Abstract
Though medicines that target mRNA are under active investigation, there has been little or no effort to develop mRNA itself as a medicine. Here, we report the synthesis of a 130-nt mRNA sequence encoding a 33-amino-acid peptide that includes the sequence of glucagon-like peptide-1, a peptide that stimulates glucose-dependent insulin secretion from the pancreas. The synthesis method used, which had previously been developed in our laboratory, was based on the use of 2-cyanoethoxymethyl as the 2′-hydroxy protecting group. We also developed novel, highly reactive phosphotriester pyrophosphorylating reagents to pyrophosphorylate the 5′-end of the 130-mer RNA in preparation for capping. We completed the synthesis of the artificial mRNA by the enzymatic addition of a 5′-cap and a 3′-poly(A) tail to the pyrophosphorylated 130-mer and showed that the resulting mRNA supported protein synthesis in a cell-free system and in whole cells. As far as we know, this is the first time that mRNA has been prepared from a chemically synthesized RNA sequence. As well as providing a research tool for the intracellular expression of peptides, the technology described here may be used for the production of mRNA for medical applications.
Introduction
RNA vaccines against influenza viruses, unknown viruses or cancer are leading to new medical applications (1–3). Although peptide vaccines induce humoral immunity through major histocompatibility complex (MHC) class II proteins, mRNA-based vaccines can induce the cell-mediated immunity that is important for cancer vaccines through MHC class I proteins. This is because mRNA can direct the synthesis of antigenic proteins directly in the cell, whereas peptide vaccines must introduce antigenic proteins into the cell from the outside (4). There is also the possibility of developing mRNA itself as a new class of therapeutic agent without toxicity. For medical applications, it is essential to have a technology for producing mRNA as a single molecular species in highly pure form by a method that can be scaled up as necessary. As well as being a useful tool for the intracellular expression of oligopeptides, such a technology may also be used in basic and applied research on non-coding RNA.
mRNA is usually prepared by enzymatic synthesis with RNA polymerase from a DNA template followed by enzymatic addition of the 5′-cap and the 3′-poly(A) tail (5). However, mRNA prepared in this way may contain transcription errors (6), can only be produced on a laboratory scale, and is not pure enough for use in the development of medicines. Furthermore, enzymatic synthesis cannot be used to incorporate modified nucleotides into desired positions in the mRNA sequence, which may be important to improve nuclease resistance without affecting function or to enhance other properties of the mRNA such as membrane penetration (7). The drawbacks of enzymatic synthesis can be overcome by chemically synthesizing the mRNA sequence before adding the 5′-cap and the 3′-poly(A) tail. Chemical synthesis of the RNA sequence also allows the artificial mRNAs to be freely designed. An example of a synthetic capped 42-mer mRNA has been reported (8), but it was synthesized in three fragments that had to be enzymatically ligated. In addition, it had no 3′-poly(A) tail.
With these considerations in mind, we designed an artificial mRNA for chemical synthesis in a single synthesizer run followed by enzymatic addition of a 5′-cap and a 3′-poly(A) tail. Before the addition of the cap, the 5′-end of the RNA must be pyrophosphorylated. We devised a scheme in which the artificial mRNA is prepared by the following steps: (i) chemical synthesis of the mRNA sequence in the solid phase, (ii) pyrophosphorylation of the synthetic RNA at the 5′-end on the solid support, (iii) cleavage of the 5′-pyrophosphorylated RNA from the solid support followed by enzymatic capping and (iv) enzymatic polyadenylation of the 5′-capped RNA at the 3′-end. At the outset of this study, it was not known whether an RNA oligomer of the necessary length could be chemically synthesized or whether the 5′-end of such a long RNA oligomer could be pyrophosphorylated.
We have previously developed a method for synthesizing very long RNA oligomers with 2-cyanoethoxymethyl (CEM) as the 2′-hydroxy protecting group (9). By the CEM method, RNA can be synthesized more simply and easily than by the conventional TBDMS method described by Usman et al. (10). We have used the CEM method to synthesize a 110-nt RNA oligomer with the sequence of a candidate precursor microRNA and shown that it had biological activity (11). To our knowledge, this 110-mer is the longest RNA reported to have been chemically synthesized to date. In the present study, we used the CEM method to synthesize the 130- and 170-mer sequences required for our designed artificial mRNAs.
After the mRNA sequence has been synthesized, it is first monophosphorylated and then pyrophosphorylated at the 5′-end. Monophosphorylation of RNA proceeds almost quantitatively with commercially available phosphorylating reagents (12). However, existing phosphorylating reagents for the pyrophosphorylation of RNA are not very reactive (12,13). Because reagents have only poor accessibility to the internal pores of a solid support filled with long RNA oligomers, the low reactivity of existing pyrophosphorylating reagents presents a real problem for pyrophosphorylation of the 5′-end of very long RNA oligomers on the solid support. Therefore a more reactive pyrophosphorylating reagent is necessary.
Here, we report the synthesis by the CEM method of a 130-nt mRNA sequence encoding a 33-amino-acid peptide that includes the sequence of glucagon-like peptide-1 (GLP-1), a peptide that stimulates glucose-dependent insulin secretion from the pancreas (14–16). We also developed novel, highly reactive pyrophosphorylating reagents and used them to pyrophosphorylate the 130-mer RNA at the 5′-end. We completed the synthesis of the artificial mRNA by enzymatically adding a 5′-cap and a 3′-poly(A) tail. We showed that our artificial mRNA supported GLP-1 production in a cell-free protein synthesis system and in Chinese hamster ovary (CHO) cells.
MATERIALS AND METHODS
General methods
UV spectra were recorded with a Hitachi U-2810 double-beam spectrophotometer. 31P-NMR spectra were recorded on a Bruker DRX 500 spectrometer with 3-trimethylsilylpropionate-2,2,3,3-d4 sodium salt or H3PO4 as the standard. Electrospray ionization (ESI) mass spectra were recorded on a JEOL JMS-SX 102 mass spectrometer. Analytical high performance liquid chromatography (HPLC) was performed on a Shimadzu HPLC system equipped with a DNAPac PA100 (4 × 250 mm, Dionex) or Mightysil RP-18 GP (4.6 × 150 mm, Kanto Kagaku) column. Matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectra were obtained with a Bruker Autoflex (Bruker Daltonics, Billerica, MA) or a Voyager-DE STR (Applied Biosystems, Foster City, CA, USA) spectrometer. Reagents and solvents were purchased from commercial suppliers and used without further purification. Anhydrous solvents were from Wako Pure Chemical Industries (Osaka, Japan) and tetrabutylammonium fluoride (TBAF) was from Nacalai Tesque (Kyoto, Japan). The following HPLC solvent systems were used: for reverse-phase HPLC, buffer A [5% acetonitrile in 50 mM triethylammonium acetate (TEAA), pH 7] and buffer B (90% acetonitrile in 50 mM TEAA, pH 7); and for anion-exchange HPLC, buffer C (10% acetonitrile in 25 mM Tris–HCl, pH 8.0) and buffer D (10% acetonitrile containing 700 mM NaClO4 in 25 mM Tris–HCl, pH 8.0). Dialysis was carried out with a 1000-Da molecular-weight-cutoff Spectra/Pore® membrane (Spectrum Laboratories, Laguna Hills, CA, USA).
Synthesis of 130-mer RNA
Commercially available controlled-pore glass (CPG) solid support with a pore size of 2000Å derivatized with N6-benzoyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-(t-butyldimethylsilyl) adenosine was placed in a column fitted with a filter and the column was installed in an Applied Biosystems Expedite Model 8909 nucleic acid synthesizer. The RNA was synthesized on a 0.6-μmol scale from the CEM amidites (CEM-A phosphoramidite, N6-acetyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-(2-CEM) adenosine 3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite; CEM-C phosphoramidite, N4-acetyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-(2-CEM) cytidine 3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite; CEM-G phosphoramidite, N2-phenoxyacetyl-5′-O-(4,4′-dimethoxytrityl)-2′-O-(2-CEM) guanosine 3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite; CEM-U phosphoramidite, 5′-O-(4,4′-dimethoxytrityl)-2′-O-(2-CEM) uridine 3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite) were prepared as described earlier (11). The detritylation reagent was 3% trichloroacetic acid in dichloromethane (CH2Cl2). For the coupling reaction, amidites were used as 0.05 M solutions in acetonitrile (CH3CN), and the activator was 5-benzylmercaptotetrazole (BMT). The capping reagent was a mixture of phenoxyacetic anhydride (Pac2O) in tetrahydrofuran (THF) and N-methylimidazole (NMI) and 2,6-lutidine in THF, and the oxidizing agent was 0.1 M iodine (I2) in THF/pyridine/water.
Monophosphorylation of 130-mer RNA at the 5′-end
Monophosphorylation of the 130-mer RNA was carried out with the 130-mer RNA still attached to the solid support. The solid support with the attached 130-mer RNA (65.3 mg, 0.5 μmol loading) was placed in a Libra tube (HiPep Laboratories, Kyoto, Japan), washed three times with dry CH3CN (3 × 3 ml) under mild argon pressure from an argon-filled balloon attached to the Libra tube, and dried in vacuo for 1 h. To the Libra tube BMT (30.8 mg, 160 μmol) and 0.1 M (2-cyanoethoxy)-2-[2′-O-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethoxy-N,N-diisopropylaminophosphine in CH3CN (400 μl, 40 μmol) were added and the mixture was left to stand for 10 min. The liquid was removed under mild argon pressure, BMT (30.8 mg, 160 μmol) and 0.1 M (2-cyanoethoxy)-2-[2′-O-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethoxy-N,N-diisopropylaminophosphine in CH3CN (400 μl, 40 μmol) were added again, and the mixture was again left for 10 min. For subsequent treatment, the following protocol was used: (i) washing with dry pyridine (3 × 1 ml), (ii) oxidation with 0.05 M I2 solution in pyridine/water (9:1, v/v; 3 × 1 ml for 15 s), (iii) washing with dry pyridine (3 × 1 ml), (iv) washing with dry CH2Cl2 (3 × 1 ml), (v) capping with 0.2 M Pac2O in CH3CN (0.5 ml) and 20% NMI and 30% 2,6-lutidine in CH3CN (0.5 ml) for 2 min, (vi) washing with dry CH2Cl2 (3 × 1 ml), (vii) treatment with 3% dichloroacetic acid in toluene (3 × 2 ml; 15 s), (viii) washing with dry CH2Cl2 (3 × 1 ml), (ix) treatment with N,O-bis(trimethylsilyl)acetamide (BSA)-pyridine (1:1, v/v; 360 μl) for 20 min, (x) addition of 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) (72 μl) for 10 min, (xi) washing with dry pyridine (3 × 1 ml), (xii) washing with dry CH2Cl2 (3 × 1 ml), (xiii) treatment with MeOH/triethylamine (Et3N) (9:1, v/v; 2 ml) for 3 h, (xiv) washing with dry CH3CN (3 × 2 ml) and (xv) drying in vacuo for 1 h.
A portion (0.1 of 0.5 μmol loading) of the solid support with the attached 130-mer RNA monophosphorylated at the 5′-end was treated with 28% aqueous ammonia solution and EtOH (3:1, v/v; 2 ml) for 15 h at 35°C to cleave the phosphorylated RNA from the resin and to remove the base-protecting groups. The reaction mixture was filtered and the filtrate was concentrated to dryness under reduced pressure. The residue was dissolved in dry dimethyl sulfoxide (DMSO; 0.5 ml), then CH3NO2 as an acrylonitrile scavenger (10 μl) and 1.0 M TBAF in DMSO (1.0 ml) were added to remove the CEM group. The reaction mixture was stirred for 5 h at room temperature, the reaction was quenched at 0°C with 1.0 M Tris–HCl (1.0 ml), and the RNA oligomer was precipitated with EtOH (40 ml).
A small amount of the precipitate was treated with MazF to generate fragments from the 5′-end for analysis by MALDI–TOF mass spectrometry. Monophosphorylation of 130-mer RNA at the 5′-end was confirmed to be quantitative. MALDI–TOF–MS calcd for the 11-mer RNA fragment monophosphorylated at the 5′-end [M+H]+, 3653.46; found, 3654.68. MALDI–TOF–MS calcd for the 25-mer RNA fragment monophosphorylated at the 5′-end [M+H]+, 8269.12; found, 8271.38.
Pyrophosphorylation of 130-mer RNA at the 5′-end
Pyrophosphorylation of the 130-mer RNA was carried out with the 5′-monophosphorylated 130-mer RNA still attached to the solid support. Pyrophosphorylating reagent was prepared by drying 1-hydroxy-6-(trifluoromethyl)benzotriazole (CF3-HOBt; 65.0 mg, 320 μmol) in vacuo for 40 min and adding 0.1 M (2-cyanoethoxy)-2-[2′-O-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethoxy-N,N-diisopropylaminophosphine in CH3CN (640 μl, 64 μmol). The mixture was stirred at room temperature for 2 h. While the pyrophosphorylating reagent was being prepared, the solid support was washed with CH3CN and dried in vacuo for 15 min. Then half of the freshly prepared pyrophosphorylating reagent was added to the solid support with the attached 5′-monophosphorylated 130-mer RNA (25.8 mg, 0.2 μmol) in a Libra tube. NMP (36 μl) was then added to the solid support and the mixture was left for 30 min. The liquid was removed under mild argon pressure and the remaining half of the pyrophosphorylating mixture was added to the solid support. A further portion of NMP (36 μl) was added and the mixture was left for 30 min. For subsequent treatment, the following protocol was used: (i) washing with dry pyridine (3 × 1 ml), (ii) washing with dry CH2Cl2 (3 × 1 ml), (iii) treatment with 3% dichloroacetic acid in toluene (3 × 2 ml; 15 s), (iv) washing with dry CH2Cl2 (3 × 1 ml), (v) treatment with BSA-pyridine (1:1, v/v; 360 μl) for 20 min, (vi) addition of DBU (72 μl) and leaving for 10 min, (vii) washing with dry pyridine (3 × 1 ml), (viii) washing with dry CH2Cl2 (3 × 1 ml) and (ix) drying in vacuo for 15 min.
The solid support (0.2 μmol equiv.) with the attached 130-mer RNA pyrophosphorylated at the 5′-end was treated with 28% aqueous ammonia solution and EtOH (3:1, v/v; 2 ml) for 15 h at 35°C. The reaction mixture was filtered and concentrated to dryness under reduced pressure. The residue was dissolved in dry DMSO (0.5 ml), then CH3NO2 (10 μl) and 1.0 M TBAF in DMSO (1.0 ml) were added. The reaction mixture was stirred for 5 h at room temperature, the reaction was quenched at 0°C with 1.0 M Tris–HCl (1.0 ml), and the RNA oligomer was precipitated with EtOH (40 ml).
The precipitate was purified by semipreparative anion-exchange HPLC (DNAPac PA100, 9 × 250 mm, Dionex). Elution was carried out with a 5–45% linear gradient of buffer D in buffer C. Dialysis was carried out with a 1000-Da molecular-weight-cutoff Spectra/Pore® membrane. The solution was concentrated to dryness under reduced pressure and the residue was dissolved in water (1.0 ml) to give 130-mer RNA pyrophosphorylated at the 5′-end (21.2 ODU; 15.9 nmol, 8.0%).
A sample of the 5′-pyrophosphorylated 130-mer was treated with MazF to generate fragments from the 5′-end for analysis by MALDI–TOF mass spectrometry. Pyrophosphorylation of 130-mer RNA at the 5′-end was confirmed to have proceeded. MALDI–TOF–MS calcd for the 11-mer RNA fragment pyrophosphorylated at the 5′-end [M+H]+, 3733.42; found, 3734.76. MALDI–TOF–MS calcd for the fragment with 25-mer RNA pyrophosphorylated at the 5′-end [M+H]+, 8349.08; found, 8352.06.
Synthesis, monophosphorylation and pyrophosphorylation of 170-mer RNA
A 170-mer RNA that includes a 40-mer poly(A) tail as a part of the synthetic sequence was twice synthesized on a 0.6-μmol scale to give 1.2 μmol of 170-mer. Of this, 1.1 μmol was monophosphorylated at the 5′-end and 1.0 μmol of the monophosphorylated product was pyrophosphorylated. All procedures used were similar to those described above for 130-mer RNA. The final yield of 5′-pyrophosphorylated product was 85.7 ODU (47.8 nmol, 4.8%).
A sample of the 5′-pyrophosphorylated 170-mer was treated with MazF to generate fragments from the 5′-end for analysis by MALDI–TOF mass spectrometry. Pyrophosphorylation of 170-mer RNA at the 5′-end was confirmed to have proceeded. MALDI–TOF–MS calcd for the 11-mer RNA fragment pyrophosphorylated at the 5′-end [M+Na]+, 3755.40; found, 3757.96. MALDI–TOF–MS calcd for the 25-mer RNA fragment pyrophosphorylated at the 5′-end [M+Na]+, 8371.06; found, 8375.65.
Capping and poly(A)-tailing reactions
The capping and poly(A)-tailing reactions were carried out with the mScript™ mRNA Production System from Epicentre Biotechnologies (Madison, WI, USA). First, the cap 1 structure, m7GpppNm, which includes a 5′–5′-triphosphate linkage, was attached to the 5′-end of the chemically synthesized, 5′-pyrophosphorylated 130-mer or 170-mer RNA by incubating the RNA (300 μg) with capping enzyme (480 U) and 2′-O-methyltransferase (48 U) in 50 mM Tris–HCl, pH 8.0, containing 6 mM KCl, 1.25 mM MgCl2, 1 mM GTP, 0.2 mM S-adenosylmethionine and RNase inhibitor (960 U) in a total volume of 1.2 ml at 37°C for 2 h. The capped RNA was extracted with phenol/chloroform, concentrated by ethanol precipitation, and resuspended in RNase-free water. Then the poly(A) tail was added by incubating the capped 130-mer RNA with poly(A) polymerase (150 U) in 50 mM Tris–HCl, pH 8.0, containing 250 mM NaCl, 10 mM MgCl2, 1 mM ATP and RNase inhibitor (300 U) in a total volume of 1.8 ml at 37°C for 4 h. The capped, poly(A)-tailed RNA was extracted with phenol/chloroform, concentrated by ethanol precipitation and resuspended in RNase-free water. Finally, the artificial mRNA was purified on a MicroSpin G-25 Column (GE Healthcare, Buckinghamshire, England). Samples were analyzed by electrophoresis on a 5% denaturing polyacrylamide gel.
Analysis of MazF digestion products of RNA
MazF is an endoribonuclease that cleaves specifically at the 5′-end of ACA sequences (17). RNA (10 μg) was incubated with MazF (80 U; Takara Bio, Otsu, Japan) in 40 mM sodium phosphate, pH 7.5, containing 0.01% Tween 20 in a total volume of 20 μl at 37°C for 1 h. The digestion products were extracted with phenol/chloroform, concentrated by ethanol precipitation, and resuspended in RNase-free water. For MALDI–TOF mass spectrometry, the digestion products were further purified on ZipTip C18 reverse-phase microcolumns (Millipore, Bedford, MA). MALDI–TOF mass spectra were acquired in positive-ion mode with a matrix solution consisting of 10 mg/ml 3-hydroxypicolinic acid and 1 mg/ml diammonium citrate. The potential for linear acceleration was 19 kV and the potential for reflector acceleration was 20 kV. The IS/2 potential was set at 16.6 kV. Ion extraction was delayed by 130 ns, and at least 150 shots were accumulated for each sample.
Expression of GLP-1 peptide in a wheat germ lysate
In vitro expression of GLP-1 peptide with an additional two N-terminal amino acids, MGHAEGTFTSDVSSYLEGQAAKEFIAWLVKGRG, was performed with an RTS 100 Wheat Germ CECF Kit (Roche Diagnostics, Mannheim, Germany) according to the supplier’s instructions.
Cell culture and transfection
CHO-K1 cells were grown in Ham’s F-12K medium (Sigma) supplemented with 2 mM l-glutamate, 1.5 g/l sodium bicarbonate, and 10% heat-inactivated fetal bovine serum at 37°C in a humidified incubator in an atmosphere of 5% CO2 and 95% air. RNA transfections were performed with Nucleofector Reagent (Amaxa Biosystems, Cologne, Germany) according to the supplier’s instructions.
Sandwich ELISA
GLP-1 in wheat germ lysate or cell lysate was determined with a microplate coated with goat anti-mouse IgG (Pierce, Rockford, IL, USA) used in combination with a GLP-1 (Active) ELISA Kit (Linco Research, St Charles, MO, USA) according to the supplier’s instructions. The goat anti-mouse IgG-coated plate was rinsed with 3 × 200 μl of TBS-T (50 mM Tris–HCl, pH 7.5, containing 0.05% Tween 20), after which 0.1 μg/ml mouse anti-GLP-1 antibody HYB147-12 (100 μl; Abcam, Cambridge, MA, USA) in TBS-T was added. The plate was incubated at 37°C for 1 h with shaking, each well was washed with wash buffer (3 × 300 μl), and assay buffer and GLP-1 sample (100 μl) were added. The plate was incubated at 4°C for 24 h, each well was again washed with wash buffer (3 × 300 μl), and GLP-1 detection conjugate was added. The plate was then incubated at room temperature for 2 h. Finally, each well was washed with wash buffer (3 × 300 μl), substrate solution was added, and the fluorescence intensity was measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm with a Wallac ARVO HTS 1420 Multilabel Counter (Perkin Elmer Life Sciences).
RESULTS AND DISCUSSION
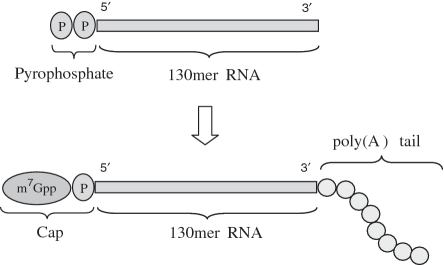
We synthesized artificial mRNA by (i) solid-phase synthesis of the RNA sequence from CEM amidites, (ii) monophosphorylation and pyrophosphorylation of the 5′-end of the RNA still attached to the solid support, (iii) cleavage of the pyrophosphorylated RNA from the solid support followed by deprotection and purification, (iv) addition of the 5′-cap to the purified, pyrophosphorylated RNA with capping enzyme and (v) polyadenylation of the 3′-end of the 5′-capped RNA with poly(A) polymerase (Figure 1). We also synthesized a 170-mer RNA that includes a chemically synthesized 40-mer poly(A) tail as a part of the synthetic sequence. Including a chemically synthesized poly(A) tail would reduce the number of enzymatic steps needed, and the chemically synthesized product would also have the being a single molecular species, in contrast to the distribution of poly(A) tail lengths obtained with the use of poly(A) polymerase.
Figure 1.
Enzymatic conversion of 130-mer to artificial mRNA.
Design of the synthetic sequence
We designed the synthetic sequence of a 130-mer RNA with a 5′-untranslated region (5′-UTR), a GLP-1-coding region, and a 3′-UTR (Figure 2). The 20-nt 5′-UTR is followed by a region encoding the 33 amino acids of GLP-1 from the AUG start codon to the final GGA codon. To increase the efficiency of translation, the 5′-UTR and the start of the coding region were designed to include a Kozak consensus sequence, which is characterized by a purine three bases upstream of the start codon and a guanine immediately downstream (18). The synthetic sequence of the 130-mer includes three MazF cleavage sites, one in the 5′-UTR and two near the start of the coding region (one of the latter was generated by introducing a silent C→A mutation at position 41). These cleavage sites facilitated analysis of the 5′-cap by MALDI–TOF mass spectrometry after MazF digestion. The 8-nt 3′-UTR following the stop codon corresponds to the first 8 nt of the 3′-UTR of the glucagon gene. Both the 5′- and the 3′-UTR were shortened from the respective untranslated regions of the glucagon gene to give RNA of a manageable length for chemical synthesis.
Figure 2.
(a) Sequence of artificial mRNA. Underline, start codon; italics, stop codon; bold, untranslated regions; blue, Kozak consensus sequence; red, MazF cleavage sites; enclosed nucleoside, C→A silent point mutation. (b) Peptide encoded by artificial mRNA. Green, GLP-1 (residues 7–37 of preproglucagon).
Synthesis of 130-mer RNA and monophosphorylation of the 5′-end
A 130-mer RNA oligomer was synthesized on a 0.6-μmol scale by the phosphoramidite method modified for CEM chemistry (Scheme 1). As in our previous synthesis of a 110-mer (10), the solid support was CPG with a pore size of 2000 Å derivatized with N4-benzoyl-2′-O-TBDMS-riboadenosine as the leader nucleoside. (In our experience, a large-pore resin is necessary to obtain good yields in the synthesis of very long oligomers.) The detritylation reagent was trichloroacetic acid in dichloromethane. For the coupling reaction, amidites were used as 0.05 M solutions in acetonitrile, and the activator was 5-benzylmercaptotetrazole rather than the 5-ethylthio-1H-tetrazole used in the original method (9) because it was found to be the most suitable for use with the CEM method. The capping reagent was a mixture of Pac2O in THF and NMI and 2,6-lutidine in THF, and the oxidizing agent was 0.1 M I2 in THF/pyridine/water. A small amount of the 130-mer synthesized as described earlier was cleaved from the solid support, deprotected and treated with MazF to generate fragments from the 5′-end for analysis by MALDI–TOF mass spectrometry (Supplementary Figure 3).
Scheme 1.
Solid-phase synthesis of RNA by the CEM method (i) and monophosphorylation of RNA at the 5′-end with 1 or 2 (ii).
The remainder of the 130-mer RNA, while still on the solid support, was monophosphorylated with phosphorylating reagent 1 or 2 (Scheme 1). Phosphorylation proceeded almost quantitatively with either reagent. After oxidation and capping, the phosphate-protecting groups (the cyanoethyl and sulfonylethyl groups) were removed by treatment with 1,8-diazabicyclo[5.4.0]undec-7-ene-bis(trimethylsilyl)acetamide (DBU–BSA) (12,19). Finally, 130-mer monophosphorylated at the 5′-end was converted to the triethylammonium salt by treatment with triethylamine-MeOH and used for the pyrophosphorylation studies described below.
Tests of reagents for pyrophosphorylation of RNA at the 5′-end
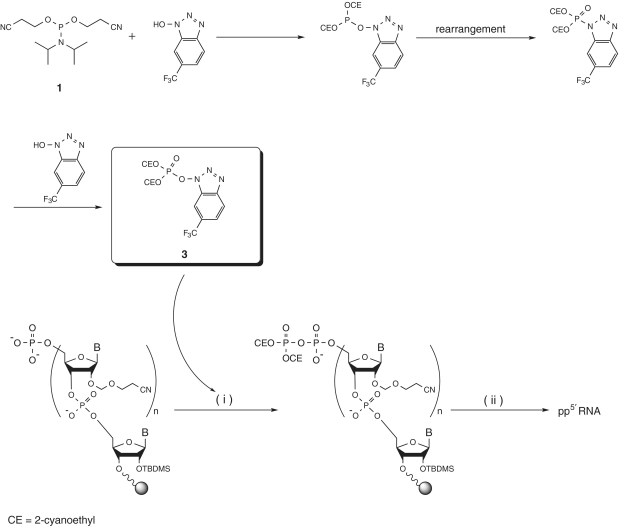
In the cell, the addition of the 5′-cap to the nascent mRNA involves the removal of one phosphate group of the original 5′-triphosphate terminus by a phosphatase followed by condensation between GTP and the remaining 5′-pyrophosphate terminus catalyzed by the capping enzyme guanylyl transferase. With a view to using capping enzyme to prepare capped RNA from RNA pyrophosphorylated at the 5′-end, we investigated the pyrophosphorylation of 130-mer RNA at the 5′-end. However, existing phosphorylating reagents for pyrophosphorylation are only poorly reactive (12,13). Because they are particularly unsuitable for pyrophosphorylation of the 5′-end of a poorly accessible, very long RNA oligomer attached to the internal pores of the solid support, we decided to develop more reactive pyrophosphorylating reagents suitable for use with very long RNA oligomers. Accordingly, we developed pyrophosphorylating reagents with a phosphotriester structure, which is much more reactive than the phosphodiester structure of existing reagents. We developed the phosphotriester reagents by reference to a method for pyrophosphate bond formation reported by Sekine and coworkers (20). Pyrophosphorylating reagent 3 was prepared from phosphorylating reagent 1 and five equivalents of 1-hydroxy-6-(trifluoromethyl)benzotriazole (CF3-HOBt; Scheme 2). The reaction presumably proceeded via an O-N phosphoryl rearrangement. Reagent 3 (observed in the 31P NMR spectrum at −1.2 ppm) was prepared in situ for use in pyrophosphorylation studies without post-processing or purification (Supplementary Figure 1).
Scheme 2.
Synthesis of pyrophosphorylating reagent 3. Reagents and conditions: 1 (80–160 equiv.), CF3-HOBt (400–800 equiv.), CH3CN, rt, 1 h. Pyrophosphorylation of RNA at the 5′-end. Reagents and conditions: (i) (a) 3 (80–160 equiv., crude mixture), CH3CN-NMP (9:1, v/v), rt, 10 min, (b) washing: pyridine, CH2Cl2, (ii) (a) 16.7% DBU/pyridine-BSA (1:1, v/v), rt, 10 min, (b) washing: pyridine, CH2Cl2, (c) conc. NH3-EtOH (3:1, v/v), 35°C, 15 h, (d) 0.67 M TBAF, 0.67% nitromethane/DMSO, rt, 5 h.
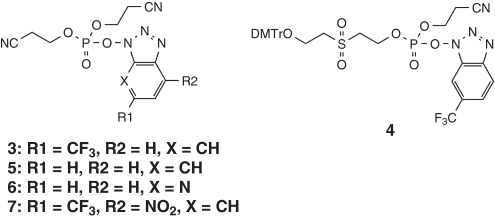
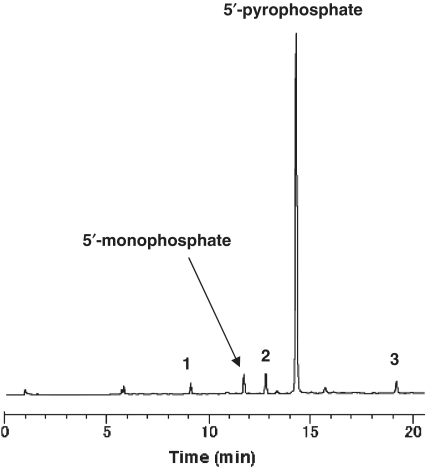
We first used trimer RNA as a model substrate for studies of our new pyrophosphorylating reagents (Scheme 3 and Table 1). Trimer RNA monophosphate prepared as described above was used as a substrate for pyrophosphorylation on a CPG solid support (Supplementary Figure 2). Reagent 3 was prepared by reaction of 1 and CF3-HOBt at room temperature for 1 h. Pyrophosphorylation with 3 gave trimer RNA pyrophosphate in 84% yield (Figure 3). Because the amount of trimer RNA sample obtained by cleavage from the CPG solid support and removal of the protecting groups was almost the same for the monophosphate and the pyrophosphate, the pyrophosphorylation yield was calculated by dividing the purity of the trimer RNA pyrophosphate by that of the trimer RNA monophosphate. Reagent 4, which was prepared from 2, gave a yield of pyrophosphorylated product that was a little lower than that obtained with 3. In addition, the yield obtained with 4 was sensitive to moisture. Pyrophosphorylating reagents 5–7 were prepared with a modified trifluoromethylbenzotriazole moiety. Pyrophosphorylation with 5, which lacks a trifluoromethyl group on the benzotriazole ring system, gave trimer RNA pyrophosphate in 61% yield. Because about 26% of the starting material remained, the reactivity of 5 was judged to be lower than that of 3. Pyrophosphorylation with 6, which has a 7-azabenzotriazole moiety, gave a yield similar to that obtained with 3. However, pyrophosphorylation with 7, which was expected to be highly reactive, gave only a poor yield. The 4-nitro-6-(trifluoromethyl)benzotriazole moiety is expected to be a good leaving group, and we suspect that 7 was so reactive that it had largely decomposed before reaction with trimer RNA monophosphate could occur.
Scheme 3.
Structures of pyrophosphorylating reagents.
Table 1.
Product distribution observed after 5′-pyrophosphorylation of trimer RNA
| Entry | Reagent | Reaction time (h) | HPLC peak area (%) |
Yield (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| 5′-Pyrophosphate (14.3 min)a | 5′-Monophosphate (11.7 min) | By-products |
|||||||
| 1 (9.1 min) | 2 (12.8 min) | 3 (19.2 min) | Others | ||||||
| 1 | 3 | 1 | 76.0 | 3.6 | 1.9 | 4.0 | 2.6 | 11.9 | 84 |
| 2 | 4 | 2 | 65.0 | 7.5 | 4.9 | 7.9 | 4.7 | 10.0 | 72 |
| 3 | 5 | 1 | 55.5 | 26.4 | 5.0 | 0.8 | 2.0 | 10.3 | 61 |
| 4 | 6 | 1 | 71.3 | 5.0 | 7.0 | 1.2 | 5.4 | 10.1 | 78 |
| 5 | 7 | 0.25 | 2.7 | 6.9 | 35.1 | 2.6 | 26.9 | 25.8 | 3 |
aThe numbers in parentheses are the HPLC retention times.
Figure 3.
Anion-exchange HPLC of the crude mixture obtained by 5′-pyrophosphorylation of trimer RNA with 3. See Entry 1, Table 1. The numbers 1–3 correspond to the by-products 1–3 shown in Table 1.
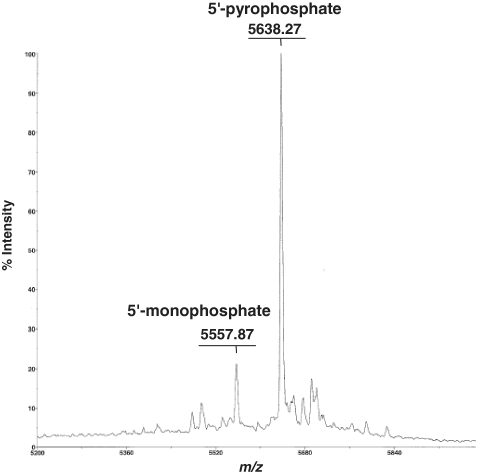
We next studied the pyrophosphorylation of very long RNA oligomers on the 5′-end. Pyrophosphorylation of 122-mer RNA monophosphate with 3 gave the RNA pyrophosphate. However, by-products were observed on HPLC, even though we used the best reaction conditions found for the pyrophosphorylation of trimer RNA monophosphate, conditions under which these by-products were hardly observed. The by-products may have been generated by overreaction during pyrophosphorylation. When 5, which is less reactive than 3, was used for the pyrophosphorylation of 122-mer RNA monophosphate, the pyrophosphate was obtained in reasonable yield without the generation of by-products. Reagent 5 is therefore the more suitable for the pyrophosphorylation of very long RNA oligomers. Pyrophosphorylation of the 122-mer was confirmed by MALDI–TOF mass spectrometric detection of the expected 5′-end fragment generated by MazF treatment (Figure 4).
Figure 4.
MALDI–TOF mass spectrum of MazF digestion products of the crude mixture obtained by 5′-pyrophosphorylation of 122-mer RNA with 5.
Reagent 3 may be especially suitable for the pyrophosphorylation of RNA of up to about 20 nt, such as siRNA strands, while 5 is more suitable for the pyrophosphorylation of RNA longer than about 50 nt. We assume that most of the by-products shown in Entry 1 of Table 1 (for the use of reagent 3) result from reaction at internucleotide linkages. These by-products are hardly noticeable for short oligomers but are very noticeable for long oligomers. Reagent 5 should be used for the 5′-pyrophosphorylation of long oligomers because the side reaction will not occur easily due to the lower reactivity of the reagent. It is possible that the phosphotriester pyrophosphorylating reagents that we have developed can be used for the pyrophosphorylation not only of nucleic acids but also of small molecules, peptides and sugars.
Preparation of 5′-pyrophosphorylated 130-mer and 170-mer RNA
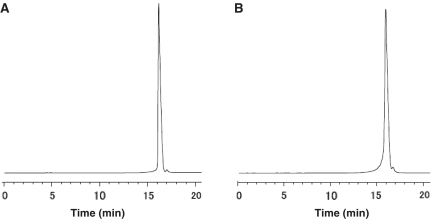
Pyrophosphorylation of 130-mer and 170-mer RNA monophosphates was carried out with reagents 3, 4 and 5. After pyrophosphorylation, the protecting groups of the phosphate group at the 5′-end (the cyanoethyl and sulfonylethyl groups) were removed by treatment with DBU–BSA. Cleavage of the oligomers from the CPG solid support and removal of the base-protecting groups was carried out by treatment with a mixture of 28% aqueous ammonia and EtOH (3:1, v/v) at 35°C for 15 h. The CEM protecting group was removed by treatment with 0.67 M TBAF in DMSO, with 0.67% nitromethane as an acrylonitrile scavenger, at room temperature for 5 h (Supplementary Figure 4) (21). Purification was carried out by semipreparative HPLC on a DNAPac PA 100 column and pure fractions were desalted by dialysis. The total yields of 130-mer and 170-mer 5′-pyrophosphorylated oligomers, including both the synthetic and the pyrophosphorylation steps, were 8.0 and 4.8%, respectively, corresponding to respective average coupling yields of 98.1 and 98.2% (Figure 5).
Figure 5.
Anion-exchange HPLC of purified 5′-pyrophosphorylated RNA 130-mer (A) and 170-mer (B).
Enzymatic steps in the production of artificial mRNA
To attach the cap 1 structure to the 5′-end of the 5′-pyrophosphorylated 130-mer or 170-mer RNA oligomer, we used a combination of a capping system and 2′-O-methyltransferase. The capping system catalyzes the formation of the cap 0 structure from GTP and 5′-pyrophosphorylated RNA, while 2′-O-methyltransferase completes the formation of the cap 1 structure by adding a methyl group at the 2′-O-position of the second nucleotide from the 5′-end of the RNA molecule (the first nucleotide before the addition of cap 0). Cap 1 rather than cap 0 was used because it confers a higher translation efficiency (22).
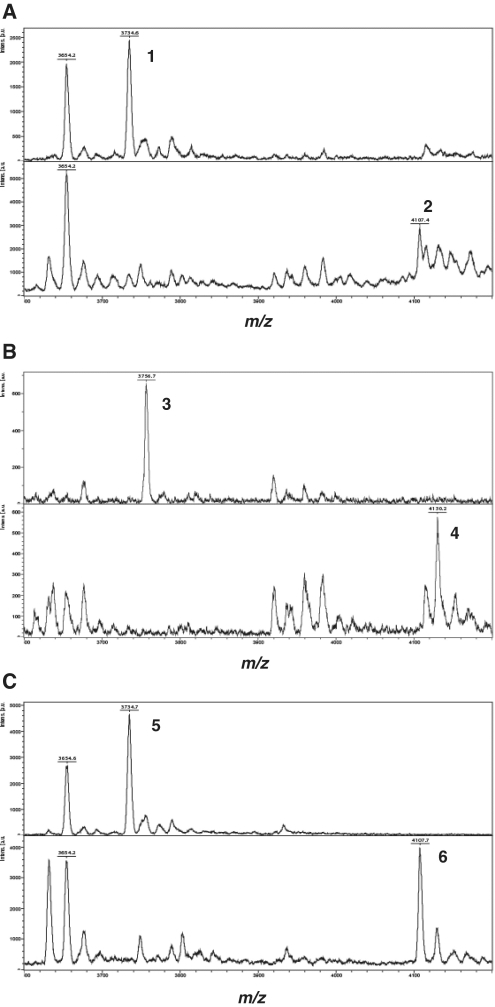
Whereas the 170-mer RNA has a chemically synthesized 40-mer poly(A) tail, the 130-mer RNA does not have a poly(A) tail. To add a poly(A) tail to the 130-mer, we used poly(A) polymerase, which catalyzes the template-independent addition of poly(A) to the 3′-end of the RNA with ATP as a substrate. To confirm that cap 1 had been correctly added, both RNA oligomers were digested with MazF and the digestion products were analyzed by MALDI–TOF mass spectrometry. MazF digestion of uncapped 130-mer produced, as expected, an 11-nt fragment of molecular weight 3734 Da that included the 5′-end (Figure 6A, upper panel), while capped 130-mer produced the expected 12-nt fragment of molecular weight 4107 Da (Figure 6A, lower panel). The difference in molecular weight, 373 Da, corresponds exactly to the molecular weight of the cap 1 structure (Table 2). Similar results were obtained for the 170-mer RNA (Figure 6B). These experiments demonstrate that cap 1 was correctly attached to both the 130-mer and the 170-mer RNA. A MazF digestion product of molecular weight 3654 Da corresponds to the 5′-end fragment derived from 5′-monophosphorylated RNA, which cannot act as a substrate in the capping system. Accordingly, this fragment showed no change after treatment of 5′-monophosphorylated RNA with capping enzyme and 2′-O-methyltransferase (Figure 6A).
Figure 6.
MALDI–TOF mass spectrometry of MazF digestion products of RNA oligomers. (A) 130-mer, (B) 170-mer and (C) 127-mer. The upper panels show the mass spectra of the MazF digestion products of uncapped RNA and the lower panels those of capped RNA.
Table 2.
Masses of 5′-end fragments produced by MazF digestion
| RNA molecule | Peak no.a | Pyrophosphate |
Peak no.a | Cap 1 |
Mass difference (b−a) | ||
|---|---|---|---|---|---|---|---|
| Calculated mass | Observed mass (a) | Calculated mass | Observed mass (b) | ||||
| 130-mer | 1 | 3734.6 | 2 | 4107.4 | 372.8 | ||
| 170-mer | 3 | 3734.2 | 3756.7 | 4 | 4107.4 | 4130.2 | 373.5 |
| 127-mer | 5 | 3734.7 | 6 | 4107.7 | 373 | ||
aPeak numbers correspond to the numbered fragments shown in Figure 6.
To confirm that the poly(A) tail had been added, 130-mer RNA that had been treated with poly(A) polymerase in the presence of ATP was analyzed by electrophoresis on a denaturing 5% polyacrylamide gel (Figure 7A). The 130-mer was converted to a higher-molecular-weight form consistent with the addition of a poly(A) tail of ∼70 nt. However, unchanged 130-mer remained that had not been polyadenylated. Polyadenylation may have been incomplete because some of the 130-mer retained the TBDMS group used to protect the 3′-end nucleoside. The deprotection conditions were specifically designed for the removal of CEM, and TBDMS is known to be incompletely removed by TBAF (23–25). A poly(A) tail presumably cannot be attached to RNA that retains the TBDMS group at the 3′-end because such RNA may be a poor substrate for poly(A) polymerase. When CEM was used as the 3′-end protecting group instead of TBDMS, the 130-mer was fully polyadenylated (Figure 7D), consistent with the fact that the CEM group is easily removed by TBAF treatment. We cannot quote the yield of the final purified artificial mRNA because of the distribution of poly(A) tail lengths.
Figure 7.
Polyacrylamide gel electrophoresis of the poly(A)-tailing reaction. RNA oligomers were analyzed on a 5% denaturing polyacrylamide gel stained with ethidium bromide. 130-mer, 170-mer and 127-mer: chemically synthesized RNA. 130-mer mRNA, 170-mer mRNA and 127-mer mRNA: enzymatically capped and poly(A)-tailed RNA (except for the 170-mer, which included a chemically synthesized poly(A) tail). 130-mer mRNA (CEM): enzymatically capped and poly(A)-tailed RNA in which CEM instead of TBDMS was used as the 3′-end protecting group.
Biological activity of artificial mRNA
Chemically synthesized 130-mer and 170-mer RNA encoding GLP-1 peptide was enzymatically treated as described earlier to produce a GLP-1 artificial mRNA. To assess the biological activity of this artificial mRNA, we performed GLP-1-expression experiments in a cell-free protein synthesis system and in whole cells and used sandwich ELISA to determine GLP-1 produced.
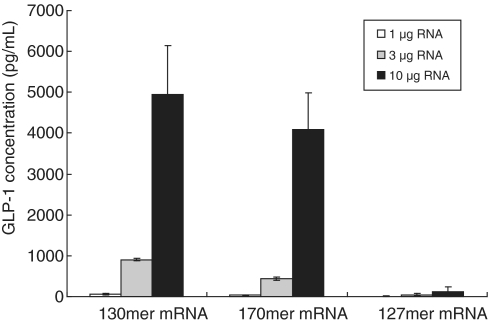
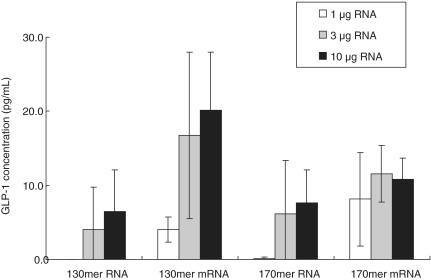
A eukaryotic cell-free protein synthesis system based on wheat germ lysate was used to express GLP-1 peptide. As a negative control, an RNA that would not be expected to support translation was synthesized. This negative control was a 127-mer that was identical to the 130-mer except that it lacked the second codon, GGA, so that it had a disrupted Kozak sequence. We showed that mRNA derived from the 127-mer had the cap 1 structure and a poly(A) tail consisting of ∼60 nt (Figures 6C and 7C). As expected, this mRNA did not support the expression of GLP-1, whereas mRNA derived from both the 130-mer and the 170-mer supported GLP-1 expression in a dose-dependent manner (Figure 8). These results demonstrate that artificial mRNA derived from chemically synthesized RNA functioned in vitro. In addition, the length of the 5′-UTR and of the poly(A) tail affected the translation efficiency, as reported earlier (26,27). Thus, mRNA derived from a 122-mer RNA with a 12-nt 5′-UTR supported only a low level of GLP-1 expression compared with mRNA derived from the 130-mer, which had a 20-nt 5′-UTR (data not shown). And mRNA derived from the 130-mer, which had a longer average length of poly(A) tail than the 170-mer mRNA (Figure 7A and B), supported the expression of GLP-1 slightly better than did the 170-mer (Figure 8).
Figure 8.
GLP-1 expression in vitro. Artificial mRNA was added to a wheat germ cell-free protein synthesis system and GLP-1 expression was determined by sandwich ELISA. Each value is the average of three independent experiments, and the error bars indicate the standard deviation.
To test the ability of the artificial mRNA to support GLP-1 expression in whole cells, we transfected CHO cells with artificial mRNA by electroporation. Twelve hours after transfection, cells were harvested and GLP-1 was determined by sandwich ELISA. Control RNA that had not been subjected to enzymatic treatment to generate cap 1 or a poly(A) tail did not support GLP-1 expression, whereas mRNA derived from both the 130-mer and the 170-mer RNA supported weak GLP-1 expression (Figure 9). The weak GLP-1 expression and the lack of dose dependence may be explained by factors such as the stability of the mRNA in the cell culture medium, the transfection efficiency and the length of both the 5′-UTR and the poly(A) tail. Cationic liposomes were also tried as a method to deliver the mRNA to the cells, but the results were no better than those obtained with electroporation. Further work is required to establish the utility of artificial mRNA in whole cells.
Figure 9.
GLP-1 expression in whole cells. Artificial mRNA was transfected into CHO cells by electroporation and GLP-1 expression was determined by sandwich ELISA. Each value is the average of three independent experiments, and the error bars indicate the standard deviation.
In the present study, we have shown that mRNA derived from a chemically synthesized 170-mer RNA encoding a 33-amino-acid peptide supported the expression of the peptide in a cell-free system and in whole cells. Biologically active peptides are usually translated as long precursors that undergo processing in the endoplasmic reticulum (ER) (28,29). There are several reports of peptides being translated from mRNA with an open reading frame (ORF) of fewer than 100 amino acids. A 36-amino-acid peptide was shown to be translated in Saccharomyces cerevisiae from mRNA with a short ORF (30), and a similar result was obtained for an 11-amino-acid peptide in Drosophila (31). These short peptides translated from short ORFs are thought to be localized in the cytosol during their biosynthesis and not moved to the ER because they do not need to undergo processing there. The functions of these short peptides are presumably distinct from those of peptides that mature in the ER. Non-coding RNA with a relatively long poly(A) tail at the 3′-end, known as mRNA-like non-coding RNA, also exists (32,33). It includes bifunctional RNA (34) transcribed from genes such as oskar in Drosophila, which acts as a non-coding RNA during early oogenesis and as an mRNA during the embryonic stage (35). The technology for synthesizing mRNA reported in the present paper can provide access to such bifunctional and multifunctional RNAs, allowing expansion of basic and applied research in this area.
CONCLUSIONS
We have developed novel, highly active pyrophosphorylating reagents based on a phosphotriester structure that are suitable for the pyrophosphorylation of short and long RNA oligomers. In particular, our phosphotriester pyrophosphorylating reagents solve the problem of the pyrophosphorylation of long RNA oligomers on a solid support. We have designed mRNA sequences up to 170 nt long and used the CEM method to chemically synthesize each of them in a single synthesizer run. This extends the length of RNA oligomer shown to be capable of being handled by the CEM method from the previously reported maximum of 110. Synthetic 130-mer and 170-mer RNA oligomers were monophosphorylated, pyrophosphorylated with our new reagents, and enzymatically capped and poly(A)-tailed. Electrophoretic and mass-spectrometric analysis of the products showed that both synthetic oligomers had been correctly converted to mRNA that supported protein synthesis in a cell-free system and in whole cells. As far as we know, this is the first report of the preparation of a mature mRNA from a chemically synthesized sequence. The technology for preparing mRNA described in this paper is expected to be useful in basic and applied research on new functional RNA as well as in the development of RNA vaccines and mRNA medicines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was supported in part by grants from the New Energy and Industrial Technology Development Organization (NEDO) of Japan for its Functional RNA Project.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Gerald E. Smyth, Discovery Research Laboratories, Nippon Shinyaku Co. Ltd, for helpful discussions and suggestions during the preparation of the article.
REFERENCES
- 1.Pascolo S. Vaccination with messenger RNA (mRNA) Handb. Exp. Pharmacol. 2008;183:221–235. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]
- 2.Weide B, Garbe C, Rammensee HG, Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol. Lett. 2008;115:33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Kofler RM, Aberle JH, Aberle SW, Allison SL, Heinz FX, Mandl CW. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl Acad. Sci. USA. 2004;101:1951–1956. doi: 10.1073/pnas.0307145101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mockey M, Bourseau E, Chandrashekhar V, Chaudhuri A, Lafosse S, Le Cam E, Quesniaux VFJ, Ryffel B, Pichon C, Midoux P. mRNA-based cancer vaccine: prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007;14:802–814. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- 5.Peyrane F, Selisko B, Decroly E, Vasseur JJ, Benarroch D, Canard B, Alvarez K. High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2′O positions. Nucleic Acids Res. 2007;35:e26. doi: 10.1093/nar/gkl1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helm M, Brulé H, Giegé R, Florentz C. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall AHS, Wan J, Shaughnessy EE, Shaw BR, Alexander KA. RNA interference using boranophosphate siRNAs: structure–activity relationships. Nucleic Acids Res. 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwase R, Maeda M, Fujiwara T, Sekine M, Hata T, Miura K. Molecular design of eukaryotic messenger RNA and its chemical synthesis. Nucleic Acids Res. 1992;20:1643–1648. doi: 10.1093/nar/20.7.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohgi T, Masutomi Y, Ishiyama K, Kitagawa H, Shiba Y, Yano J. A new RNA synthetic method with a 2′-O-(2-cyanoethoxymethyl) protecting group. Org. Lett. 2005;7:3477–3480. doi: 10.1021/ol051151f. [DOI] [PubMed] [Google Scholar]
- 10.Usman N, Ogilvie KK, Jiang M-Y, Cedergren RJ. Automated chemical synthesis of long oligoribonucleotides using 2′-O-silylated ribonucleoside 3′-O-phosphoramidites on a controlled-pore glass support: Synthesis of a 43-nucleotide sequence similar to the 3′-half molecule of an Escherichia coli formylmethionine tRNA. J. Am. Chem. Soc. 1987;109:7845–7854. [Google Scholar]
- 11.Shiba Y, Masuda H, Watanabe N, Ego T, Takagaki K, Ishiyama K, Ohgi T, Yano J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2′-O-protecting group: structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007;35:3287–3296. doi: 10.1093/nar/gkm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadokura M, Wada T, Seio K, Moriguchi T, Huber J, Lührmann R, Sekine M. Solid-phase synthesis of a 5′-terminal TMG-capped trinucleotide block of U1 snRNA. Tetrahedron Lett. 2001;42:8853–8856. [Google Scholar]
- 13.Kore AR, Shanmugasundaram M, Vlassov AV. Synthesis and application of a new 2′,3′-isopropylidene guanosine substituted cap analog. Bioorg. Med. Chem. Lett. 2008;18:4828–4832. doi: 10.1016/j.bmcl.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 14.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 15.Holst JJ. The physiology of glucagon-like peptide 1. Physiol. Rev. 2007;87:1409–1439. doi: 10.1152/physrev.00034.2006. [DOI] [PubMed] [Google Scholar]
- 16.Perfetti R, Merkel P. Glucagon-like peptide-1: a major regulator of pancreatic β-cell function. Eur. J. Endocrinol. 2000;143:717–725. doi: 10.1530/eje.0.1430717. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine M, Tsuruoka H, Iimura S, Wada T. Simultaneous removal of two 2-cyano-1,1-dimethylethyl groups from bis(2-cyano-1,1-dimethylethyl) thymidine 3′-phosphate derivatives by the use of DBU/BSA. Nat. Prod. Lett. 1994;5:41–46. [Google Scholar]
- 20.Ohkubo A, Aoki K, Seio K, Sekine M. A new approach for pyrophosphate bond formation starting from phosphoramidite derivatives by use of 6-trifluoromethyl-1-hydroxybenzotriazole-mediated O-N phosphoryl migration. Tetrahedron Lett. 2004;45:979–982. [Google Scholar]
- 21.Umemoto T, Wada T. Nitromethane as a scavenger of acrylonitrile in the deprotection of synthetic oligonucleotides. Tetrahedron Lett. 2005;46:4251–4253. [Google Scholar]
- 22.Kuge H, Brownlee GG, Gershon PD, Richter JD. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sproat BS, Calonna F, Mullah B, Tsou D, Andrus A, Hampel A, Vinayak R. An efficient method for the isolation and purification of oligoribonucleotides. Nucleosides Nucleotides. 1995;14:255–273. [Google Scholar]
- 24.Gasparutto D, Livache T, Bazin H, Duplaa A-M, Guy A, Khorlin A, Molko D, Roget A, Téoule R. Chemical synthesis of a biologically active natural tRNA with its minor bases. Nucleic Acids Res. 1992;20:5159–5166. doi: 10.1093/nar/20.19.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westman E, Strömberg R. Removal of t-butyldimethylsilyl protection in RNA-synthesis. Triethylamine trihydrofluoride (TEA, 3HF) is a more reliable alternative to tetrabutylammonium fluoride (TBAF) Nucleic Acids Res. 1994;22:2430–2431. doi: 10.1093/nar/22.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallie DR, Ling J, Niepel M, Morley SJ, Pain VM. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehfeld JF, Bardram L, Cantor P, Cerman J, Hilsted L, Johnsen AH, Mogensen N, Odum L. Peptide hormone expression and precursor processing. Acta Oncol. 1989;28:315–318. doi: 10.3109/02841868909111199. [DOI] [PubMed] [Google Scholar]
- 29.Rholam M, Fahy C. Processing of peptide and hormone precursors at the dibasic cleavage sites. Cell Mol. Life Sci. 2009;66:2075–2091. doi: 10.1007/s00018-009-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huyer G, Kistler A, Nouvet FJ, George CM, Boyle ML, Michaelis S. Saccharomyces cerevisiae a-factor mutants reveal residues critical for processing, activity, and export. Eukaryot. Cell. 2006;5:1560–1570. doi: 10.1128/EC.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 32.Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki S, Numata K, Kondo T, Tomita M, Yasuda K, Kanai A, Kageyama Y. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells. 2005;10:1163–1173. doi: 10.1111/j.1365-2443.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 34.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenny A, Hachet O, Závorszky P, Cyrklaff A, Weston MDJ, St Johnston D, Erdélyi M, Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.