.
Luciferase assays
HEK-293 cells plated in 12-well plates were transfected with 0.4 µg of either the empty pGL3-Control vector or constructs containing miRNA target sites, the indicated pre-miRNAs (100 nM final, Ambion) and 20 ng of Renilla luciferase control vector (pRL-TK from Promega). Assays were performed 24 h after transfection using the Dual Luciferase Reporter Assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. As a control, we used the pre-miR Precursor Molecule-Negative Control #1 (referred to here as control RNA) from Ambion. TGFβ signaling in HepG2 cells was tested in sixplicates in 24-well plates using the Cignal SMAD Reporter Assay Kit (Luciferase) from Super Array Bioscience Corporation. This kit is designed to monitor the activity of the canonical TGFβ-induced signal transduction pathway, i.e. to give a measure of the rate of SMAD2 and/or SMAD3 nuclear transcriptional activity in conjunction with SMAD4 on a reporter construct containing a functional SMAD-binding site.
Western blots
Cells transfected and/or treated as needed were lysed 24 h or 48 h after transfection. Anti-p21/CDKN1A and -α-tubulin antibodies were from Cell Signaling Technology. Anti-c-Myc and anti-Drosha antibodies were from Abcam. Anti-E2F1, anti-pan-Ras, anti-GAM/ZNF512B, anti-Dicer, anti-DGCR8 and anti β-Actin antibodies were from Santacruz.
Apoptosis and proliferation assays
Apoptosis and proliferation assays were conducted using the Mitochondrial Permeability Transition Detection Kit from Immunochemistry Technologies and the XTT Cell Proliferation Kit from Cayman, respectively, according to the manufacturer's instructions.
RNA and miRNA quantitative real-time PCR
RNAs extracted with TRIzol (Invitrogen) were subsequently subjected to DNase digestion (Turbo-DNase-Ambion). GAM and Drosha quantitative real-time PCRs (qRT–PCRs) were conducted after reverse transcription with the commercial ready-to-use primers/probe mix of Applied Biosystems. Values were normalized using GAPDH or β-actin. MiRNA qRT–PCR were performed using TaqMan MicroRNA Assays (Applied Biosystems). Values were normalized using RNU-44. Real-time PCRs were run in triplicates from three different cDNAs. As miR-92a-1 and -92a-2 sequences are 100% identical, they are collectively referred to as miR-92a in qRT–PCR experiments.
RNase-protection assays
Sense oligonucleotides used for the preparation of double-strand DNA templates for T7 RNA polymerase were as follows: miR-17: 5′-CAAAGTGCTTACAGTGCAGGTAG(T)13; miR-18a: 5′-TAAGGTGCATCTAGTGCAGATAG(T)13; miR-20a: 5′-TAAAGTGCTTATAGTGCAGGTAG(T)13; RNAs containing miR-92a1: 5′-GTTTCTGTATGGTATTGCACTTGTCCCGGCCTGTTGAGTTTG (the position of miR-92a-1 is given by bolded underlined letters); U6snRNA: 5′-CGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGG. Antisense RNA probes were synthesized using the mirVana miRNA Probe Construction Kit from Ambion in the presence of α32P-CTP (3000 Ci/mmol) and subsequently purified onto a denaturing TBE-Urea 15% polyacrylamide gel. The resulting antisense probes are 10-nt longer than the protected fragments. RNAs extracted with TRIzol (Invitrogen) or yeast tRNA were incubated overnight at 42°C in the presence of 8 × 104 cpm of the antisense RNA probes in the hybridization buffer provided by the mirVana miRNA Detection Kit from Ambion. RNase-digestion were done for 30 min at 37°C using a 1/50 dilution of the RNAse A/RNase T1 solution provided by the same kit. Samples were subsequently separated on a denaturing TBE-Urea 15% polyacrylamide gel in the presence of radiolabeled Ambion Decade Markers (not shown on the figures due to the necessity to overexpose the gels to allow for the detection of the miR-17-92 miRNAs). Gels were then subjected to autoradiography at −80°C in the presence of an amplifying screen.
Promoter activity studies
MCF7 cells were transfected with the empty pGL3-Basic (Promega) or a construct in pGL3-Basic containing different fragments of the miR-17-92 cluster promoter inserted upstream of the Luciferase gene, together with the pRL-TK vector for internal control, along with either pCMV-HA, pCMV-HA-GAM, pCMV-c-Myc, or their combinations, or with a Control RNA or with either siGAM and/or siMyc. The Firefly luciferase activity was measured 48 h post-transfection and normalized to Renilla luciferase activity.
Protein interaction assays
HEK-293 cells were transfected with either an empty control vector or a construct expressing the tagged factor of interest. Cells were lysed 24 h after transfection in 1 ml of NP-40 lysis buffer supplemented with 0.5 mM ZnCl2. Supernatants were incubated overnight with anti-HA (Covance) or anti-Flag (Sigma) antibody. HA- or Flag-expressing lysates were further incubated with protein A/G sepharose (Amersham). The bound proteins were subjected to three washes of 30 min each and then eluted by boiling for 5 min in the loading buffer. Whenever needed, RNase-digestion was conducted at 4°C for 30 min before addition of the anti-HA antibody using RNase A from Qiagen (200 µg/ml). Immunocomplexes were analyzed by SDS–PAGE electrophoresis followed by immunoblotting with anti-Drosha, anti-Flag or anti-HA.
Statistical analysis and quantification
Comparisons with the respective controls were done using the Student’s t-test, and P-values are provided in the figure legends. Band quantification was done using the Adobe Photoshop software. The relative intensities of the bands of interest, prealably normalized to either α−tubulin (western blots) or U6snRNA (RNase-protection assays), are given under each panel in percent of the respective control sample.
RESULTS
GAM transcripts are targeted by miR-17-92 and let-7 miRNAs
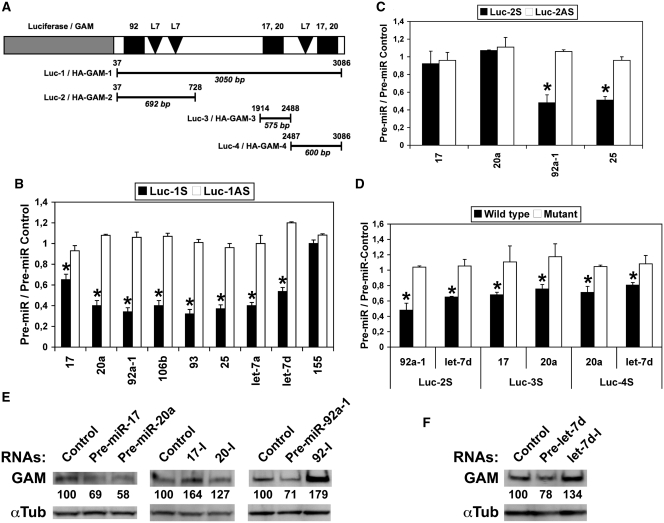
In silico analyzes using the TargetScan software (http://www.targetscan.org) showed that the 3′-UTR of human GAM contains conserved putative target sites for ∼150 different miRNAs, including three sites for miR-17-92 miRNAs (one for miR-92a-1 and two for miR-17/20a) or their respective orthologs and three for let-7 miRNAs (Figure 1A and Supplementary Tables S1 and S2). In order to validate these target sites, the 3′-UTR of human GAM was inserted in both orientations downstream of the luciferase gene in the pGL3-Control vector, providing us with sense (Luc-1S) and antisense (Luc-1AS) constructs collectively referred to as Luc-1 on Figure 1A. Three shorter clones (Luc-2, -3 and -4) were prepared similarly. Transfecting HEK-293 cells with pre-miR-17, -20a and -92a-1 or their respective orthologs pre-miR-106b, -93 and -25 significantly decreased the expression of Luc-1S while remaining without effects on Luc-1AS (Figure 1B). Similar results were obtained with two let-7 pre-miRNAs, namely pre-let-7a and -let-7d (Figure 1B). In contrast, miR-155 remained without effects, in accordance with the fact that GAM 3′-UTR contains no miR-155 target site. The expression of Luc-2S was also reduced by either pre-miR-92a-1 or -25, but neither by pre-miR-17 nor -miR-20a, due to the lack of their common target sites (Figure 1C). Luc-2S was also targeted by let-7d (Figure 1D). Furthermore, miR-17, -20a, -106b and -93 targeted Luc-3S and/or Luc-4S, the latter being also targeted by let-7d (Supplementary Figure S1A and B). Finally, mutating the different target sites (Supplementary Tables S1 and S2) abolished the effects of the different pre-miRNAs (Figure 1D).
Figure 1.
GAM transcripts are direct targets of miR-17-92 and let-7 miRNAs. (A) Schematic representation of GAM 3′-UTR constructs in pGL3-Control (Luc-1, -2, -3 and -4) and pCMV-HA (HA-GAM-1, -2, -3 and -4). Conserved putative target sites for miR-17/20a (17,20), miR-92a-1 (92) and let-7 (L7) miRNAs are indicated. The positions of the 5′- and 3′-limits of the inserts with respect to GAM 3′-UTR are indicated over the lines, and their respective lengths are given in bp below the lines. (B–D) Luciferase assays were done with Luc-1 to Luc-4 as indicated, both in sense (S) and antisense (AS) orientation. Mutations in constructs used in D are given in Supplementary Tables S1 (miR-17-92 miRNA target sites) and S2 (let-7 miRNA target sites). Bars show the ratios of the Firefly luciferase (normalized to Renilla luciferase) activity measured following transfection with the indicated pre-miRNAs to that obtained following transfection with the pre-miR Control for the same construct. Values represent the mean ± SD (n = 4). (B and C) *P < 0.001, significantly different from pre-miR Control. (D) *P < 0.005, significantly different from wild type construct. (E and F) Endogenous GAM levels in HEK-293 cells transfected with either a pre-miR-Control, the indicated pre-miRNAs or antisense inhibitory RNAs to miR-17, miR-20a or miR-92a (17-I, 20-I and 92-I, respectively) were determined by western blotting.
To determine if the targeting of GAM 3′-UTR by the above miRNAs would also reduce GAM protein accumulation, a human GAM cDNA encoding the same protein as GenBank AB033022 sequence except for a conservative proline to leucine change was inserted upstream of the different parts of GAM 3′-UTR in the pCMV-HA expression vector (Figure 1A). As expected, the levels of HA-GAM-1 and HA-GAM-2 were reduced by either pre-miR-17, -20a, -92a-1 or their respective orthologs (Supplementary Figure S2A), and the downregulation of HA-GAM-4 by pre-miR-20a was impaired by mutating the corresponding miR-17/20a target site (Supplementary Figure S2B). Finally, an anti-GAM antibody allowed us to definitely ascertain the downregulating effects of pre-miR-17, -20a, -92a-1 and -let-7d (Figure 1E and F, respectively). In contrast, as one would expect, transfecting HEK-293 cells with either antisense miR-17, -20a, -92a or let-7d inhibitory RNAs increased GAM levels (Figure 1E and F), which shows that both miR-17-92 and let-7 miRNAs regulate GAM expression in vivo. Collectively, these results give strong evidence that these two families of miRNAs should actually target the 3′-UTR of GAM transcripts in cell.
GAM regulates E2F1 and Ras levels as well as cell homeostasis
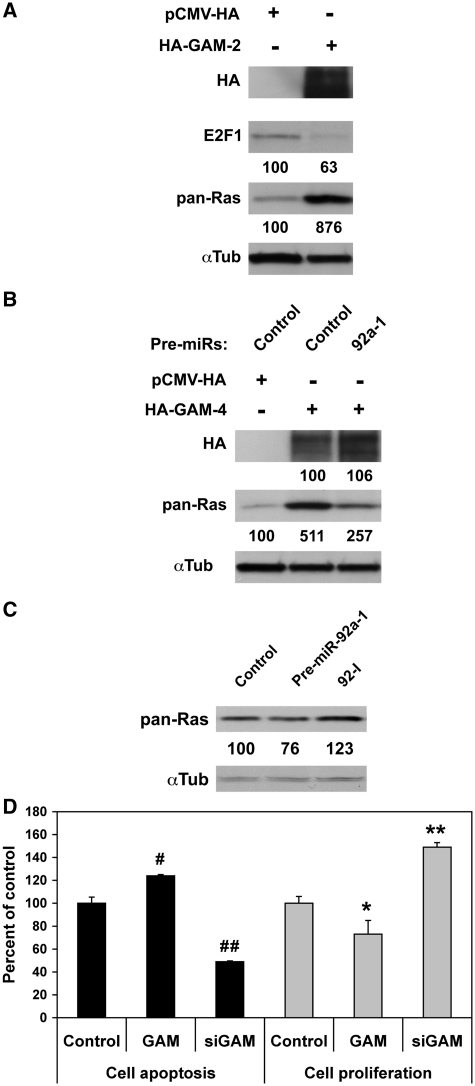
As E2F1 was previously shown to be targeted by both miR-17 and miR-20a (10,12), the blots from the middle and right panels of Supplementary Figure S2A were reprobed with an anti-E2F1 antibody. Expectedly, pre-miR-20a and -106b downregulated E2F1 and HA-GAM-1. Given that the two corresponding transcripts contain target sites for miR-17/20a, this provides good evidence that the effects of pre-miR-17-92 miRNAs on HA-GAM levels were due to the production of fully functional miRNAs (Supplementary Figure S2A). In contrast, while downregulating HA-GAM-1 and HA-GAM-2, pre-miR-92a-1 and its ortholog pre-miR-25 increased E2F1 levels, suggesting that they may normally work to limit certain effects of miR-17 and miR-20a, specially those related to the regulation of E2F1 expression.
Given the opposite effects of miR-17, -20a and -92a-1 on E2F1 levels despite their common downregulating effects on endogenous GAM (Supplementary Figure S2A and Figure 1E, respectively), we then checked if increasing GAM levels may have any consequences on E2F1 accumulation. Overexpressing HA-GAM-2 reduced E2F1 level by ∼40% (Figure 2A). In contrast, siRNAs directed against GAM transcripts (hereafter referred to as siGAM), which significantly decreased both GAM transcript and GAM protein levels (Supplementary Figure S3A–D), increased E2F1 levels by ∼50% (Figure 5D). This suggests that GAM may somehow interfere with the effects of E2F1 on cell proliferation. To determine whether GAM may also affect other regulators of cell homeostasis, the blot of Figure 2A was reprobed with a pan-Ras antibody. Namely, Ras proteins function as intracellular switches in signal transduction cascades and, due to their ability to modulate transcription, they control cell growth and proliferation as well as other aspects of cell biology including senescence/cell-cycle arrest, differentiation and survival (18). Furthermore, like GAM transcripts, Ras transcripts represent validated targets of let-7 miRNAs (15). In sharp contrast with E2F1, overexpressing HA-GAM-2 increased Ras level ∼9-fold (Figure 2A), while siGAM showed opposite effects (Supplementary Figure S4). Of note, while HA-GAM-4 and Ras 3′-UTRs do not contain any miR-92 target site, cotransfecting HEK-293 cells with pre-miR-92a-1 and HA-GAM-4 reduced the upregulation of Ras by GAM by ∼50% (Figure 2B). This prompted us to look for the effects of miR-92a-1 on Ras expression. As shown in Figure 2C, overexpressing miR-92a-1 slightly decreased Ras levels. In contrast, transfecting HEK-293 cells with a miR-92a antisense inhibitory RNA increased Ras accumulation. Thus, miR-92a-1 can downregulate Ras by targeting either endogenous GAM transcripts or other transcripts encoding positive regulators of Ras.
Figure 2.
GAM modulates E2F1 and Ras levels, increases cell apoptosis and reduces cell proliferation. (A) HEK-293 cells were transfected with either pCMV-HA or HA-GAM-2. GAM effects on E2F1 and Ras were analyzed by western blotting. (B) HEK-293 cells were transfected with either pCMV-HA or HA-GAM-4 along with a control RNA or pre-miR-92a-1. GAM effects on Ras were analyzed by western blotting. (C) Ras levels in HEK-293 cells transfected with either a pre-miR-control, pre-miR-92a-1 or a miR-92a antisense inhibitory RNA (92-I) were determined by western blotting. (D) HEK-293 cells transfected as indicated were assayed 24 h later for apoptosis or proliferation. Values represent the mean ± SD (n = 3). # and ##, Significantly different from apoptosis control, #P = 0.063, ##P < 0.005. * and **, Significantly different from proliferation control, *P < 0.02, **P < 0.005.
Figure 5.
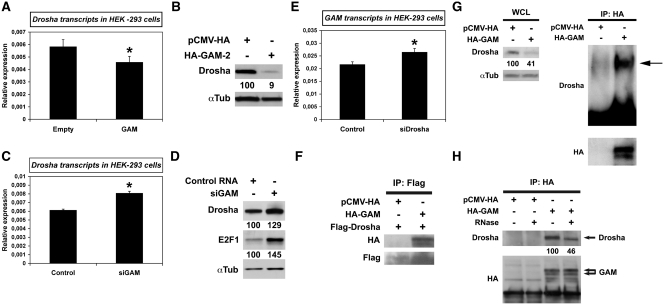
GAM downregulates Drosha, a critical effector of miRNA maturation, and GAM-Drosha interaction is RNA-dependent. (A and C)The relative levels of Drosha transcripts in HEK-293 transfected with either the empty pCMV-HA vector or HA-GAM (A), or with either a Control RNA or siGAM (C), were determined by qRT–PCR. Values represent the mean ± SD (n = 3). *P < 0.05, significantly different from control RNA. (B and D) Drosha levels in extracts prepared from HEK-293 cells transfected with either the empty pCMV-HA vector or HA-GAM-2 (B), or with either a Control RNA or siGAM (D), were analyzed by western blotting. The blots in B were from the same experiment as the blots of Figure 2A. (E) The relative levels of GAM transcripts in HEK-293 transfected with either the Control RNA or siRNAs directed against Drosha transcripts (siDrosha) were determined by qRT–PCR. Values represent the mean ± SD (n = 3). *P < 0.001, significantly different from Control RNA. (F–H) HEK-293 cells were transfected with the empty pCMV-HA vector or pCMV-HA-GAM. In (F), cells were cotransfected with a Flag-Drosha construct. In (H), extracts were submitted to RNase digestion before immunoprecipitation as indicated. Immunoprecipitations were conducted using anti-Flag (F) or anti-HA antibody (G and H). Immunocomplexes were analyzed by western blotting with the indicated antibodies. In (G), the left blot (WCL) shows the levels of endogenous Drosha in whole cell lysates of HEK-293 cells transfected as indicated, before immunoprecipitation (these blots were prepared using 1/25th of the whole lysate subsequently used for immunoprecipitation).
Finally, as mentioned earlier, miR-17-92 and let-7 miRNAs are respectively considered as oncogenes and tumor suppressor genes, due to their opposite effects on cell proliferation, apoptosis and more generally on cell homeostasis. GAM transcripts being a target of these two functionally opposite groups of miRNAs, and GAM modulating E2F1 and Ras levels, we hypothesized that it might act as a molecular sensor aimed at maintaining cell homeostasis. Indeed, overexpressing GAM increased cell apoptosis while reducing cell proliferation, with siGAM having opposite effects (Figure 2D).
GAM decreases the expression of miR-17-92 miRNAs differentially
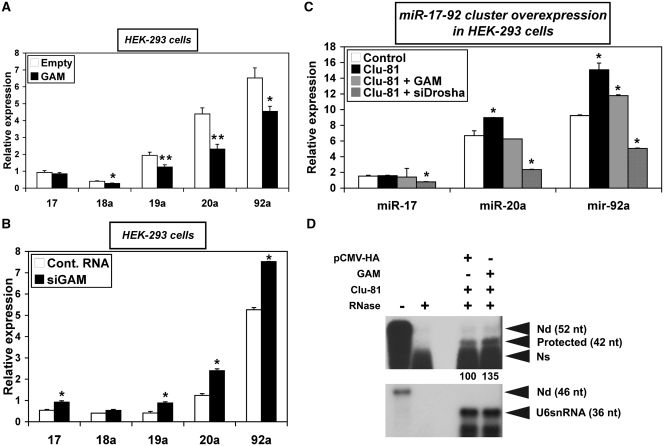
Aimed at regulating the expression of their target genes, miRNAs tend to build feedback regulatory loops with their regulators (4). Thus, as GAM transcripts were targeted by miR-17-92 miRNAs, and given that GAM and miR-92a-1 had opposite effects on both E2F1 and Ras expression (Supplementary Figure S2A and Figure 2A–C, respectively), we therefore hypothesized that GAM in turn may regulate miR-17-92 miRNA levels. Indeed, overexpressing GAM in HEK-293 (Figure 3A) or in K562 lymphoid cells (Supplementary Figure S5) decreased the levels of miR-17-92 miRNAs. In contrast, the levels of these miRNAs increased following the transfection of HEK-293 (Figure 3B), HepG2 (Figure 6B) or MCF-7 breast cancer cells (Supplementary Figure S6) with siGAM. Finally, GAM overexpression also decreased the levels of miR-17, -20a and -92a in HEK-293 cells transfected with Clu-81, a 3837-bp expression construct containing the whole miR-17-92 cluster (Figure 3C). This indicates that GAM generally works to impair any surge of miR-17-92 miRNA levels. It also suggests that both the upregulation of Ras and the downregulation of E2F1 by GAM (see hereabove) might be due at least in part to GAM downregulating effects on miR-92a-1 levels.
Figure 3.
GAM downregulates miR-17-92 miRNAs. (A and B). The relative levels of miR-17-92 miRNAs in HEK-293 cells transfected with either the empty pCMV-HA vector or pCMV-HA-GAM (A), or with either a control RNA or siGAM (B), were determined by qRT–PCR. Values represent the mean ± SD (n = 3). (A) * and **, Significantly different from empty control vector, *P < 0.004, **P < 0.0004. (B) *P < 0.005, significantly different from control RNA. (C) HEK-293 cells were transfected with either a control pCMV-HA/Control RNA mix or a construct in the pCMV vector expressing the whole miR-17-92 cluster (Clu-81) in combination with either a pCMV-HA/Control RNA mix, a pCMV-HA-GAM/Control RNA mix, or a pCMV-HA/siDrosha mix. Clu-81 contains a 3837-bp insert bearing the whole miR-17-92 cluster sequence (787 bp) plus 216-bp upstream of the first nucleotide of pre-miR-17 and 2834-bp downstream of the last nucleotide of pre-miR-92a-1. The relative levels of the indicated miRNAs were determined by qRT–PCR. Values represent the mean ± SD (n = 3). *P < 0.01, significantly different from control mix. (D) HEK-293 cells were transfected with Clu-81 along with either the empty pCMV-HA vector or pCMV-HA-GAM. The relative levels of RNAs containing unprocessed miR-92a-1 (42-nt long protected band) were determined by RNase-protection assays using 2 µg total RNA per assay. The efficiency of RNase digestion was assessed in parallel following the incubation of each probe with the same amount of yeast tRNAs (two left lanes). ‘-RNase’ controls correspond to a 15-fold dilution of the undigested sample. The relative intensities of the protected fragments, calculated using the U6snRNA as an internal control, are given in percent of the pCMV-HA control. Nd, non-digested, Ns, non-specific.
Figure 6.
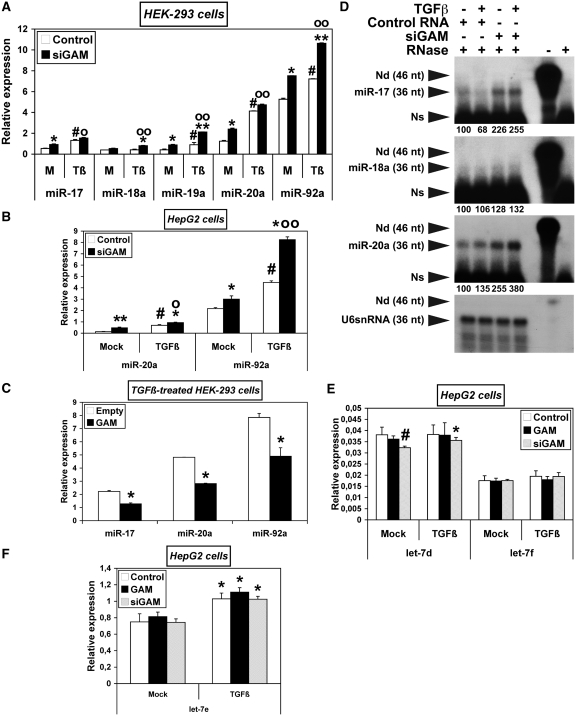
GAM opposes the upregulation of miR-17-92 miRNAs by TGFβ effectors. (A–C) HEK-293 (A and C) or HepG2 cells (B) were transfected with either a control RNA or siGAM (A and B), or with the empty pCMV-HA vector or pCMV-HA-GAM (C). After 34 h, they were either mock-treated (A and B) or treated with TGFβ (A–C) as indicated. The relative levels of the indicated miRNAs were determined by qRT–PCR. Values represent the mean ± SD (n = 3). The results obtained with mock-treated samples (M) in (A) are the same as those in Figure 3B. They are presented again along with the results obtained with TGFβ-treated samples (Tβ) for easier comparison, as they were drawn from the same experiment. Panel A: * and **, siGAM significantly different from Control RNA for each treatment, *, P < 0.005, **, P < 0.0005; #, TGFβ-treated Control RNA significantly different from mock-treated Control RNA, P < 0.0002; o and oo, TGFβ-treated siGAM significantly different from mock-treated siGAM, o, P < 0.003, oo, P < 0.0004. Panel B: * and **, siGAM significantly different from Control RNA for each treatment, *, P < 0.05, **, P < 0.002; #, TGFβ-treated Control RNA significantly different from mock-treated Control RNA, P < 0.02; o and oo, TGFβ-treated siGAM significantly different from mock-treated siGAM, o, P < 0.02, oo, P < 0.01. Panel C: *, Significantly different from Empty control vector, P < 0.05. (D) The relative levels of miR-17, miR-18a and miR-20a were determined by RNase-protection assays. Total RNAs extracted from HepG2 cells (10 µg per assay) treated and transfected as indicated (four left lanes) were hybridized with a radiolabelled RNA antisense probe complementary to the indicated miRNAs. The lengths of the antisense miR-17, miR-18a and miR-20a probes were made equal to that of the U6snRNA antisense probe by addition of a 3′-poly(A)-tail. Namely, the RNases used in the assay (RNase 1 and RNase T1) are not able to cut phosphodiester bonds following a non-paired A nt. The relative intensities of the protected fragments, given under each panel in percent of the control sample (lane 1), were calculated using the U6snRNA as an internal control. The efficiency of RNase digestion was assessed in parallel following the incubation of each probe with 10 µg yeast tRNAs (two rights lanes). ‘-RNase’ controls correspond to a 15-fold dilution of the undigested samples. Exposure time at −80°C was 36 h for miR-17, miR-18a and miR-20a probes, and 2 h for U6snRNA probe. Nd, non-digested, Ns, non-specific. (E,F) HepG2 cells were transfected with either a pCMV-HA/Control RNA mix (Control), a pCMV-HA-GAM/Control RNA mix (GAM) or a pCMV-HA/siGAM mix (siGAM). 34 h later, they were either mock-treated or treated with TGFβ. The relative levels of let-7d and let-7f (E) and of let-7e (F) were determined by qRT–PCR. Values represent the mean ± SD (n = 3). Panel E: #, mock-treated siGAM significantly different from mock-treated Control, P = 0.01; *, TGFβ-treated siGAM significantly different from mock-treated siGAM, P = 0.03. Panel F: *, Significantly different from the corresponding mock-treated sample, *, P < 0.02.
Of note, and although qRT–PCR assays cannot discriminate between miR-92a-1 and miR-92a-2, the relative levels of expression of miR-17-92 miRNAs located 3′ of the cluster were generally higher than those with a more 5′ position (Figures 3A and B, 6A and B and Supplementary Figures S5 and 6). These results indicate that miR-17-92 miRNAs are processed with a differential efficiency from transcripts produced from the miR-17-92 cluster host gene, i.e. MIRHG1 or C13orf25. This was further confirmed by the fact that transfecting HEK-293 cells with Clu-81 also lead to the differential expression of miR-17, -20a and -92a (Figure 3C), which suggests that HEK-293 cells may contain limiting amounts of unidentified factors required for the processing of each of the miR-17-92 miRNAs.
Furthermore, in the four cell lines under study, GAM and/or siGAM affected the levels of the tested miR-17-92 miRNAs differentially (Figures 3A–C, 6A and B and Supplementary Figures S5 and 6), suggesting that GAM may somehow affect their processing. Unfortunately, nothing is known about the processing of miR-17-92 miRNAs beyond the fact that the production of miR-18a requires the presence of hnRNPA1 (19,20). It is also not known whether transcripts starting at either the TSS1 (transcription start site 1, i.e. the TSS of the C13orf25 host gene) (13), the TSS2 (21), or both TSSs may function as miR-17-92 primary RNAs, nor if any of these transcripts allows the simultaneous production of the six mature miR-17-92 miRNAs, or not. Gaining definitive answers to these questions will obviously require a number of experiments and is well beyond the scope of this article. Nevertheless, to gain further evidence of GAM effects on miR-17-92 miRNAs processing, we used RNase-protection assays to follow the levels of RNAs containing unprocessed miR-92a-1 in HEK-293 cells transfected with Clu-81 along with either an empty pCMV-HA control vector or a construct overexpressing GAM. Indeed, GAM overexpression increased the relative levels of RNAs containing unprocessed miR-92a-1 by ∼35% (Figure 3D). Given the facts that GAM is located in the nucleus [(17) and Tili et al., unpublished results] and that GAM overexpression did not change the levels of Dicer, which further processes the pre-miRNAs in the cytoplasm (Supplementary Figure S7), the result of Figure 3D gives an independent indication that GAM may somehow decrease, directly or indirectly, the processing of miR-17-92 primary transcripts in the nucleus.
GAM impairs the activation of the miR-17-92 cluster by c-Myc
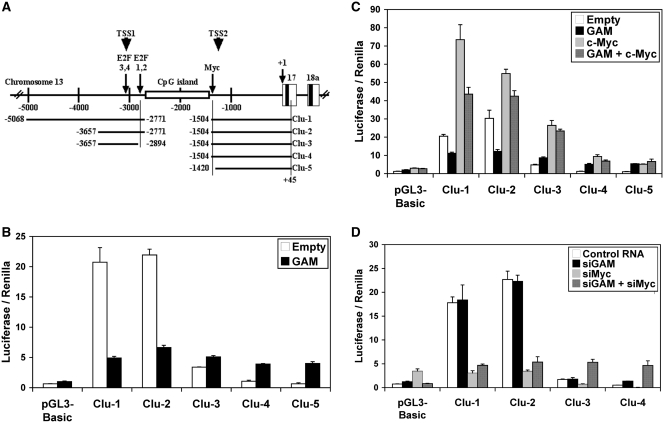
The expression of miRNAs can be regulated both transcriptionally and post-transcriptionally. The above results suggested that GAM downregulating effects on the expression of miR-17-92 miRNAs come at least in part from post-transcriptional effects. To determine if the downregulation of miR-17-92 miRNAs by GAM may also arise from effects on the miR-17-92 cluster expression, we used five promoter constructs containing miR-17-92 TSS2 (21) (Figure 4A). Four of them, but Clu-5, contain the Myc-binding site previously shown to be bound by c-Myc to transactivate the miR-17-92 cluster (10). Clu-1, Clu-2 and Clu-3 were generated by fusing Clu-4 to more upstream sequences containing the TSS1 (13). These constructs were tested in MCF7 cells, where both miR-17-92 miRNAs (22) and c-Myc (23) are well expressed. A high level of luciferase activity was produced from Clu-1 and Clu-2, but the lack of a 123-bp fragment containing E2F-binding sites 1 and 2 (12) in Clu-3 decreased the luciferase activity by ∼75% (Figure 4B). This indicates that this 123-bp fragment is required for a robust expression of the cluster, in agreement with the fact that E2F factors have been shown to bind E2F-binding sites 1–4 and activate the transcription from the TSS1 (12,13). The activity of Clu-4 was still lower, and that of Clu-5 similar to the empty control vector. Overexpressing GAM decreased four times the activation of Clu-1 and Clu-2, while remaining without significant effects on Clu-3 (Figure 4B). In contrast, it increased the expression of Clu-4 and Clu-5, which was probably due to off-target effects allowing the activation of the TSS2 through modifications of the levels of some of the transcriptional activators and repressors whose target sequences are known to be present in the pGL3-Basic vector. Regardless, these results suggest that GAM can decrease the transcriptional activation of the C13orf25 host gene from the TSS1.
Figure 4.
GAM impairs the activation of the miR-17-92 promoter. (A) Schematic representation of the clones in pGL3-Basic containing different parts of the genomic region upstream of the miR-17-92 cluster. The first nucleotide of pre-miR-17 was arbitrarily used as nucleotide +1 for convenience. The ends of the DNA fragments used to generate the Clu-1 to Clu-5 constructs are indicated. The positions of one c-Myc-binding site (10) and of E2F-binding sites 1, 2 and 3, 4 (12) are indicated by an arrow. The approximate locations of TSS1 (transcription start site 1) (13) and TSS2 (21) are indicated by arrowheads. (B and C) Effects of GAM on the miR-17-92 promoter either alone (B) or in conjunction with c-Myc (C). MCF7 cells were transfected with the empty pGL3-Basic vector or the Clu-1 to Clu-5 constructs as indicated along with the empty pCMV-HA vector or a construct expressing GAM and/or c-Myc. The Firefly luciferase activity was measured 48 h after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± SD (n = 4). (D) Effects of siRNAs directed against GAM (siGAM) or against c-Myc transcripts (siMyc) on the miR-17-92 promoter expression either alone or in conjunction. MCF7 cells were transfected with the empty pGL3-Basic vector or the Clu-1 to Clu-4 constructs as indicated along with a control RNA, siGAM and/or siMyc. The Firefly luciferase activity was measured 48 h after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± SD (n = 4).
In agreement with previously published results (10), overexpressing c-Myc lead to a further activation of Clu-1 to Clu-4 (Figure 4C). The highest levels of expression were again obtained with Clu-1 and Clu-2, indicating that transcription factors other than c-Myc, whose target sequences should be present both within and upstream the 123-bp fragment missing in Clu-3, were working along c-Myc to activate the expression of the reporter constructs. In contrast, siRNAs directed against c-Myc transcripts (siMyc) reduced the levels of expression of Clu-1 and Clu-2 to that of the empty pGL3-Basic vector, showing that the presence of c-Myc activity is mandatory for the activation of the miR-17-92 cluster promoter from the TSS1, at least in the conditions of the experiment (Figure 4D). Coexpressing GAM along with c-Myc decreased the activating effects of c-Myc on Clu-1 and Clu-2 (Figure 4C). Of note, neither GAM overexpression nor siGAM affected c-Myc protein levels (Supplementary Figure S8), indicating that GAM activity interferes, directly or indirectly, with the transcriptional activation of the miR-17-92 cluster by c-Myc. However, GAM overexpression did not reduce the activating effects of c-Myc on Clu-3, Clu-4 and Clu-5 (Figure 4C), suggesting that GAM effects might not be directed toward c-Myc itself but rather toward some of the unidentified transcriptional activators acting on the 123-bp fragment deleted in Clu-3 and/or on sequences located upstream of Clu-2. To determine if GAM repressing effects were dependent on E2F sites 1 and 2, we then used constructs derived from Clu-2 whose either E2F site 2 (Clu-2-ME2) or both E2F sites 1 and 2 (Clu-2-ME1,2) had been mutated (Supplementary Figure S9). These mutations did not change the levels of expression of Clu-2, indicating that the effects of E2F factors on this construct rather take place through E2F sites 3 and/or 4. Of note, these mutations also did not impair GAM repressing activity, indicating that GAM opposes the activating effects of some factors working on this 123-bp fragment either beside or along E2F factors. Finally, the lack of effects of siGAM showed that endogenous GAM activity is normally too low to impair the activation of the miR-17-92 promoter (Figure 4D). Collectively, these results provide good evidence that c-Myc is critical for the miR-17-92 cluster activation, and that GAM impairs the miR-17-92 promoter activation by reducing, either directly or indirectly, the positive transcriptional effects of cMyc and those of unidentified factors acting on the 123-bp fragment lacking in Clu-3 construct or on sequences located upstream of Clu-2, at least in the conditions of the experiment. Similar experiments showed that GAM also impairs the activation of the miR-17-92 cluster in HEK-293 and HepG2 cells (data not shown).
Of note, the activation of Clu-4 and Clu-5 following GAM overexpression (Figure 4B), of Clu-5 following c-Myc and/or GAM transfection (Figure 4C), of the empty pGL3-Basic vector following siMyc transfection (Figure 4D), as well as of Clu-3 and Clu-4 following transfection with both siGAM and siMyc (Figure 4D) most probably arose from off-target effects, known to occur with the pGL3-Basic vector.
GAM and Drosha modulate each other's levels
The differential effects of GAM on miR-17-92 miRNA levels (Figures 3A–C, 6A and B and Supplementary Figures S5 and S6) suggested that GAM may somehow affect the processing of miR-17-92 primary RNAs. This could possibly be done either by modulating, directly or indirectly, the levels of Drosha or those of factors required for the differential processing of miR-17-92 miRNAs, or by interfering, directly or indirectly, with Drosha activity. Interestingly, GAM overexpression in HEK-293 cells decreased the levels of Drosha transcripts (Figure 5A) and, as shown by reprobing the blots from Figure 2A with an anti-Drosha antibody, of Drosha protein (Figure 5B). Accordingly, siGAM lead to an increase of both Drosha transcripts and Drosha protein levels (Figure 5C and D). Of note, GAM overexpression did not affect the level of DGCR8, the general partner of Drosha (Supplementary Figure S7). On the other hand, transfecting the cells with siRNAs directed against Drosha transcripts (siDrosha), which decreased Drosha transcripts and Drosha protein levels by ∼60% (Supplementary Figure S3E and F), showed to increase GAM transcripts levels (Figure 5E), most probably by decreasing the levels of several of the miRNAs potentially targeting GAM transcripts. Thus, GAM and Drosha work within the same gene regulatory network and control, directly or indirectly, each other's expression.
The fact that GAM downregulates Drosha suggested that GAM may decrease the levels of most, if not all, miRNAs by decreasing the processing of their primary transcripts by Drosha. However, GAM overexpression in HepG2 did not decrease the levels of either let-7d, let-7e or let-7f and siGAM in contrast slightly but significantly downregulated let-7d (Figure 6E and F). This indicates that the repressing effects of GAM are not global but rather miRNA-specific. Accordingly, qRT–PCRs from RNAs extracted of SW480 colon cells as well as microarrays from RNAs prepared from THP-1 monocytic cells showed that GAM can upregulate or downregulate some miRNAs while leaving the levels of the others unchanged (Tili et al., unpublished results). Thus, GAM-specific effects on miRNA expression should require the presence of specific trans-regulators of miRNA processing. Finally, as siGAM did not increase Drosha levels in HEK-293 cells by >30% (Figure 5D), and given than Drosha is rather an abundant protein in these cells, as shown by proteomic analyses (Tili et al., unpublished results), the differential downregulation of miR-17-92 miRNAs by endogenous GAM cannot solely come from its capability to downregulate Drosha, and should also arise from GAM effects on at least some of those unidentified factors regulating the differential processing of miR-17-92 primary RNAs by Drosha.
GAM interacts with Drosha in a RNA-dependent manner
As GAM affected both the transcription and the processing of miR-17-92 miRNAs, and given that these two processes are generally coupled, we hypothesized that in addition to modulating the expression of Drosha, GAM may also possibly interfere with the processing of pri-miRNA by Drosha. Furthermore, a massive yeast two-hybrid screen suggested that GAM might directly interact with Drosha (24). Indeed, a Flag-tagged full-size Drosha allowed us to immunoprecipitate HA-tagged full-size GAM in HEK-293 cells using an anti-Flag antibody (Figure 5F). Accordingly, we were able to immunoprecipitate endogenous Drosha along with HA-GAM, confirming that GAM and Drosha may actually interact in vivo (Figure 5G). To gain further insights into the possible consequences of GAM-Drosha interaction, we then pre-treated the cellular extracts with RNase A before immunoprecipitation. This reduced the amount of endogenous Drosha interacting with GAM by ∼50% (Figure 5H), indicating that Drosha-GAM interactions take place when Drosha is engaged in pri-miRNA processing. However, transfecting HEK-293 cells with Clu-81 did not allow us to precipitate Drosha more efficiently than in presence of an empty control vector (results not shown), suggesting that GAM differential effects on miR-17-92 miRNAs processing require the presence of other factors present in limiting amount, most probably the very same factors responsible for the differential processing of miR-17-92 miRNAs in untransfected cells (see hereabove). This would also explain why GAM was never reported to be part of the microprocessor complex. Finally, whether GAM competes with some of these miR-17-92 processing factors for binding to Drosha and/or downregulates factors needed for the processing of miRNAs such as miR-20a or miR-92a-1 remains to be determined.
GAM opposes the upregulation of miR-17-92 miRNAs by TGFβ
Usually, TGFβ maintains tissue homeostasis and prevents incipient tumor formation by regulating cellular proliferation, differentiation, survival and adhesion as well as the cellular microenvironment. However, pathological forms of TGFβ signaling can promote tumor growth and invasion, evasion of immune surveillance, cancer cell dissemination and metastasis (25). Importantly, miRNAs have recently been shown to regulate the TGFβ signaling pathway. For example, the expression of miR-21 in breast cancers predicts for elevated TGFβ1 expression and a poor clinical prognosis (26), while that in gliomas results in the suppression of multiple components of the TGFβ signaling system, including its ligands (i.e. TGFβs 1 and 3), its receptors (i.e. TβR-II and TβR-III), and its effector molecules (i.e. Smad3, Daxx, and PDCD4) (27,28). Interestingly, the two-hybrid screen referred to here above also showed that GAM may interact with Smad1 (an infrequent transducer of TGFβ signaling), and that siRNAs directed against GAM transcripts impaired the upregulation of endogenous PAI-1 (Plasminogen Activator Inhibitor-1) by TGFβ (24). Therefore, as the misexpression of miR-17-92 miRNAs has been associated with cancers (5,9), we then looked for the effects of TGFβ1 (herafter referred to as TGFβ) on miR-17-92 miRNA expression, and checked if these effects may be affected by GAM levels.
TGFβ increased the levels of miR-17-92 miRNAs in HEK-293 (Figure 6A), HepG2 (Figure 6B) and MCF7 cells (Supplementary Figure S6). These levels generally increased further when cells were transfected with siGAM, indicating that GAM opposes the upregulation of miR-17-92 miRNAs by TGFβ. This was confirmed by the fact that GAM overexpression reduced the levels of miR-17, 20a and -92a in HEK-293 cells treated with TGFβ (Figure 6C). Interestingly, the upregulation of miR-17-92 miRNAs was not uniform, suggesting that TGFβ, as well as GAM, may somehow affect the maturation of miR-17-92 miRNAs. The differential effects of TGFβ and siGAM on miR-17-92 miRNA expression were further confirmed in HepG2 cells by RNase-protection assays using probes specific of miR-17, -18a and -20a (Figure 6D). Together, these results show that, in HEK-293, HepG2 and MCF7 cells, the differential upregulation of miR-17-92 miRNAs by TGFβ is differentially buffered by endogenous GAM activity. Of note, the upregulation of miR-17-92 miRNAs by TGFβ effectors was not due to its inhibition of the downregulation of Drosha by endogenous GAM: namely, the TGFβ treatment did not significantly affect Drosha transcripts nor Drosha protein levels (Supplementary Figure S10). This was further confirmed by the fact that, while also upregulating let-7e, TGFβ did not change the levels of let-7d or let-7f (Figure 6E and F). Thus, like those of GAM, TGFβ effects are miRNA-specific, and TGFβ effectors most probably impact the levels of the same unidentified regulators of miR-17-92 miRNA processing (see hereabove).
Finally, siGAM also increased the levels of miR-106b and -93 in MCF7 cells, with or without TGFβ treatment (Supplementary Figure S11), pointing to possible GAM effects on the two miR-17-92 ortholog clusters.
GAM, miR-17-92a miRNAs and TGFβ effectors work within the same gene regulatory network
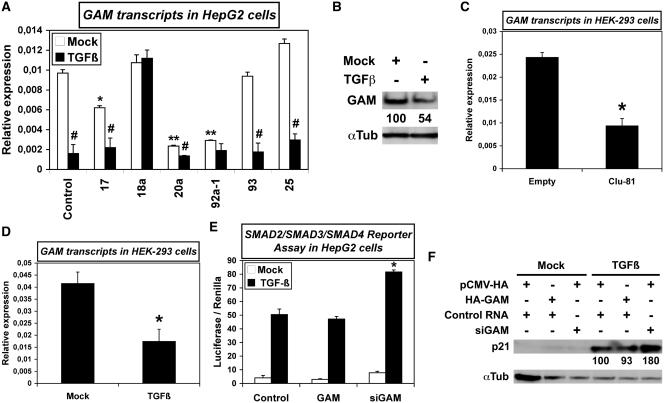
Beside reducing the translation of their target transcripts, miRNAs also often drive their degradation (29). Therefore, we then checked for possible effects of miR-17-92 miRNAs on GAM transcript levels. In HepG2 cells, pre-miR-17 downregulated GAM transcript levels by 40% (Figure 7A). Of note, while pre-miR-20a and -92a-1 reduced the levels of GAM transcripts by ∼80%, their respective orthologs pre-miR-93 and -25 did not, showing that miRNAs with comparable effects on mRNA translation may well work differently on mRNA stability (Figure 7A). In accordance with the fact that no miR-18a target site is found in GAM transcripts, pre-miR-18a did not affect GAM transcript levels. The targeting of GAM transcripts by miR-17-92 miRNAs was further confirmed in HEK-293 cells overexpressing Clu-81 (Figure 7C).
Figure 7.
TGFβ and miR-17-92 miRNAs downregulate GAM transcripts, while GAM impairs the canonical TGFβ signaling pathway. (A) The levels of GAM transcripts in HepG2 cells transfected with a pre-miR Control (Control) or the indicated pre-miRNAs, and either mock-treated or treated with TGFβ, were determined by qRT–PCR. Values represent the mean ± SD calculated from triplicate qRT–PCR reactions of the same cDNAs. #, TGFβ-treated sample significantly different from the corresponding mock-treated sample, P < 0.003. * and **, Pre-miR mock-treated significantly different from mock-treated pre-miR Control, *, P < 0.002, **, P < 0.001. (B) GAM levels in Hep-G2 cells mock-treated or treated with TGFβ were analyzed by western blotting. (C,D) The relative levels of GAM transcripts in HEK-293 cells transfected with either the empty pCMV vector (Empty) or Clu-81 (C), or either mock-treated or treated with TGFβ (D) were determined by qRT–PCR. Values represent the mean ± SD calculated from three independent reverse transcription reactions, each analyzed by qRT–PCR in triplicates. Panel C: *, Significantly different from Empty control vector, P < 0.001. Panel D: *, Significantly different from mock-treatment, P < 0.001. (E) After cotransfection with the SMAD2/SMAD3/SMAD4 luciferase reporter plasmid along with either a pCMV-HA/Control RNA mix, a pCMV-HA-GAM/Control RNA mix or a pCMV-HA/siGAM mix, HepG2 cells were mock-treated or treated with TGFβ. The Firefly luciferase activity was measured 48 h after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± SD (n = 6). *, Significantly different from Control, P < 0.005. (F) The effects of GAM or siGAM on p21/CDKN1A expression in HepG2 cells mock-treated or treated with TGFβ were analyzed by western blotting.
We then checked if TGFβ may modulate the effects of miR-17-92 miRNAs on GAM expression. TGFβ alone reduced GAM transcript as well as GAM protein levels (Figure 7A and B). A similar downregulation of GAM transcript level by TGFβ was also observed in HEK-293 (Figure 7D) and in MCF7 cells (Supplementary Figure S12). Of note, transfecting HepG2 cells with either pre-miR-17, -20a, -92a-1, -93 or -25 did not affect the downregulation of GAM transcripts by TGFβ (Figure 7A). This was probably due to the capability of TGFβ to upregulate endogenous miR-17-92 miRNAs, and possibly also other miRNAs targeting GAM transcripts. However, we do not rule out the possibility that TGFβ signaling may also trigger miRNA-independent repression of GAM. Surprisingly, miR-18a impaired the downregulation of GAM by TGFβ (Figure 7A), possibly by targeting SMAD2, a transducer of TGFβ signaling whose 3′-UTR contains a miR-18 target site, and/or some other repressors of GAM. Overall, these results suggest that the downregulation of GAM by the effectors of TGFβ signaling is at least in part dependent of miR-17-92 miRNA expression, and conversely that the upregulation of miR-17-92 miRNAs by TGFβ might arise, at least in part, from its repressing effects on GAM expression. Of note, the conjugate effects of GAM and TGFβ on miRNA expression also appear to be miRNA-specific. Namely, TGFβ unexpectedly impaired the downregulation of let-7d by siGAM (Figure 6E), suggesting that GAM and TGFβ effectors, while having opposite effects on miR-17-92 miRNA expression, may work together to keep let-7d levels above a certain threshold, in accordance with the fact that GAM and TGFβ were previously shown to collaborate and upregulate endogenous PAI-1 (24).
Given the above interactions between GAM and TGFβ signaling, we further looked whether GAM may have broader effects on TGFβ canonical signaling pathway. In this pathway, the active TGFβ dimer signals by bringing together two pairs of receptor serine/threonine kinases known as the type I and type II receptors, respectively. On binding TGFβ, the type II receptors phosphorylate and activate the type I receptors that then propagate the signal by phosphorylating SMAD transcription factors, namely SMAD2 and SMAD3. Once activated, SMAD2 and SMAD3 form a complex with SMAD4, a binding partner common to all regulatory SMADs (30), and then shuttle to the nucleus. While GAM overexpression in HepG2 cells transfected with a luciferase reporter construct containing SMAD2-4 responsive elements remained without significant effects, targeting endogenous GAM transcripts with siGAM increased the luciferase activity by ∼60% (Figure 7E). Accordingly, while GAM overexpression did not significantly impair the well established upregulation by TGFβ of the cell-cycle regulator p21/CDKN1A, siGAM lead to a 80% increase of p21 level (Figure 7F). Thus, one possible way for GAM to reduce the upregulation of miR-17-92 miRNAs by TGFβ may be to impair SMAD activity. Furthermore, these results suggest that another important role for GAM may be to lower the plateau of activation reached by TGFβ responsive genes under TGFβ signaling.
Together, and taking also into account that miR-20a has been shown to target TGFβ type II receptors (22), our results give strong evidence that GAM, miR-17-92 miRNAs and the effectors of the canonical TGFβ signaling pathway work within the same gene regulatory network.
DISCUSSION
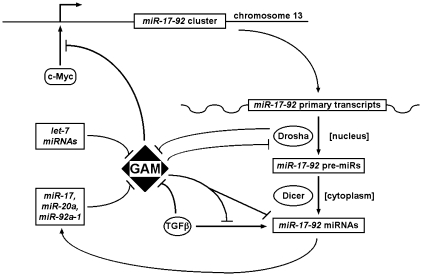
Our study provides the very first data concerning the role of GAM, a previously uncharacterized factor, in human cells, in addition to shedding a new light on the regulation and functions of miR-17-92 miRNAs. Altogether, our results show that GAM is central to a gene regulatory network, illustrated on the working model of Figure 8, through which: (i) miR-17, -20a, -92a-1 and let-7 miRNAs reduce GAM expression by targeting GAM transcripts; (ii) GAM downregulates miR-17-92 miRNAs both by impairing the transcriptional activation of the miR-17-92 cluster by c-Myc and by differentially affecting the maturation of miR-17-92 miRNAs; (iii) GAM also impairs the upregulation of miR-17-92 miRNAs by TGFβ effectors, possibly by decreasing the transcriptional activity of SMADs following the activation of the TGFβ canonical pathway; (iv) GAM increases cell apoptosis while reducing cell proliferation, and modulates, directly or indirectly, the levels of E2F1 and Ras; and (v) GAM decreases the levels of and interacts in a RNase-dependent manner with Drosha, the main effector of miRNA maturation in the nucleus.
Figure 8.
Working model depicting the intricate interaction loops between GAM, TGFβ effectors, Drosha, miR-17-92 and let-7 miRNAs. For clarity, only the main interactions have been represented. Arrows indicate activating effects, and T-barred arrows inhibitory effects. The effects of c-Myc on the miR-17-92 cluster were previously described (10). See text for more details.
Using synthetic pre-miRNAs as well as antisense inhibitory RNAs for luciferase assays, qRT–PCRs and western blotting analyses, we provided strong evidence that endogenous miR-17-92 and let-7 miRNAs target GAM transcripts. Importantly, our experiments also showed that, while miR-17, miR-20a and miR-92a-1 work together to downregulate GAM transcripts, miR-92a-1 upregulates E2F1 in contrast to miR-17 and miR-20a. As miR-92a-1 is the only miRNA of the cluster with orthologs in non-vertebrates, it is possible that, following gene duplications in vertebrate lineage, evolution has kept the different miR-17-92 miRNAs in cluster to establish a yet poorly understood mechanism of differential maturation, thus allowing for the fine tuning of their effects on their hundreds of respective target genes.
Our promoter studies confirmed the previously established activating effects of c-Myc on the miR-17-92 cluster (10), and showed that GAM limits, directly or indirectly, c-Myc trancriptional activation of the miR-17-92 cluster through TSS1 without affecting the levels of endogenous c-Myc, although the mechanistic bases of GAM/c-Myc interference remains to be elucidated. They revealed that the miR-17-92 cluster constitutes a very complex transcriptional unit with regulatory sequences spanning over several thousands base pairs, from the site bound by c-Myc just upstream of the TSS2 (10) to an upstream 123-bp fragment containing E2F1 sites 1 and 2 previously shown to allow the transcriptional activation of the cluster by E2F factors (12,13) to sequences located upstream of the TSS1 and the 5′-end of Clu-2 (this article). In any case, it can be expected that the transcription of a cluster containing miRNAs targeting key regulators of cell proliferation would be regulated in a number of different ways. Of note, we have previously shown that chicken GAM interacts with the heterogenous nuclear ribonucleoprotein U (hnRNPU/SAF-A) (17), which is known to interact with different transcription factors, including the glucocorticoid receptor, a member of the superfamily of hormone nuclear receptors. Through this interaction, hnRNPU represses glucocorticoid-induced activation by sequestrating the glucocorticoid receptors on the nuclear matrix (31). Further experiments would be required to determine whether interacting with hnRNPU might allow GAM to block the accession of c-Myc or other transcriptional regulators to the miR-17-92 cluster promoter.
However, transcriptional regulation cannot explain why each of the miR-17-92 miRNAs is processed differentially within a certain type of cells based on qRT–PCRs and RNase-protection assays. This form of differential expression of miR-17-92 miRNAs seems similar to a certain degree in the different cell lines studied, which suggests that it may represent a more general phenomenon. Namely, it was previously shown that miR-17-92 miRNA levels in mouse embryos are not uniform and change differentially according to the stage of development (32,33). Therefore, there is not much doubt that these miRNAs are processed with a differential efficiency from the miR-17-92 primary transcripts. Interestingly, the relative levels of miR-20a and even more of miR-92a remained consistently higher than those of the miRNAs in a more 5′-position, whether cells were submitted to the different treatments, or not. One can hypothesize that the differential processing of miR-17-92 miRNAs depends on their relative position in the cluster. Addressing this question will require to determine the levels of the different miRNAs after shuffling their respective pre-miRNA sequences within the expression constructs.
We further show that GAM and TGFβ respectively downregulates and upregulates miR-17-92 miRNAs in HEK-293, HepG2 and MCF7 cells, and that GAM also downregulates them in K562 lymphoid cells. In each case, the above effects were miRNA-specific. This suggests that GAM as well as TGFβ effectors can modulate the levels of the factors which control the specific processing of each of miR-17-92 miRNAs. To date, nothing is known about these regulators, except that hnRNPA1 is required to allow the specific processing of miR-18a (19,20). Nevertheless, the fact that miR-18a levels remained consistently low in the different cell lines, with or without treatment, indicate that these cells contain factors able to specifically bias the processing of miR-17-92 primary RNAs toward the production of miRNAs such as miR-17, miR-20a and miR-92a-1. Furthermore, overexpressing the whole miR-17-92 cluster using a high expression pCMV vector did not change miR-17 expression, while increasing the levels of miR-20a and miR-92a by no more than about 30 and 60%, respectively. This suggests that the factors controlling the processing of miR-17-92 primary transcripts by Drosha are present in limiting amounts, thus precluding the in vitro analysis of the consequences of GAM-Drosha interaction on the activity of a purified Microprocessor complex following GAM overexpression. Interestingly, GAM was previously shown to interact not only with hnRNPU/SAF-A, but also with hnRNP-M and Matrin 3, a component of the nuclear matrix (17). As hnRNPU and hnRNP-M4 have been found in the Microprocessor (34–36), and as hnRNPU has also been shown to interact with Matrin 3 (37), this raises the question of whether GAM might impair the access of Drosha to some of its target pri-miRNAs by relocating it in specific domains of the nuclear matrix.
Furthermore, while GAM downregulated both Drosha and miR-17-92 miRNAs, TGFβ signaling upregulated the same miRNAs without changing Drosha levels. Of note, the upregulation of miR-17-92 miRNAs by TGFβ may possibly help to understand how TGFβ signaling can promote tumor growth and metastasis (25), given that miR-17-92 miRNAs tend to increase cell proliferation and are usually considered as oncomiRs (5). Furthermore, the effects of both GAM and TGFβ effectors were miRNA-specific. For example, TGFβ upregulated let-7e but did not change let-7d or let-7f levels in HepG2 cells. On the other hand, while overexpressing GAM or transfecting cells with siGAM did not affect the levels of let-7e or let-7f miRNAs, siGAM slightly but significantly decreased let-7d levels, an effect unexpectedly impaired by TGFβ. Accordingly, qRT–PCR experiments in SW480 cells as well as miRNA microarrays in THP-1 monocytic cells showed that the levels of a number of miRNAs remain unchanged following transfection of these cells with either siGAM or a construct overexpressing GAM, while other miRNAs are either upregulated or downregulated (Tili et al., unpublished results). Therefore, neither the downregulation of Drosha by GAM nor GAM-Drosha interaction leads to a general decrease of miRNA levels. Furthermore, neither GAM nor TGFβ exert global effects on the activity of the miRNA processing machinery. In contrast, it is more likely that the miRNA-specific effects of both GAM and TGFβ effectors may arise from their respective effects on the expression and/or the activity of factors controlling the processing of specific pri-miRNAs, especially that of miR-17-92 primary transcripts. Of note, it has been previously shown that, depending on BMP or TGFβ signaling, SMAD1, SMAD5 or SMAD3 can control Drosha-mediated miRNA maturation through their binding to DEAD box RNA helicase p68 (27). It would thus be very interesting to determine if SMAD2 and/or SMAD3 might similarly interfere with the processing of miR-17-92 pri-miRNAs by Drosha under TGFβ signaling, and, if this is the case, if GAM would be able to impair this interaction between SMAD2 or SMAD3 and Drosha. However, while we were able to confirm the previously established GAM–SMAD1 interaction (24), we did not found any GAM-SMAD2 or GAM–SMAD3 interaction (Tili et al., unpublished results). Ultimately, elucidating how the differential processing of miR-17-92 miRNAs is controlled and identifying their key target transcripts may be critical to understand the role of the miR-17-92 cluster in tumor formation.
Importantly, in addition to opposing the upregulation of miR-17-92 miRNAs by TGFβ, GAM impaired the activation of TGFβ responsive genes, while TGFβ in turn downregulated GAM and GAM transcripts levels. This suggests that GAM may be a bona fide TGFβ effector, potentially implicated in many aspects of TGFβ signaling. It will thus be interesting to check whether GAM might differentially affect the cytostatic and the pro-metastatic activities of TGFβ.
We also show that GAM modulates the levels of E2F1 and Ras at least in part through its repressing effects on endogenous miR-92a-1, while increasing cell apoptosis and reducing cell proliferation. The precise effects of GAM on such complex phenotypes will not be easy to determine. As GAM seems to regulate the levels of many miRNAs, and in particular miRNAs which potentially target hundreds of transcripts (e.g. ∼1.000 for miR-17 and miR-20a, and ∼700 for miR-92a-1), GAM activity is likely to affect cell homeostasis in a number of different ways. Of note, it has also been shown that TGFβ1 can overcome Ras mitogenic effects (38–40), and that Ras can counteract TGFβ signaling by altering the expression of TGFβ type II receptor (41). It would thus also be interesting to look whether GAM may play a role in these reciprocal interactions. Finally, the available public data show that GAM expression is higher in brain, kidney and lung tumors but lower in breast, colon, pancreas and prostate cancers, and that the expression of the miR-17-92 cluster negatively correlates with that of GAM except in lung cells (http://www.cgl.ucsf.edu/Research/genentech/gepis/index.html). Given the above results, one can speculate that, while GAM has not yet been connected to a particular pathology, GAM misexpression may impair the proper balance between pro- and anti-proliferation factors, thus increasing the probability of tumor formation.
As a last remark, the emergence of the vertebrate lineage coincided with successive gene duplications, the apparition of new transcriptional regulators and new signaling molecules, and the acquisition of new functionalities such as a complex and more performing brain or adaptative immunity. It is thus probable that the new level of molecular complexity reached by vertebrates has been made possible by the development of robust genetic circuitries required to maintain cell homeostasis while allowing the organism to dynamically respond to an ever changing environment. Like GAM, TGFβ as well as the miR-17-92 cluster also appeared in vertebrates. It is therefore not surprising that they may be implicated in the control of cell homostasis and interact with each other in so many different ways. As GAM transcripts are potentially targeted by ∼150 miRNAs, GAM is likely to participate in feedback regulatory loops with many other miRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Comprehensive Cancer Center of the Ohio State University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 2.Tili E, Michaille J-J, Costinean S, Croce CM. MicroRNAs, the immune system and rheumatic disease. Nat. Clin. Pract. Rheumatol. 2008;4:534–541. doi: 10.1038/ncprheum0885. [DOI] [PubMed] [Google Scholar]
- 3.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 4.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MiRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Tili E, Michaille J-J, Gandhi V, Plunkett W, Sampath D, Calin GA. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;3:521–537. doi: 10.2217/14796694.3.5.521. [DOI] [PubMed] [Google Scholar]
- 7.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 11.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 13.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 14.Büssing I, Slack FJ, Großhans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 17.Michaille J-J, Tili E, Calin GA, Garin J, Louwagie M, Croce CM. Cloning and characterization of cDNAs expressed during chick development and encoding different isoforms of a putative zinc finger transcriptional regulator. Biochimie. 2005;87:939–949. doi: 10.1016/j.biochi.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Rojas JM, Santos E. Ras genes and human cancer: different implications and different roles. Curr. Genomics. 2002;3:295–311. [Google Scholar]
- 19.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNPA1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 20.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benz CC, Scott GK, Santos GF, Smith HS. Expression of c-myc, c-Ha-ras1, and c-erbB-2 proto-oncogenes in normal and malignant human breast epithelial cells. J. Natl Cancer Inst. 1989;81:1704–1709. doi: 10.1093/jnci/81.22.1704. [DOI] [PubMed] [Google Scholar]
- 24.Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massagué J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-β1. Breast Cancer Res. Treat. 2008;117:131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 27.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 29.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 31.Eggert H, Schulz M, Fackelmayer FO, Renkawitz R, Eggert M. Effects of the heterogenous nuclear ribonucleoprotein U (hnRNPU/SAF-A) on glucocorticoid-dependent transcription in vivo. J. Steroid Mol. Biol. 2001;78:59–65. doi: 10.1016/s0960-0760(01)00074-7. [DOI] [PubMed] [Google Scholar]
- 32.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Thomson JM, Wang HYF, Hammond SM, Hogan BLM. Transgenic over-expression of the miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory RI, Yan K-p, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda T, Kaoru Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 36.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malyavantham KS, Bhattacharya S, Barbeitos M, Mukherjee L, Xu J, Fackelmayer FO, Berezney R. Identifying functional neighborhoods within the cell nucleus: Proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA Polymerase II, HP1γ, Matrin 3 and SAF-A. J. Cell. Biochem. 2008;105:391–403. doi: 10.1002/jcb.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFβ/Smad signalling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park BJ, Park JI, Byun DS, Park JH, Chi SG. Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signalling pathway in human prostate cancer. Cancer Res. 2000;60:3031–3038. [PubMed] [Google Scholar]
- 40.Iglesias M, Frontelo P, Gamallo C, Quintanilla M. Blockade of Smad4 in transformed keratinocytes containing a Ras oncogene leads to hyperactivation of the Ras-dependent Erk signalling pathway associated with progression to undifferentiated carcinomas. Oncogene. 2000;19:4134–4145. doi: 10.1038/sj.onc.1203764. [DOI] [PubMed] [Google Scholar]
- 41.Alcock RA, Dey S, Chendil D, Inayat MS, Mohiuddin M, Hartman G, Chatfield LK, Gallicchio VS, Ahmed MM. Farnesyltransferase inhibitor (L-744,832) restores TGF-beta type II receptor expression and enhances radiation sensitivity in K-ras mutant pancreatic cancer cell line MIA PaCa-2. Oncogene. 2002;21:7883–7890. doi: 10.1038/sj.onc.1205948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.