Abstract
Spatial chromatin organization is emerging as an important mechanism to regulate the expression of genes. However, very little is known about genome architecture at high-resolution in vivo. Here, we mapped the three-dimensional organization of the human Hox clusters with chromosome conformation capture (3C) technology. We show that computational modeling of 3C data sets can identify candidate regulatory proteins of chromatin architecture and gene expression. Hox genes encode evolutionarily conserved master regulators of development which strict control has fascinated biologists for over 25 years. Proper transcriptional silencing is key to Hox function since premature expression can lead to developmental defects or human disease. We now show that the HoxA cluster is organized into multiple chromatin loops that are dependent on transcription activity. Long-range contacts were found in all four silent clusters but looping patterns were specific to each cluster. In contrast to the Drosophila homeotic bithorax complex (BX-C), we found that Polycomb proteins are only modestly required for human cluster looping and silencing. However, computational three-dimensional Hox cluster modeling identified the insulator-binding protein CTCF as a likely candidate mediating DNA loops in all clusters. Our data suggest that Hox cluster looping may represent an evolutionarily conserved structural mechanism of transcription regulation.
INTRODUCTION
Spatial chromatin organization is an essential feature of genome function. For example, interphase chromosomes are known to occupy distinct territories that can be positioned in the nuclear space according to transcription activity (1). Chromosomes are further organized into microscopically visible substructures such as nucleoli and Cajal bodies, which are important for the production, storage and recycling of diverse molecules. An additional ultrastructural level of genome organization was recently uncovered with the chromosome conformation capture (3C) and 3C-related technologies (2–7). This organization includes long-range physical contacts between DNA sequences located within (cis) or between (trans) chromosomes.
Although very little is known about spatial chromatin organization at high-resolution, several studies show that long-range DNA contacts are important to regulate transcription. For example, enhancers can stimulate the expression of genes by physically interacting with them. This type of interaction was first described in the beta-globin locus where contacts between the locus control region (LCR) and transcribed genes are mediated by the hematopoietic transcription factor GATA-1 and co-factor FOG-1 (8,9). Long-range contacts are also important for insulator function. For example, the enhancer-blocking activity of the imprinting control region (ICR) insulator in the Igf2/H19 locus involves looping contacts with promoters and enhancers (10). Cis and trans DNA contacts have been shown to regulate genes from various cellular pathways indicating that genomes are likely organized into dynamic three-dimensional (3D) networks of physical contacts essential for transcription regulation.
Given the recent development of 3C and 3C-related technologies (2–5,7,8,11), the role of chromatin architecture in regulating key genomic loci such as the Hox clusters remains mostly unknown. The Hox clusters encode evolutionarily conserved transcription factors that function as master regulators of development. In mammals, there are 39 Hox genes organized into four genomic clusters of thirteen paralog groups. The HoxA, B, C and D clusters are located on different chromosomes and are thought to derive from tandem duplication of ancestral genes. During development, the Hox genes are involved in anterior-posterior (A/P) body patterning, and in formation of limbs and genitalia. Interestingly, Hox gene silencing is essential during development since premature expression can lead to homeotic transformation. For example, the 5′-end Hox genes (e.g. HoxA7-13) expressed at the posterior end of the developing embryo are repressed by Polycomb group (PcG) proteins until the appropriate time. PcG proteins silence developmental genes epigenetically with repressive post-translational histone modifications. Knockout of PcG proteins results in posterior transformation defects in mice and drosophila by inducing leaky 5′-end Hox expression in the middle and head part of embryos (12–14). Hox gene silencing is also important in adult tissues since inappropriate expression is associated with cancer. In fact, a number of Hox genes have been involved in the development and/or progression of a variety of solid and hematopoietic malignancies. For example, HoxA9 overexpression in mouse bone marrow leads to acute myelogenic leukemia (AML) and is a hallmark of most MLL-rearranged human leukemias (15). Therefore, understanding the silencing mechanism of Hox genes will be essential to explain their precise expression during development and their role in human health. Here we characterized the 3D organization of the human Hox clusters using 3C. Our results suggest that modeling of spatial chromatin organization may be used to predict gene expression states and potential mechanisms of Hox regulation.
MATERIALS AND METHODS
Cell culture
The NT2/D1 (NTERA2) cells are human testicular pluripotent embryonal carcinoma cells derived from the metastasis site (lung) of a 22-year-old Caucasian male (16). These cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS; HyClone). Cells were grown at 37°C in 5% CO2 atmosphere in the presence of 1% penicillin–streptomycin. All experiments presented in this study were performed using log-phase cells.
To induce Hox expression in NT2/D1, exponentially growing cells were seeded at 2 × 106 per 75 cm2 flasks in 12 ml of complete DMEM containing 10 µM all trans retinoic acid (RA; Sigma) or 0.1% DMSO control. Cells were treated continuously with RA to achieve maximal induction and passaged to maintain exponential growth. To determine the effect of transcription induction on chromatin loops (Figure 3), control and RA-induced cells were collected after 14 days for RNA extraction and 3C library preparation. To examine the role of PcG on chromatin looping and silencing of Hox clusters (Figure 4), control and RA-induced cells were collected after only 12 h to achieve partial (3′-end) rather than full induction of Hox genes.
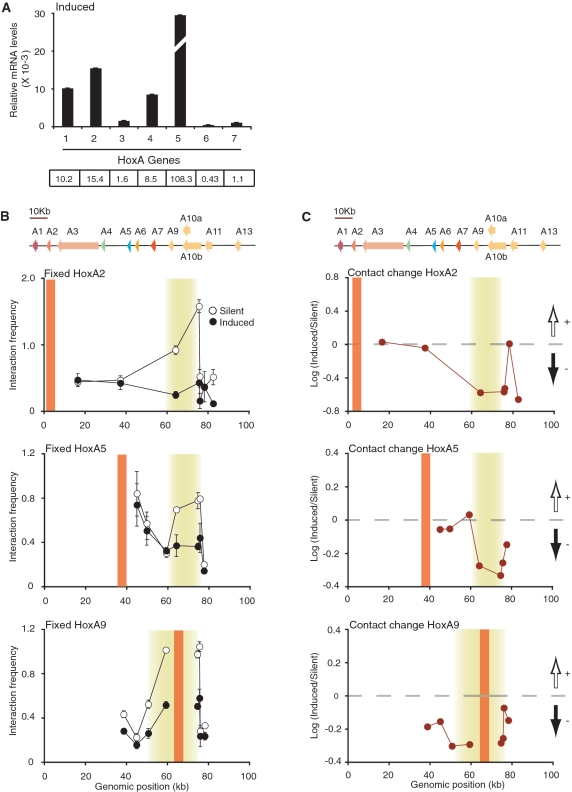
Figure 3.
The silent HoxA conformation is dependent on transcription activity. (A) Quantitative real-time PCR analysis of HoxA genes following RA induction in NT2/D1 cells. Steady-state HoxA mRNA levels were normalized relative to actin. Histogram break in HoxA5 highlights out of range value. Number below histogram bar identify HoxA paralog group. Values shown below each paralog group represent corresponding relative mRNA levels (×10−3). Quantifications are derived from at least three PCRs and error bars show SD. (B, C) Fixed 3C interaction profile of HoxA2 (top), HoxA5 (middle) and HoxA9 (bottom) genes in transcriptionally silent and RA-induced NT2/D1 cells. Schematic representation of the HoxA cluster is presented above graphs and is to scale. 3C interaction frequency is presented in (B). RA-induced chromatin contact changes are presented in (C). Black and white vertical arrows above and below horizontal dashed lines represent gain and loss of contacts in induced cells, respectively. Vertical orange lines indicate position of fixed primer and shaded yellow areas highlight looping contacts. Individual interaction frequencies were derived from at least three PCRs. Error bars show standard error of the mean (SEM).
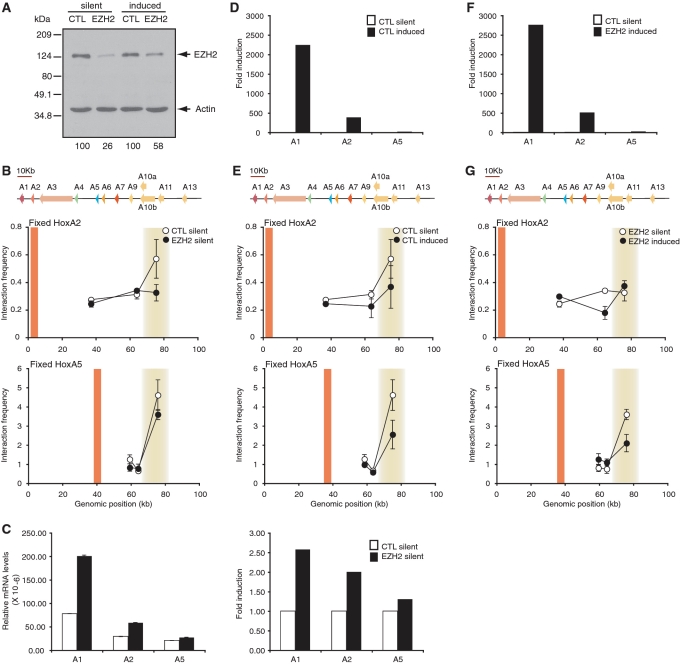
Figure 4.
PcG is partially required for chromatin looping and transcription silencing of the human HoxA cluster. (A) RNAi knockdown specifically reduces EZH2 protein levels. Extracts from control (CTL) and EZH2 siRNA-treated cells treated with DMSO or RA were analyzed by western blotting with antibodies against EZH2 (top) or actin (bottom). EZH2 levels remaining after RNAi knockdown are indicated below each lane and are normalized to actin. (B) Loss of HoxA chromatin loops in EZH2 knockdown cells. Fixed 3C interaction profiles of HoxA2 (top) and HoxA5 (bottom) in control (CTL) and EZH2 knockdown cells. A schematic representation of the HoxA cluster is presented to scale above each graph. Vertical orange lines indicate position of fixed 3C primers and shaded yellow areas highlight looping contacts. Individual interaction frequencies were derived from at least three PCRs. Error bars show SEM. (C) 3′end HoxA genes are derepressed in EZH2 knockdown cells. Quantitative real-time PCR analysis of HoxA genes in control (CTL) and EZH2 siRNA-treated cells. Steady-state HoxA mRNA levels were normalized relative to actin (left). Fold induction after EZH2 knockdown is relative to levels in corresponding siRNA control, which was set at 1 (right). (D) 3′end genes are activated in partially RA-induced HoxA clusters. Quantitative real-time PCR analysis of HoxA genes in control siRNA-treated cells treated with DMSO (CTL silent) or RA (CTL induced). Fold induction after RA induction is relative to levels in corresponding siRNA control treated with DMSO, which was set at 1. (E) Loss of chromatin loops in partially RA-induced HoxA clusters. Fixed 3C interaction profiles of control siRNA-treated cells treated with DMSO (CTL silent) or RA (CTL induced) are as described in (B). (F) 3′end genes activation in partially RA-induced HoxA clusters depleted of EZH2. Quantitative real-time PCR analysis of HoxA genes in control siRNA-treated cells treated with DMSO (CTL silent) in EZH2 knockdown cells treated with RA (EZH2 induced). Fold induction after RA induction is relative to levels in corresponding siRNA control treated with DMSO, which was set at 1. (G) Loss of chromatin loops in partially RA-induced HoxA clusters depleted of EZH2. Fixed 3C interaction profiles of EZH2 siRNA-treated cells treated with DMSO (EZH2 silent) or RA (EZH2 induced) are as described in (B).
Real-time polymerase chain reaction quantification
Total RNA was extracted from DMSO control (Silent) and RA-treated (Induced) NT2/D1 cells with the GenEluteTM Mammalian Total RNA Miniprep Kit as described by the manufacturer (Sigma). The Omniscript Reverse Transcription Kit (Qiagen) was used to perform reverse transcription with oligo(dT)20 (Invitrogen). Human Hox genes were quantified by real-time polymerase chain reaction (PCR) with a LightCycler (Roche) in the presence of SYBR Green I stain (Molecular Probes). The primer sequences used to measure human HoxA genes and actin control were described elsewhere (17). 5′-end HoxA genes (HoxA9–13) are known not to be induced by RA and were excluded from our real-time analysis (18–20). The basal expression levels of all remaining human Hox genes (HoxB, HoxC and HoxD) were characterized by real-time and endpoint PCR in undifferentiated NT2/D1 to verify very low expression levels (data available on our website (see URLs below)). In these experiments, 2-fold dilutions of total cDNA were amplified under quantitative real-time PCR conditions. Most Hox Ct values were below linear quantitative real-time PCR detection range confirming that basal Hox expression levels are very low. Endpoint PCR products were detected by ethidium bromide staining on agarose gels. A complete list of all human RT-PCR primers is available on our website.
Design and preparation of control 3C libraries
Human control 3C libraries were used to correct differences in 3C primer pair efficiency. Libraries were generated from bacterial artificial chromosomes (BACs) as previously described (17,21,22). Briefly, BAC clones covering each of the four Hox clusters and one gene desert region (ENCODE region ENr313 on chromosome 16) were mixed in equimolar ratio. Mixed BACs were digested with either BglII or HindIII and randomly ligated with T4 DNA ligase. The libraries were generated with the following BACs: RP11-1132K14, CTD-2508F13, RP11-657H18, RP11-96B9, RP11-197K24, CTD-2594L23 and RP11-1132K14. BAC clones were obtained from Invitrogen.
3C analysis
Human cellular 3C libraries were generated as previously described (22). Briefly, exponentially growing cells were fixed in 1% formaldehyde, digested with a restriction enzyme and ligated under dilute conditions to promote intermolecular ligation of crosslinked restriction fragments. 3C libraries were purified and titrated by PCR with 3C primers detecting neighboring DNA fragments in a gene desert region (human ENCODE region ENr313) and in a Hox gene cluster. The quality of cellular 3C libraries was verified systematically by generating compaction profiles in gene desert regions as described previously (21). PCRs were performed manually using amplification conditions described elsewhere (22). At least 3–42 PCRs were performed for each predicted contact. PCR products were resolved on agarose gels containing 0.5 µg/ml ethidium bromide and visualized by ultraviolet (UV) transillumination. Gel documentation was conducted with a ChemiDocTM XRS system featuring a 12-bit digital camera and quantification was performed with the Quantity One® computer software (version 4.6.3; BioRad). 3C primer sequences used in this study are available on our website.
RNA interference and western blotting
Human EZH2 knockdown was performed in NT2/D1 by reverse transfection in the presence of 5 nM siRNAs (control or EZH2) using HiPerfect transfection reagent as recommended by the manufacturer (Qiagen). Briefly, 6 × 105 cells were plated over siRNA/HiPerfect complexes in 35-mm dishes containing a final volume of 2.3 ml of complete DMEM (0 h transfection). Cells attached onto plates in the presence of siRNAs and were collected 72 h for western blotting, RNA extraction and 3C library production. Control siRNA (siGENOME Non-Targeting siRNA #2) and human EZH2 siRNA (5′-GACCUUGAAUGCAGUUGCUUU-3′) were purchased from Dharmacon.
For western blot analysis, protein samples were prepared by scraping cells directly in 1X SDS sample buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 7.5% glycerol, 5% beta-mercaptoethanol, 0.04% bromophenol blue). Samples were transferred to eppendorfs, sonicated twice for 15 s and heated at 95°C for 5 min. Twenty microliters of each sample was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto 0.45 µM nitrocellulose membrane for 45 min at 100 V. Immunoblotting was performed with anti-human EZH2 mouse monoclonal antibody AC22 (Cell Signalling) and anti-actin rabbit polyclonal antibody (Cell Signalling) as recommended by the manufacturer. Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Signals were visualized by chemiluminescence (Perkin Elmer LAS, Inc.) followed by autoradiography. Films were scanned with the ChemiDocTM XRS Imaging system and signals were quantified with the Quantity One® software. Detailed protocols for sample preparation and western blotting analysis can be found on our website (see URLs below).
Informatics
GelToIF
We developed the ‘GelToIF’ program to calculate pair-wise interaction frequency (IF) values based on agarose gel band intensity measurements. This program minimizes the noise in 3C IFs by eliminating outliers in raw data sets before calculating IFs. During 3C, fragment pairs are usually analyzed in triplicates. GelToIF compares these measurements to apply several filters and remove outliers. First, the minimum (Min), median (Med) and maximum (Max) intensity value is identified in the triplicate of a given pair. The following rules are then applied: (i) If (Med-Min)/(Max-Med) is between 1/5 and 5, then all three measurements are retained; otherwise (ii) if (Med–Min) > 0.5 * Med, then Min is called an outlier. If (Max–Med) > 0.5 * Med, then Max is called an outlier. This outlier removal process was independently applied to the cellular and BAC 3C data. Fragment pairs with at least one retained measurement in each cellular and BAC data sets were considered further. For each retained fragment pair with k cellular measurements and l BAC measurements, the set of k*l possible cellular-to-BAC ratios were computed. The average of cellular-to-BAC ratios was reported as the IF value for each fragment pair, unless the standard deviation of the set of ratios was larger than their average. If the standard deviation was larger than the average, the measurements were deemed too noisy to be useful and no IF data was generated for that pair. GelToIF was used to calculate the IFs presented in Figures 1 and 2, Supplementary Figures S2–S5. The set of IF data thus generated was fed into the 5C3D program to generate 3D models of each Hox cluster.
Figure 1.
The transcriptionally silent HoxA cluster is organized into multiple chromatin loops in the human genome. (A) Linear schematic representation of the HoxA cluster on human chromosome 7. Predicted BglII restriction fragments are identified from left to right by numbers. (B) 3C interaction frequency heat map of the silent HoxA cluster. Pair-wise interaction frequencies were measured by 3C in undifferentiated NT2/D1 cells. Intersecting column and row number identifies DNA contact corresponding to interaction frequency. Interaction frequencies are color-coded based on the scale presented on the left. Genomic regions involved in looping interactions are highlighted by shaded boxes. Additional information about this data set is available on our website (‘Materials and Methods’ section). (C) Two-dimensional schematic representation of the silent human HoxA cluster in vivo. Schematic is not to scale to enable two-dimensional illustration of DNA contacts. Yellow circles highlight an area encompassing strongest looping contacts in the cluster rather than the actual number of interactions.
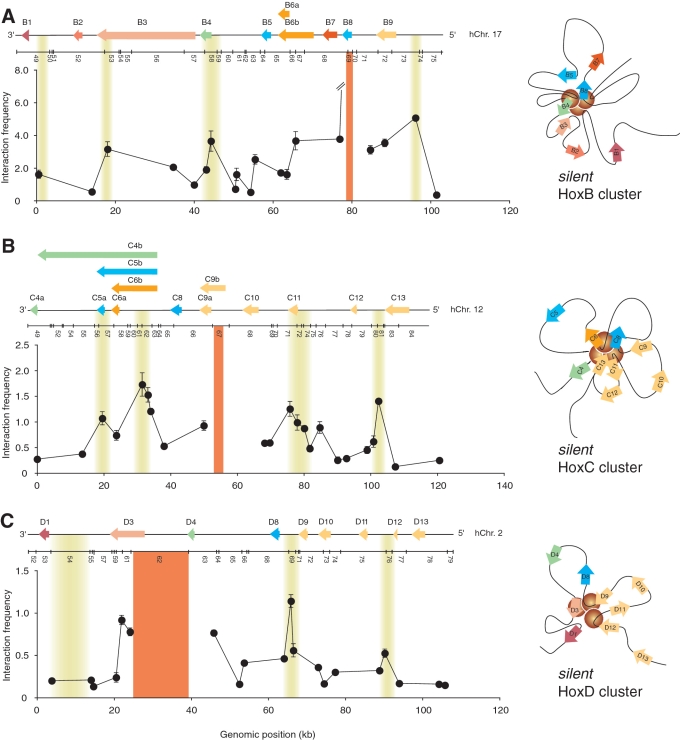
Figure 2.
Chromatin looping is a conserved feature of transcriptionally silent Hox clusters. (A) The silent HoxB cluster is organized into a distinct set of looping contacts. (left panel) Representative ‘fixed’ 3C interaction profile illustrating looping contacts. The looping interactions shown are specific to the HoxB cluster. The y-axis indicates normalized interaction frequency. The x-axis shows genomic position relative to the start of the cluster. Line break in graph highlights out of range value. A linear diagram of the HoxB cluster and the predicted BglII restriction pattern are shown to scale above the graph. Cluster features are as described in Figure 1A. Solid orange vertical line identifies the position of ‘fixed’ 3C region. Shaded yellow areas highlight the position of looping contacts. Each data point is the average of at least three PCRs. Error bars represent the standard error of the mean. (right panel) Predicted two-dimensional representation of the silent HoxB cluster. Note that the yellow circles simply highlight an area encompassing looping contacts and not the number of interactions. The model is based on the heatmap results presented in Figure S2 and is as described in Figure 1C. (B) The silent HoxC cluster is organized into multiple chromatin loops. (left panel) The representative 3C interaction profile from a ‘fixed’ HoxC region illustrates DNA looping specific to the cluster and is as described in (A). (right panel) The two-dimensional model of the transcriptionally silent HoxC cluster is based on the heatmap results presented in Figure S3 and is as described in Figure 1C. Yellow circles highlight an area containing looping interactions. (C) The silent HoxD cluster is organized into multiple looping contacts. (left panel) The 3C interaction profile from a ‘fixed’ HoxD region shows looping contacts specific to HoxD and is as described in (A). (right panel) The two-dimensional model of the transcriptionally silent HoxD cluster is based on the heatmap results presented in Figure S4 and is as described in Figure 1C. The area containing looping interactions is highlighted with yellow circles.
5C3D
The 5C3D program (17) was previously described and was used to infer 3D chromatin organization based on experimental 3C data. 5C3D predicts 3D models by converting IFs to distances and using a gradient descent approach to find best-fit models. IFs were calculated with the ‘GelToIF’ program described above.
Microcosm 1.0 and 2.0
The Microcosm 1.0 program (17) was used to determine base density along Hox clusters (panels B in Figure 5 and Supplementary Figures S6–S8). This program uses models generated with 5C3D to calculate the number of bases in a sphere with a radius of R = 0.65 centered at given features. The local base density every 10 bp along Hox clusters was calculated in 100 individual 5C3D models. Base densities at corresponding genomic positions were then averaged from the 100 models and the standard deviation calculated for each measurement.
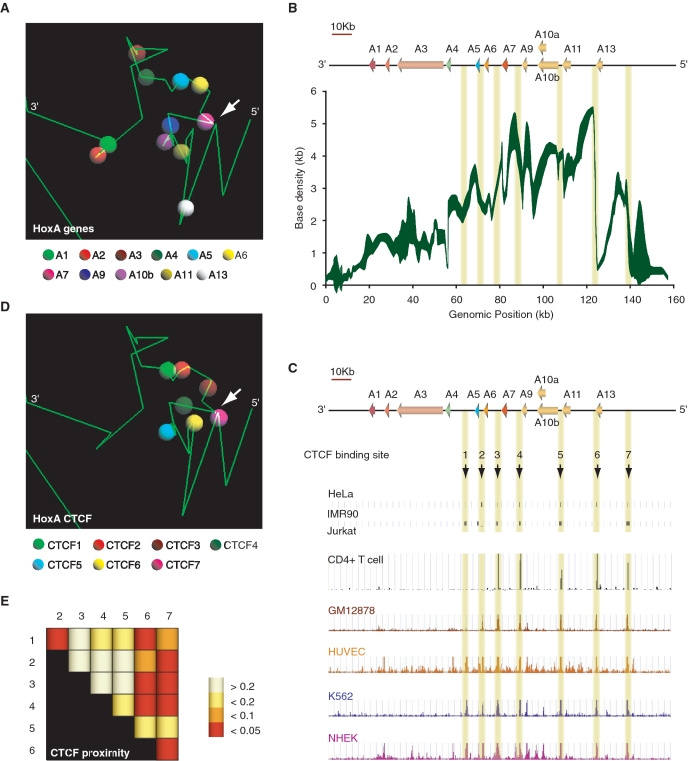
Figure 5.
Three-dimensional modeling of the silent HoxA cluster identifies CTCF as a likely candidate mediating chromatin loops. (A) Example of a 5C3D output model of the transcriptionally silent HoxA cluster. Green lines represent genomic DNA, and vertices define boundaries between consecutive restriction fragments. Colored spheres as indicated in the legend below identify the transcription start site of corresponding paralog group. (B) Three-dimensional local base density scan of the transcriptionally silent HoxA cluster. Local base densities at consecutive 10 bp was estimated in 100 possible 5C3D outputs models with Microcosm 1.0 (y-axis) and represented graphically along the corresponding genomic region (ENCODE hg18 Chr7:27079118 to 27236536) (x-axis). The weight of the trace is proportional to the standard deviation with sharper areas indicating smaller deviations. (C) CTCF binds to multiple discrete sites conserved in various cell lines at the 5′-end of the HoxA cluster. Conserved CTCF sites are highlighted by yellow vertical lines. (D) Conserved CTCF binding sites are clustered three-dimensionally at the 5′-end of the HoxA cluster. The position CTCF binding sites numbered in (C) are illustrated in the example 5C3D output model presented in (A). CTCF binding sites are represented by colored spheres as indicated in the legend below. (E) CTCF binding sites are significantly close to each other in three-dimensional models. Distances between pairs of CTCF binding sites were measured with Microcosm 2.0. and expressed as P-values summarized in a heatmap. Numbers at the top and on the left of heatmap identify CTCF binding sites. Intersecting column and row number identifies the CTCF pair. P-values are color-coded based on the scale presented on the right. P-values were calculated as described in ‘Materials and Methods’ section.
An updated version of the Microcosm program (17) (Microcosm 2.0) was designed to assess whether the Euclidean distance between two specified features in 3D models is significantly lower than expected compared to other pairs of genomic locations separated by the same linear genomic distance. Specifically, a P-value is calculated for each pair-wise distance, to assess whether the observed Euclidean distance δ is significantly low, given the genomic distance d (number of base pairs) between the two locations. This P-value is calculated as the fraction of pairs of locations (i, i + d), over all possible values of i, whose Euclidean distance in the 3D model is smaller or equal to δ.
This program was used to determine whether the proximity between pairs of predicted CTCF binding sites in models was significant (panels E in Figure 5, and Supplementary Figures S6–S8). All programs including GelToIF and updated Microcosm 2.0 are available on our website (see URLs section below).
Databases
We used the May 2004 human reference sequence (NCBI Build 35) produced by the International Human Genome Sequencing Consortium for 3C experimental design (see ‘URLs’ section below). Genome-wide CTCF binding sites in HeLa (23), IMR90 (24), Jurkat (23) and CD4+ T (25) cells were published previously and are available online on the websites listed in the URLs below. Genome-wide CTCF mapping in GM12878, HUVEC, K562 and NHEK were produced by the ENCODE Chromatin Group at Broad Institute and Massachusetts General Hospital. These data sets are available online on the ENCODE Pilot Project at UCSC website (hg18: see URLs section below) (26).
URLs
The human genome sequence is available at http://genome.ucsc.edu/. Detailed protocols, 3C support information (design and analysis) can be found on our website at http://Dostielab.biochem.mcgill.ca. Complete raw data sets and bioinformatics tools including those developed in this study are also available on our website. Tools include ‘5C3D’, ‘GelToIF’ and ‘Microcosm 2.0’. The HeLa (Acc. no. GSM325897) and Jurkat (Acc. no. GSM325899) CTCF ChIP-seq data sets were obtained from the Gene Expression Omnibus (GEO) website http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12889. The IMR90 chromatin immunoprecipitation (ChIP)–chip data set was obtained from http://licr-renlab.ucsd.edu/download.html and converted to hg18 using the ‘LiftOver’ tool available at http://genome.ucsc.edu/cgi-bin/hgLiftOver. The CD4+ T cell CTCF ChIP-seq data set was obtained from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspx.
RESULTS
The transcriptionally silent HoxA gene cluster is organized into multiple chromatin loops
To determine whether spatial chromatin organization plays a role in Hox gene silencing, we first mapped the human HoxA cluster with 3C technology (Figure 1A). 3C uses genomic libraries of pair-wise ligation products to measure physical contacts between DNA segments in vivo at high resolution (2,8). These libraries contain ligation products in quantities that are inversely proportional to original 3D distances in vivo. It should be noted however that because 3C libraries derive from large cell populations, only averaged 3D distances might be inferred from 3C data. Thus, 3C and 3C-related technologies can only yield averaged 3D models rather than true individual in vivo chromatin structures. Despite this inherent limitation, 3C nevertheless remains a very powerful approach for mapping molecular interactions in the nuclear space at very high resolution.
We generated 3C libraries from untreated human NT2/D1 cells and analyzed them by PCR (Figure 1B). To further reduce noise in our 3C data sets, we developed the ‘GelToIF’ computer program, which applies a series of filters to raw and converted measurements before producing a list of pair-wise interaction frequencies (see ‘Materials and Methods’ section). NT2/D1 cells were used in our analysis because this cell line is commonly used to study Hox cluster regulation (16,27). Detailed 3C mapping revealed that the human HoxA cluster is organized into several long-range chromatin loops when transcriptionally silent (Figure 1B and C). Looping contacts were mostly located within the cluster’s 5′-end and interacting regions were often engaged in multiple contacts. For example, HoxA13 interacted strongly with HoxA10 while A10 displayed extensive looping along the cluster including A5 and A11. HoxA5 also interacted extensively but with equal intensities throughout the cluster indicating that it might be located at its center. Looping was not restricted to the cluster 5′-end as HoxA2 displayed weaker but distinct binding to the A10 region. Since untreated NT2/D1 cells express very low levels of HoxA genes (27) (see ‘Materials and Methods’ section), this result indicates that the cluster is likely folded onto itself when transcriptionally silent.
An important result from this analysis is the extensive looping observed between regions surrounding each of the HoxA9, 10, 11 and 13 genes. This result suggests that genes at the HoxA 5′-end are clustered together by a series of loops when not expressed. We previously found a similar looping network at the 5′-end of the HoxA cluster in the human THP-1 differentiation system (17). THP-1 can be differentiated into monocyte/macrophages with phorbol myristate acetate (PMA). 5′-end HoxA genes are repressed during THP-1 differentiation and we found that repression is associated with formation of discrete contacts. In fact, looping contacts between downregulated genes were highly conserved in both differentiation systems (Supplementary Figure S1). However, loops were stronger in NT2/D1 cells, which correlates with the more repressed transcriptional state of HoxA genes in these cells (17). Thus, 5′-end HoxA gene clustering appears independent of cell type but dependent on transcription activity.
Interestingly, our results are reminiscent of the higher-order structures described in the Drosophila bithorax complex (BX-C) (28). The BX-C complex corresponds to the 5′-end of mammalian Hox clusters including paralog groups 9–13. Although the general linear organization of Hox genes is similar in both species, BX-C is over 10 times larger and exceeds 340 kb in length. The repressed BX-C was found engaged into several chromatin loops between promoters and Polycomb response elements (PREs). Our results therefore indicate that multiple chromatin loops might represent an evolutionarily conserved feature of transcriptionally silent Hox clusters. However, in this study, as in the ones mentioned above, data generated by 3C and other related technologies can only yield averaged 3D chromatin models and not individual structures. Thus, although we detect multiple chromatin loops at the silent HoxA cluster, we cannot resolve whether they occur individually or simultaneously at the locus, and the two-dimensional HoxA model presented in Figure 1C remains one interpretation of the data.
Chromatin looping is a conserved but distinct feature in silent Hox clusters
Given the conserved linear structure of mammalian clusters and the folding similarity of HoxA and BX-C, we wondered whether all silent human Hox clusters were organized into multiple chromatin loops. To determine the 3D organization of the HoxB, C and D clusters, we generated a high-coverage 3C interaction matrix for each genomic region in untreated NT2/D1 cells (Figure 2, and Supplementary Figures S2–S5). Interestingly, we found that HoxB, C and D clusters are also organized into multiple chromatin loops but that the pattern of long-range contacts is specific to each cluster. As with HoxA, whether these molecular interactions occur simultaneously or individually at the clusters cannot be distinguished due to inherent 3C limitations. Our data nevertheless demonstrate that looping is a conserved feature of transcriptionally silent Hox clusters, and suggest that a distinct structure-based control mechanism might regulate Hox genes in each cluster.
The HoxA cluster 3D organization is dependent on transcription activity
RA treatment of NT2/D1 cells recapitulates the induction pattern of Hox genes in developing axial systems. Therefore, 3′end HoxA genes including A1 to A5 can be specifically induced in a dose-dependent manner with RA. The NT2/D1 system is thus diametrically opposite of the THP-1 system where PMA-induction represses 5′-end HoxA genes. Considering the conserved looping and clustering pattern of silent HoxA genes in both lines, we tested whether RA-induced Hox transcription is also accompanied with changes in chromatin architecture. We compared the general organization of silent and fully RA-induced HoxA clusters with 3C (Figure 3). As predicted, RA treatment considerably induced 3′end genes (A1–A5) and not HoxA6 or HoxA7 (Figure 3A). We found that RA-induced transcription disrupted looping contacts under these conditions. For example, interaction between HoxA2 and A10 or A11 decreased over 5-fold (Figure 3B and C, top). Similarly, the interaction between HoxA5 and A9 or A10 was reduced ∼3-fold (middle). These results suggest that RA activation induces conformational changes reducing interactions between the 3′ and 5′-end of the cluster. Surprisingly, the activation of 3′end genes was accompanied by a general decondensation and loss of contacts at the 5′-end of the cluster (bottom). Since the 5′-end HoxA9-13 genes are insensitive to RA, chromatin remodeling does not appear restricted to the region induced by RA, unlike in PMA-treated THP-1 cells where conformational changes were mainly observed surrounding regulated genes.
Our findings are consistent with the conservation of RA-induced spatial chromatin remodeling between clusters and across species. Indeed, previous low-resolution in situ hybridization studies show that mouse HoxB and D activation with RA is accompanied by chromatin decondensation and looping out from the chromosome territory (29,30). Furthermore, the Drosophila BX-C 3D structures were also dependent on its transcriptional state (28). As PcG proteins appear to mediate the higher chromatin structures in Drosophila BX-C, might PcG also play a role in chromatin looping in the human Hox clusters?
PcG is only partially required for looping and silencing of the HoxA cluster
Transcriptional silencing of Hox clusters by PcG complexes remains poorly understood. In mammals, Polycomb-repressive complex 1 (PRC1) and PRC2 regulate genes epigenetically in part by modifying histones with repressive marks. It was recently demonstrated that PcG proteins could also form higher-order chromosome conformations in human cells (31). Therefore, PcG complexes might also be required for the 3D organization of silent human Hox clusters. We examined HoxA in samples depleted of the PRC2 subunit EZH2 to determine the role of PcG in looping and silencing of human Hox clusters (Figure 4). We found that EZH2 depletion (panel A) slightly disrupted long-range interaction between HoxA2 and the cluster 5′-end when transcriptionally silent (panel B, top). Binding between A5 and the 5′-end was very slightly reduced under these conditions (panel B, bottom). Importantly, EZH2 knockdown led to derepression of 3′end genes in a pattern consistent with changes in DNA contacts and mimicking RA induction (panel C). Indeed, although gene expression was much higher in RA-induced cells, the induction and looping patterns were very comparable between EZH2 knockdown and RA-induced HoxA clusters (compare induction in panels C and D, and looping in B and E). Thus, EZH2 is only partially required for DNA looping and silencing of the human HoxA cluster. Furthermore, loss of contact between A2 and the cluster 5′-end was not proportional to the gene induction level (compare panels B and E). EZH2 knockdown also did not affect the induction pattern of HoxA genes by RA and only resulted in slightly higher induction and lower long-range contacts (compare D and F, E and G). Therefore, our data suggest that at least in the NT2/D1 model system, unfolding of the chromatin is not sufficient to completely induce HoxA transcription, and that other protein(s) likely participate in cluster folding.
3D modeling identifies CTCF as a candidate regulatory protein of Hox cluster architecture and gene expression
Our results indicate that PcG participate in chromatin looping in human clusters. However, their rather broad distribution along clusters instead of localized binding at peak contacts is inconsistent with their role in mediating the discrete chromatin loops (18,27). Thus, the proteins responsible for DNA looping in mammalian Hox clusters remain unknown. To help identify candidate proteins mediating long-range contacts in silent Hox clusters, we modeled the 3C data sets three-dimensionally with the 5C3D computer program and examined a set of metrics in predicted models (Figure 5 and Supplementary Figures S6–S8). We previously developed 5C3D to predict averaged 3D models from high-resolution 3C-Carbon Copy (5C) data sets (17). 5C3D reduces the incidence of false positives in interaction data sets by integrating all compatible contacts into individual possible ‘structures’. Several features of the 5C3D HoxA models were recapitulated from the initial two-dimensional 3C heatmap models (Figure 1). For example, 5′-end HoxA genes were close to each other and to the 3′-end A1 and A2 genes, and the cluster appeared folded onto itself. 5C3D modeling of the HoxB, C and D clusters also recapitulated most features identified from two-dimensional analysis of 3C heatmaps (Supplementary Figures S6A–S8A). These features include a potential core mediated by B4 and B8 in HoxB, extensive folding of internal sequences and gene clustering in HoxC, and 3′-5′ communication in HoxD.
We previously developed the ‘Microcosm 1.0’ program to help robustly identify differences between 3D models generated with 5C3D (17). Microcosm uses 5C3D output models to calculate local chromatin densities surrounding given genomic positions. Here, we modified Microcosm 1.0 to convert 5C3D information into manageable two-dimensional genomic density scans. We estimated the local base density every 10 bp along Hox cluster regions (Figure 5B and Supplementary Figures S6B, S7B and S8B) and aligned these scans against existing genome annotation tracks to identify candidate regulatory DNA sequences or binding proteins. Consistent with the two-dimensional 3C heatmap analysis, we found that base density was much higher at the HoxA 5′-end where genes are clustered together by a looping network (Figure 5B). We then compared the HoxA base density scan with several genome-wide histone modification and binding protein profiles available at the UCSC website or from various publications to identify candidate sequences or proteins responsible for DNA looping in the cluster (see ‘Materials and Methods’ section). We found that the 5′-end region where base density was high contained several discrete CTCF binding sites (Figure 5C). CTCF is an insulator-binding protein known to regulate the expression of genes by mediating chromatin loops (32). Several of these binding sites were conserved in other cell lines. These binding sites were found clustered together in 3D models (Figure 5D). Interestingly, some binding sites were associated with base density peaks and found at contacts in HoxA 3D models. For example, the CTCF4, 6 and 7 binding sites localized to base density and peak contacts in 3D models (compare panels A and B). Furthermore, the CTCF7 binding site was found in very close proximity to the HoxA7 TSS, suggesting that it might regulate the expression of that gene directly (compare white arrows in panels A and D).
CTCF binding sites were also found in the HoxB, C and D clusters and conserved in different cell lines (panels C in Supplementary Figures S6–S8). Like HoxA, the sites were clustered together in 3D models (panels D), and often associated with base density and peak contacts (compare panels B and A in Supplementary Figures S6–S8). For example, CTCF1 and 5 were associated with base density peaks and contacts in HoxB models. These sites are located in regions found to interact by two-dimensional 3C heatmap analysis (Supplementary Figure S2 and S6A). The CTCF3 and CTCF5 binding sites in HoxC were also associated with base density peaks and contacts in 3D models and two-dimensional 3C heatmap models (Supplementary Figure S3 and S7B).
CTCF is known to dimerize (32) and could therefore be responsible for Hox cluster looping through self-association. To determine whether the proximity of CTCF binding sites in models was significant, we developed the ‘Microcosm 2.0’ program to compute P-values for each predicted 3D distance between CTCF binding site pairs. We found several CTCF binding sites significantly close to each other in each cluster, suggesting that it likely mediates DNA looping in all four Hox clusters. (panels E, Figures 5 and Supplementary Figures S6–S8; see ‘Materials and Methods’ section). Interestingly, CTCF was recently shown to regulate gene expression at the imprinted Igf2/H19 locus by recruiting PRC2 through direct binding with its SUZ12 component (33). Unlike in Drosophila where PREs have been identified, targeting of PcG proteins to mammalian chromatin is not entirely clear. The localization of CTCF at the HoxA cluster 5′-end and the elevated base density at that region, possibly through CTCF dimerization and looping, could explain why PRC2 complexes bind throughout the 5′-end of transcriptionally silent HoxA. Together, these results indicate that CTCF might mediate the correct 3D architecture of Hox clusters through dimerization.
DISCUSSION
Very little is known about genome organization at high-resolution in vivo. Here, we mapped the 3D organization of the human Hox clusters and show that each silent cluster is folded into a distinct pattern of chromatin loops. Although 3C data does not distinguish between simultaneous and individual looping at the clusters, we previously found that transcription repression of 5′-end HoxA genes is accompanied by increased packaging of the locus and clustering of downregulated genes (17). In that study, the 3D organization of the HoxA cluster was mapped with 5C technology during differentiation of a leukemia cell line. Although a cursory 3C analysis restricted to the HoxA 5′-end identified a network of physical contacts responsible for the clustering of genes, looping contacts throughout the cluster could not be resolved in these experiments since the maximum interaction coverage achievable per 5C library is 50%.
Together with the observation that multiple loops are present in all human clusters and in the Drosophila BX-C, our results suggest that chromatin looping is a conserved feature of transcriptionally silent Hox clusters. What might be the purpose of DNA looping in silent Hox clusters? This chromatin organization might prevent improper Hox activation by irrelevant cellular pathways. Alternatively, specific looping patterns might be important for the temporal colinearity of Hox clusters. The molecular mechanisms governing this process are poorly understood. At the microscopic level, Hox induction was shown to involve extensive nuclear reorganization such as decondensation and extrusion from the chromosome territory (29,30). Although distinct mechanisms appear to regulate clusters in different developing systems, progressive looping out from the chromosome territory was proposed to induce sequential transcription activation along clusters in RA treated mouse ES cells. A distinct set of DNA loops in each cluster may represent the underlying structural mechanism of this process. Thus, looping contacts may be required to regulate temporal colinear Hox induction in vivo.
We found that looping is dependent on transcription activity. Physical DNA contacts can be regulated by changes in binding protein levels, through post-translational modification of histones and/or binding proteins, or by modification of DNA sequences. PcG proteins were shown to maintain higher-order structures in BX-C and to mediate long-range contacts responsible for transcription downregulation in human cells. Although Hox genes were not included in the latter study, long-range contacts were lost following RA differentiation and EZH2 depletion also resulted in disruption of long-range contacts. In our study, EZH2 was found to be required for looping and silencing of HoxA. However, given its broad distribution at the cluster’s 5′-end, it seems likely that EZH2 may strengthen the silent structure but that it is not responsible for DNA looping. This mechanism is in contrast to Drosophila BX-C where PcG proteins were shown to bridge PREs and promoters and where gene activity is pronounced in PcG depleted cells (28).
Alignment of base density scans with genome annotation tracks helped us identify CTCF as candidate mediator of DNA loops. Unlike PcG in human, CTCF is known to bind directly to DNA and consensus-binding sites have been identified (23,34,35). Several genome-wide CTCF ChIP–Chip and ChIP-seq datasets indicate that binding sites are largely invariant across cell types and thus independent of gene expression. CTCF was previously suggested to play a role in the organization of genome into higher-order chromatin domains that allow for the normal regulation of gene expression (36). We propose that CTCF may be the pivotal player in weaving the 3D architecture that mediates silencing of the Hox clusters and that PcG act downstream of CTCF binding. How might CTCF regulate Hox gene expression and in which cellular context? Is CTCF required to maintain the 3D architecture of silent Hox clusters and to recruit regulatory proteins such as PcGs? It will be interesting to define the role of CTCF in spatial chromatin remodeling and temporal colinear induction of Hox genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Canadian Institutes of Health Research (CIHR DC0190GP to J.D.); Canadian Cancer Society Research Institute (CCSRI 019252 to J.D.); Discovery Grant from the National Sciences and Engineering Research Council of Canada (NSERC) (to M.B.). CCSRI, fellowship (to M.A.F.); NSERC (to M.R.). M.B. is an Alfred P. Sloan Fellow. J.D. is a CIHR New Investigator and FRSQ Research Scholar. Funding for open access charge: Canadian Institutes of Health Research (CIHR DC0190GP).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our laboratory for stimulating discussions. We are grateful to Drs. J. Teodoro, J. Pelletier, and W. Bickmore for critical reading and comments on this manuscript.
REFERENCES
- 1.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.Würtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Singh Sandhu K, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 5.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 6.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 7.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 9.Vakoc C, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 10.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 11.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 12.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 13.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell. Mol. Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adler PN, Charlton J, Brunk B. Genetic interactions of the suppressor 2 of zeste region genes. Dev Genet. 1989;10:249–260. doi: 10.1002/dvg.1020100314. [DOI] [PubMed] [Google Scholar]
- 15.Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: clinical and biological aspects. Cancer Res. 2008;68:10024–10027. doi: 10.1158/0008-5472.CAN-08-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, Dracopoli NC, Fogh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- 17.Fraser J, Rousseau M, Shenker S, Ferraiuolo MA, Hayashizaki Y, Blanchette M, Dostie J. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10:R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boncinelli E, Simeone A, Acampora D, Mavilio F. HOX gene activation by retinoic acid. Trends Genet. 1991;7:329–334. doi: 10.1016/0168-9525(91)90423-n. [DOI] [PubMed] [Google Scholar]
- 20.Simeone A, Acampora D, Nigro V, Faiella A, D'Esposito M, Stornaiuolo A, Mavilio F, Boncinelli E. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech. Dev. 1991;33:215–227. doi: 10.1016/0925-4773(91)90029-6. [DOI] [PubMed] [Google Scholar]
- 21.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Vol. Supplement 74. Hoboken, N.J.: John Wiley & Sons; 2006. pp. 21.11.1–21.11.20. [DOI] [PubMed] [Google Scholar]
- 22.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 23.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.ENCODE-consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell. Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 29.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 31.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.