Abstract
microRNAs are widely expressed, ∼22-nt-long regulatory RNAs. They are first transcribed as much longer primary transcripts, which then undergo a series of processing steps to yield the single-stranded, mature microRNAs, although the mechanisms are incompletely understood. Here, we show that the terminal loop region of human primary microRNA transcripts is an important determinant of microRNA biogenesis. Mutations that restrain the terminal loop region inhibit Drosha processing of primary microRNA transcripts as well as Dicer processing of precursor microRNA transcripts in vitro. The inhibition may result from lower enzyme turnover on the mutant transcripts. Consequently, the mutations reduce miRNA maturation in transfected human cells. We conclude that a flexible terminal loop region is critical for microRNA processing.
INTRODUCTION
microRNAs (miRNAs) are a class of abundant, ∼22-nt-long RNA molecules that primarily inhibit gene expression at the post-transcriptional level (1–3). miRNAs control a wide range of biological processes, such as development, metabolism, cell growth, cell death and cell fate determination (4). Furthermore, altered miRNA expression and, hence, function, have been associated with human disease (5). Thus, it is imperative to understand the mechanism governing miRNA expression.
An miRNA is initially transcribed as part of a long primary transcript, or pri-miRNA (6). The pri-miRNA is cleaved by an RNase called Drosha, along with its regulatory subunit DGCR8 in mammals, to liberate a hairpin precursor of ∼65 nt in the nucleus (7–11). A small number of miRNA precursors can also be generated independent of the Drosha holoenzyme (Drosha in short hereafter; 12–16). The precursor, or pre-miRNA, is then exported to the cytoplasm by Exportin5 (17,18). Once in the cytoplasm, the precursor is further processed by Dicer, another RNase, to produce an ∼22 -bp RNA duplex intermediate (19–22). The binding of an Argonaute protein to the duplex and subsequent rearrangements result in the retention and final production of the mature, single-stranded miRNA in the Argonaute:miRNA complex (23,24). Additional proteins such as TRBP and PACT may facilitate the selection and transfer of mature miRNAs to Argonaute (25–31).
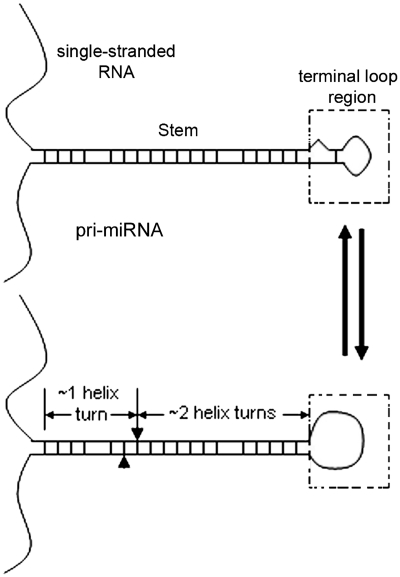
Advances in identification of the basic machinery that processes miRNA transcripts notwithstanding, how miRNAs are matured is incompletely understood. A pri-miRNA contains the following structural features (Figure 1): a terminal loop region, a mostly double-stranded RNA (dsRNA) stem encompassing the miRNA duplex and an ∼1 helical turn extension, and flanking single-stranded RNA. Analyses of a select number of miRNAs showed that disrupting the stem inhibits miRNA production, and the single-stranded RNA domain is also required for the processing including Drosha cleavage of pri-miRNAs (32–35). Some reports also suggested that the terminal loop region is important for Drosha processing and miRNA maturation (32–34,36). Here, the term ‘terminal loop region’ refers to the apical domain beyond the miRNA duplex segment. It can be folded into a small terminal loop and a short stem, as RNA folding programs typically predict, or relaxed into a largely single-stranded structure, i.e. a large terminal loop (Figure 1). We proposed that this region is flexible, with a more single-stranded conformation preferred for miRNA production (33). Han et al. (35), however, argued that the terminal loop is dispensable for Drosha processing. To clarify this discrepancy, we examined the production of select miRNAs in test tubes and cultured human cells, focusing on the role of the terminal loop region in the transcripts.
Figure 1.
Schematics of pri-miRNA structure and flexibility in the terminal loop region. A pri-miRNA consists of a central stem of ∼3 helical turns flanked by the terminal loop region at one end and single-stranded RNA at the other. The terminal loop region is structurally dynamic. The arrowhead and arrow on the RNA signify Drosha cleavage sites.
MATERIALS AND METHODS
Plasmid construction
Approximately 130 bp of DNA encoding miR-16-1 was amplified from human genomic DNA (Clontech) and inserted into the HindIII and XhoI sites of a modified pSuper vector (33). Mutations were introduced using the Quikchange method (Stratagene) and verified by sequencing.
Cell culture and transfection
293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 5% CO2 and transfected using Lipofectamine 2000 (Invitrogen). Total RNA was isolated using Trizol reagent (Invitrogen).
Drosha cleavage assays
DNA templates for RNA synthesis were generated by polymerase chain reaction (PCR) with a primer containing T7 promoter sequence. RNAs were then prepared by in vitro transcription (Promega) in the presence of [α-32P] CTP. DNA size markers (Promega) were labeled at their 5′-ends with [γ-32P] ATP by T4 polynucleotide kinase (New England BioLabs). Poly (dI:dC) and poly (I:C) were purchased from Sigma. To purify the Drosha holoenzyme, 293T cells were co-transfected with plasmids that expressed a FLAG-tagged Drosha and HA-tagged DGCR8 (37). Two days later, cell extract was prepared in lysis buffer (20 mM Tris pH 7.5, 0.15 M NaCl, 1 mM EDTA and 0.4% NP-40) and incubated with anti-FLAG antibody conjugate agarose beads (Sigma) for ∼1 h at 4°C. Beads were washed six times with the lysis buffer containing 0.8 M NaCl. The Drosha holoenzyme (Drosha in short) was eluted with 3xFLAG peptide (Sigma) in reaction buffer (20 mM HEPES-KOH pH 7.6, 100 mM KCl, 0.2 mM EDTA and 5% glycerol). In vitro Drosha cleavage reactions were carried out as described (37) for 30 min or the indicated times. When required, poly (dI:dC) or poly (I:C) was mixed with labeled RNA prior to Drosha addition. Data were analyzed using a Storm 840 PhosphorImager (GE Healthcare) or by autoradiography.
RNase A cleavage assay
RNA substrates (sequences shown in Figure 3) were prepared by in vitro transcription using T7 RNA polymerase, de-phosphorylated, and run on a 10% denaturing gel. RNAs were purified from the gel and then labeled with [γ-32P] ATP using T4 polynucleotide kinase. The RNAs have a short 5′ end overhang to facilitate 32P labeling, but because T7 RNA polymerase tends to add a few extra nucleotides at the 3′ end, there might be 3′ end heterogeneity in the actual products. Nevertheless, these RNAs are expected to contain at most a small overhang, which is not supportive of Drosha cleavage (34). RNAs (∼10−14 mol) were mixed with purified Drosha (∼10−14–10−13 mol) at room temperature for 15 min. To further prevent Drosha cleavage, 2 mM EDTA was included and Mg2+ excluded in the reaction buffer. RNase A (1–8 ng, Invitrogen) was then added. Total volume was ∼10 µl. After 10 min, RNAs were purified and analyzed by 7 M urea/10% polyacrylamide gel electrophoresis. The 39-nt-long and 45-nt-long pri-miR-30 markers were similarly produced and labeled.
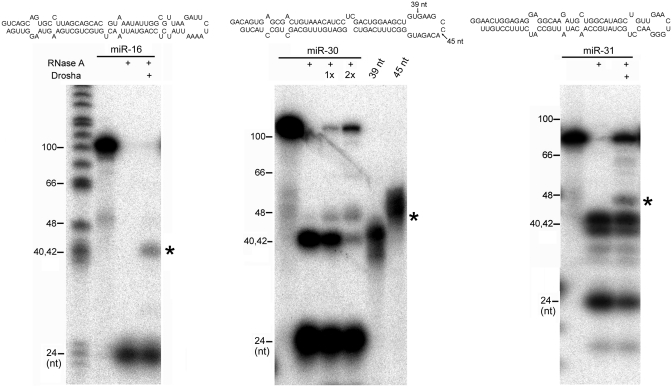
Figure 3.
Interaction between Drosha and pri-miRNAs. 5′-end labeled, pri-miR-16, pri-miR-30 and pri-miR-31 substrates were digested with RNase A with or without prior incubation with Drosha. Sequences and the predicted secondary structures of the pri-miRNAs are shown on top. Stable RNase A cleavage products in the presence of Drosha were indicated with an asterisk. DNA size markers are indicated in the left. The pri-miR-30 substrate was incubated with two concentrations of Drosha (indicated as ‘1x’ and ‘2x’) before RNase A addition, and two RNA markers of 39-nt and 45-nt-long were run on the same gel for size comparisons: their 3′ ends are indicated by arrows on top. Shown are representatives of at least three independent experiments.
Northern blotting
Approximately 20 μg of total RNAs were fractionated on a 7 M urea/15% polyacrylamide gel and transferred to a Hybond N+ membrane (GE Healthcare). The membrane was probed with 5′-end, 32P-labeled oligonucleotides, with blot stripping in between. The oligonucleotide sequences for miRNA detection are 5′-TTAACGCCAATATTTACGTGCTGCTAAGGCA-3′ (complementary to the 5′ arm of the pri-miR-16-1 stem) and 5′-TACTTCAGCAGCACAGTTAATACTGGAGATAA-3′ (complementary to the 3′ arm of the pri-miR-16-1 stem), for the small interfering RNA (siRNA) against the green fluorescent protein (GFP) mRNA, 5′-GTACACAAGAACGGCATCAAGG-3′, and for U6 snRNA: 5′-ACGAATTTGCGTGTCATCCTTGCG-3′.
Primer extension
Approximately 5 μg of total RNAs from transfected 293T cells were annealed to a primer specific for miR-16 (5′-GCATCCCGCCAATATTTACGT-3′) at 37°C for 20 min, and primer extension experiment was performed as described (32).
Dicer cleavage assays
RNA substrates were prepared by in vitro T7 transcription and labeled either with [α-32P] CTP during transcription or afterwards with [γ-32P] ATP using T4 polynucleotide kinase. RNA labeled with both methods yielded equivalent results. Pre-miRNA substrates might harbor substitutions to satisfy the requirement that T7 transcription starts from a G residue. dsRNAs were prepared by annealing two complementary RNA strands followed by native gel-purification, before labeling with [γ-32P] ATP by T4 polynucleotide kinase. Dicer cleavage assays were performed as described (38). Briefly, reaction was initiated by adding human Dicer (Invitrogen) to labeled RNA at 37°C (∼0.025 unit of Dicer/µl final reaction volume). At designated time points, 4 µl of the reaction mixture was added to 4 µl of 2× sample buffer (98% formamide, 10 mM EDTA and 0.1% bromophenol blue) on ice. After gel electrophoresis, data were analyzed using a PhosphorImager or by autoradiography. To quantify data in Figure 6B, we used cleavage percentage defined as product intensity divided by total (products and full-length substrate) intensity. This parameter is an imprecise estimate of the catalytic rates, as intermediates/products from ds50 and ds120 were Dicer substrates as well. Nevertheless, from the figure ds50 and ds120 intermediates and their ∼10-bp product, which signified complete dsRNA digestion, did not accumulate significantly. Moreover, the full-length ds50 and ds120 were always present in a large excess over the intermediates, after accounting for 32P loss in the intermediates, so the full-length ds50 and ds120 remained the major substrates during Dicer reaction. These considerations led us to conclude that the cleavage percentage shown in the y-axis is a useful measure of product formation.
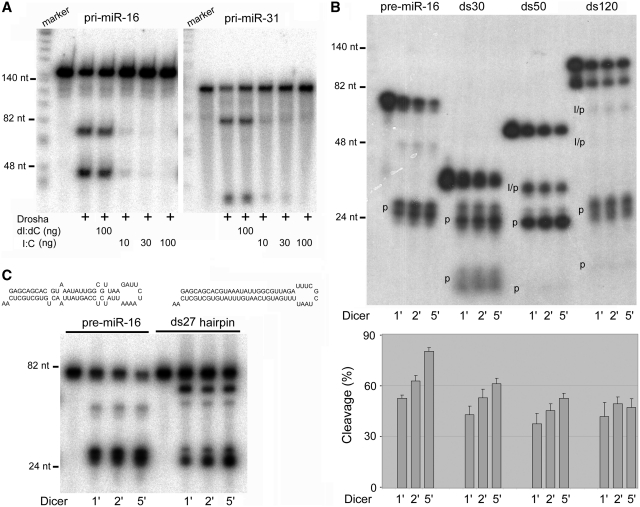
Figure 6.
dsRNA interferes with miRNA processing. (A) Poly (I:C) inhibited Drosha cleavage of pri-miR-16 and pri-miR-31. Labeled DNA markers were loaded in the right lanes. The amount of poly (dI:dC) or poly (I:C) added to the reactions is indicated at the bottom. Shown are representatives of three independent experiments. (B) Dicer cleavage of pre-miR-16 and three dsRNA substrates, ds30, ds50 and ds120. Dicer cleavage was terminated after 1, 2 or 5 min. RNAs were quantified using a PhosphorImager. Cleavage intermediates (‘I’) and products (‘p’) are indicated. Cleavage percentage is calculated as product and intermediate intensities divided by total (substrate, intermediate and products) intensities. (C) Dicer cleavage of pre-miR-16 and its mimic, ds27 hairpin. The sequences and predicted secondary structures of pre-miR-16 and ds27 hairpin are shown at the top.
RESULTS
Mutations in the terminal loop region of pri-miR-16 impact Drosha processing
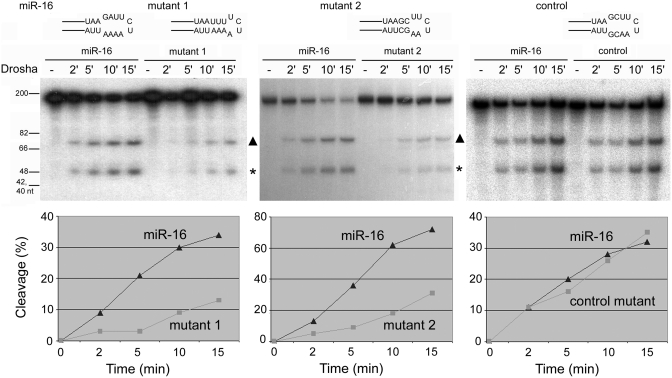
A flexible or large terminal loop has been shown to be important for the maturation of a number of human miRNAs, such as miR-18a, miR-21, miR-27a, miR-30a (miR-30 in short hereafter) and miR-31 (32–34,36). RNA folding programs tend to predict small terminal loops, e.g. ∼4- or 5-nt loops, to maximize RNA thermodynamic stability. Nevertheless, mutations enforcing such small terminal loops limited Drosha cleavage and miRNA expression (33). Han et al. (35), however, concluded that the loop did not influence Drosha processing, based largely on their analysis of pri-miR-16-1 (miR-16 in short) in test tubes. We therefore examined how mutations that minimized the terminal loop of pri-miR-16 influenced Drosha cleavage. Figure 2 shows that two mutations (as in mutants 1 and 2) that were predicted to extend the stem further into the apical region similarly reduced pre-miR-16 production by at least 60%. While the degree of reduction was modest, it was consistent and significant. On the other hand, a control pri-miR-16 mutant that had the same nucleotide composition as mutant 2 while maintaining the wild-type loop structure was cleaved by Drosha as efficiently as the wild-type pri-miR-16 (Figure 2). Clearly, a loose terminal loop facilitates processing of pri-miRNAs including pri-miR-16 by Drosha in vitro. Human miR-16, miR-30 and miR-31 sequences were arbitrarily selected as representatives for subsequent analyses.
Figure 2.
Drosha cleavage of human pri-miR-16 (wild-type and mutants, sequences and RNAfold-predicted secondary structures in the loop region are shown). Drosha cleavage reactions were stopped at 2, 5, 10 or 15 min and subjected to gel electrophoresis. Sizes of DNA markers are indicated in the left. Shown are representatives of at least two independent experiments, with the cleavage percentages plotted in the graphs below. Cleavage is calculated as the intensities of the products (pre-miR-16, marked with a triangle, and flanking sequences, marked with an asterisk) divided by the intensities of the products and remaining substrate at each time point.
Interaction between Drosha and pri-miRNAs
How the Drosha holoenzyme physically interacts with pri-miRNAs is unknown. To probe such an interaction, we performed RNase A cleavage assays (Figure 3). The most prominent band generated by RNase A digestion was ∼22 nt long and represented cleavage near the middle of the miRNA duplex (Figure 3). The addition of Drosha led to new RNase A cleavage products of ∼40 nt long (indicated by ‘*’, Figure 3). These new RNAs had their 3′ end in the terminal loop region of the pri-miRNAs, based on the their estimated size as compared to the DNA markers and to the synthetic, 5′-end, 32P-labeled, 39-nt-long and 45-nt-long pri-miR-30 fragments (Figure 3). The pri-miRNA substrates and buffer compositions were designed (Figure 3 and ‘Materials and Methods’ section) such that Drosha did not cleave the RNAs under these assay conditions (data not shown). While more work is needed to determine whether Drosha contacts the apical region directly, these results demonstrated that Drosha interaction with a pri-miRNA alters the RNase A sensitivity, or conformation, of the region, which is consistent with a putative role of the terminal loop region in pri-miRNA processing.
Mutations in the terminal loop region affect mature miR-16 production in transfected cells
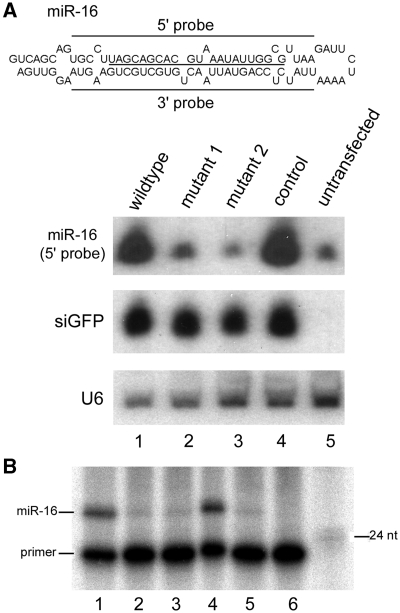
To further investigate the role of the terminal loop in miR-16 biogenesis, we transfected 293T cells with plasmids that encoded the wild-type and mutant pri-miR-16 as studied in Figure 2. A pH1-GFP plasmid that expressed an siRNA targeting GFP mRNA was used as a co-transfection control (38). Northern blot analysis revealed that mutations 1 and 2 (lanes 2 and 3, Figure 4A) greatly reduced exogenous miR-16 expression, while the control mutant was expressed similarly as the wild-type (lanes 4 and 1, Figure 4A). Northern blotting for the ectopically expressed GFP siRNA and endogenous U6 snRNA was used to demonstrate equivalent transfection and RNA loading in the different samples (Figure 4A). Mutants 1 and 2 did not produce any aberrant small RNA, as a probe complementary to the 3′ arm of pri-miR-16 failed to detect any small RNA (data not shown), and primer extension experiments did not identify any small RNA with a different 5′ end from miR-16, either (Figure 4B). Therefore, mutations 1 and 2 led to inefficient miRNA processing in cells.
Figure 4.
miR-16 expression in transfected 293T cells. (A) 293T cells were transiently co-transfected with pH1-GFP and a plasmid that encoded wild-type or the indicated mutant pri-miR-16. miR-16, GFP siRNA (siGFP), and the endogenous U6 snRNA expression was examined by northern blotting. Sequences and the predicted, secondary structure of pri-miR-16 are shown on top, with mature miR-16 underlined. Sequences complementary to the 5′ and 3′ probes are marked with bars. (B) Primer extension to detect miR-16 expression. Positions of the primer, the miR-16 extended product, and a 24-nt DNA marker are indicated. Lanes 1–5 are the same as those in (A). Lane 6: primer only.
The terminal loop region influences Dicer cleavage
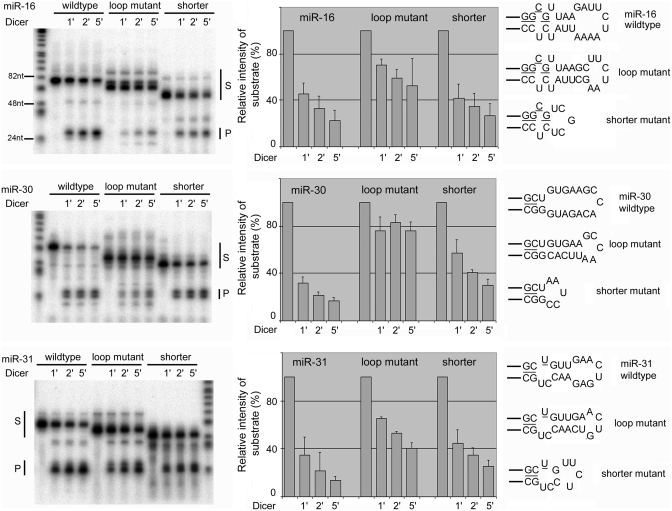
Interestingly, reduction in miR-16 expression due to loop restriction was more severe in 293T cells than revealed by the Drosha assay: mutants 1 and 2 were hardly overexpressed above the endogenous miR-16 level in transfected cells (e.g. compare lanes 2 and 3 to lane 5, Figure 4A), while their pri-miRNAs were still processed by Drosha in test tubes, albeit at a modestly lower efficiency than the wild-type (Figure 2). We therefore tested if the mutations also affected other steps in the miRNA processing pathway, e.g. Dicer cleavage of pre-miRNAs. Indeed, a pre-miR-16 mutant (corresponding to mutant 2 in Figures 2 and 4) was cleaved less efficiently than wild-type pre-miR-16 by Dicer in vitro, with the same phenomenon observed for pre-miR-30 and pre-miR-31 as well (‘loop mutant’, Figure 5). These mutations enforced smaller terminal loops but maintained the overall length of the hairpin RNAs, even though the RNAs tended to migrate faster than the wild-type during gel electrophoresis, apparently as a result of conformational changes in the terminal loop region. We then constructed another set of mutant pre-miRNAs (‘shorter’ mutants, Figure 5) that contained the stem corresponding to the miRNA duplex segment connected directly by a terminal loop similar in size to that of the ‘loop mutants’. (Because the miRNA duplex moiety in a pre-miRNA often fashions unpaired nucleotides at the ends, it is not always possible to have the above two sets of mutants possessing the same terminal loop.) As shown in Figure 5, Dicer cleaved these mutant RNAs clearly better than the loop mutants and only slightly less than the wild-type pre-miRNAs. Dicer processed all the hairpin RNAs to yield the same miRNA duplexes (pointed out by ‘P’, Figure 5), indicating that changes in the loop region did not alter cleavage site selection by Dicer, which is consistent with the notion that Dicer measures and cleaves from the end of a dsRNA (39). These findings demonstrate that Dicer processing is also influenced by the flexibility in the terminal loop region of pre-miRNAs.
Figure 5.
Dicer cleavage of pre-miRNAs and their mutants. The ‘loop mutant’ denotes RNA of the same length as the wild-type pre-miRNA but with a smaller terminal loop. The ‘shorter’ mutant denotes RNA with the miRNA duplex moiety linked directly to a terminal loop that was similar in size to that in the ‘loop mutant’. Dicer cleavage was allowed to proceed for 1, 2 or 5 min before termination. RNAs were quantified using a PhosphorImager. S, substrates; P, miRNA duplex product. The same amount of total reaction mixture was loaded in each lane, except that the lane without Dicer addition contained only 75% of the RNA in the other lanes. The disappearance of the substrates was used as a measure of Dicer cleavage, shown by the graphs in the middle with averages and standard deviations (error bars; n ≥ 3). Sequences and predicted secondary structures in and near the terminal loop region are shown in the right, with mature miRNAs underlined.
dsRNA interferes with miRNA processing by Drosha and Dicer
A compact terminal loop region due to conformation changes or mutations creates a longer dsRNA extension, which might increase Drosha and Dicer's degree of freedom along the RNA stem and impede catalysis or product release by the enzymes. Of note, mutations that restrict the terminal loop region and mutations that extend the stem near or into the single-stranded RNA segment both inhibit pri-miRNA cleavage (33–35). A common theme is that these mutations all lengthen the stem of a pri-miRNA; i.e. they increased dsRNA characteristics and hindered cleavage by Drosha in cis. It has been also shown that Dicer can stably associate with its substrates and products (40). To test if a strong dsRNA characteristic could indeed hinder the action of Drosha and Dicer, therefore, we carried out the following experiments. We first added poly (dI:dC) or poly (I:C) to Drosha cleavage reactions against pri-miR-16 and pri-miR-31. Only poly (I:C), as a long dsRNA mimic, severely inhibited pri-miRNA processing (Figure 6A). Thus, Drosha function can be inhibited by a high degree of dsRNA feature both in cis and in trans.
We then investigated Dicer cleavage of the 5′-end labeled, pre-miR-16 and three perfectly matched dsRNAs of ∼30 bp, 50 bp or 120 bp in length (ds30, ds50 and ds120, Figure 6B). These dsRNAs were designed with one end mimicking the end sequence and secondary structure of pre-miR-16. We compared how fast Dicer processed the different full-length substrates to generate the products and/or intermediates (Figure 6B). Dicer cleaved these different RNAs at similar rates by 1 min, but at the later, 2 and 5 min time points it became more apparent that pre-miR-16 was the best Dicer substrate, ds30 was the next, and the longer dsRNAs the worst substrates (Figure 6B). The data suggest that Dicer cleavage can be negatively influenced by an eminent dsRNA trait in the substrates. Similarly, Dicer cleaves a modified pre-let-7 substrate more rapidly than a 37-bp dsRNA (41). A complication is that Dicer cuts dsRNAs from both of their free ends, which made cleavage seem even faster. We therefore compared Dicer processing of pre-miR-16 with that of a pre-miR-16 mimic possessing a predicted, perfect 27-bp dsRNA stem (Figure 6C). Of the two substrates, pre-miR-16 was digested much faster by Dicer, demonstrating that imperfect basepairing in the stem region, i.e. less dsRNA feature, enhances Dicer cleavage in cis.
DISCUSSION
Identification of important features in pri-miRNAs will advance our knowledge of what constitutes a miRNA as well as shed light on the mechanisms of miRNA processing. This study establishes that the terminal loop region plays a critical role in human miRNA biogenesis. A flexible terminal loop region facilitates both Drosha processing of pri-miRNAs and Dicer processing of pre-miRNAs. The mechanism probably includes enhancing enzyme turnover on RNA substrates.
RNA folding algorithms predict that the terminal loop region of a pri-miRNA folds into a small terminal loop and a short stem for maximal stability. We consider this region is structurally dynamic and can also be relaxed into a largely single-stranded conformation or a larger terminal loop (Figure 1). Studies have shown that such a large loop enhances miRNA processing and expression. In particular, point mutations that strengthened basepairing in the terminal loop region inhibited miRNA expression in cell culture as well as Drosha cleavage of pri-miRNAs in test tubes. These mutations are relevant because they reinforce secondary structures predicted by RNA folding programs for many miRNAs. On the other hand, it had been argued that the terminal loop is dispensable for Drosha function, based largely on analysis of in vitro pri-miR-16 processing (35). Here, we demonstrated that mutations that created a smaller terminal loop in pri-miR-16 hindered Drosha cleavage (Figure 2). Drosha also altered RNase A accessibility of pri-miRNAs in the terminal loop region (Figure 3), indicative of a direct or indirect interaction between Drosha and the loop. Surprisingly, the terminal loop region further impacts Dicer action, as loop restriction likewise repressed Dicer cleavage of pre-miRNAs (Figure 5). These data might help fully explain the result that those mutations dramatically reduced miR-16 expression in cell cultures (Figure 4), as did analogous mutations in other miRNAs (33).
How does a large terminal loop facilitate miRNA maturation? Both Drosha and Dicer have RNAs with double stranded characteristics as substrates, so a possibility is that a flexible loop, by dynamically reducing dsRNA features, enhances RNA release and enzyme turnover. Consistent with this explanation, RNAs with a minimal duplex region and a small terminal loop were cleaved more efficiently by Dicer than similar RNAs with a more elongated or perfectly complementary stem (Figures 5 and 6). This finding is significant, as it is commonly assumed that only the end structure in a substrate is critical for its cleavage by Dicer. Moreover, mutations that created a longer central stem inhibited the cleavage of pri-miRNAs by Drosha in cis (33–35; Figure 2), and long dsRNA such as poly (I:C), while not a Drosha substrate, blocked pri-miRNA processing in trans (Figure 6A). Drosha would face many irrelevant RNAs that possess a hairpin or dsRNA structure in a cell, so the ability to dissociate from such RNAs might be critical for Drosha's normal function in pri-miRNA processing. Finally, it is notable that the stem in pri- and pre-miRNAs always harbors multiple mismatches. These mismatches, along with a flexible apical loop region, might have evolved in miRNAs to enable their efficient selection and cleavage by Drosha and Dicer (e.g. Figure 6B and C). Another, nonexclusive possibility is that a large, open RNA conformation in the apical region is preferred for the binding or catalysis of the RNases. For Drosha, mutant pri-miRNAs that differed from the wild-type by only having a smaller terminal loop region were poorer substrates (33,36). Natural Dicer substrates include long dsRNAs, so the requirement for a large terminal loop seems more stringent for Drosha than for Dicer function. Nevertheless, Dicer cleaved wild-type pre-miRNAs slightly faster than ‘shorter’ mutant hairpins with a minimal duplex and a small loop, at least for pre-miR-30 (Figure 5), suggesting that the terminal loop could still contribute to Dicer action.
We propose that pri-miRNAs contain three structural elements important for maturation: a terminal loop region, a central stem and a single-stranded RNA domain (Figure 1). All human pri-miRNAs contain a terminal loop, which enables efficient and correct processing. Slight variations in the junction between the loop and the stem or between the stem and single-stranded RNA modify relevant RNase cleavage sites, while disruption of any of the three traits reduces or eliminates miRNA processing (33–35). Proteins such as hnRNP A1, Lin-28 and KSRP have been shown to bind to the terminal loop region to regulate the processing of specific miRNAs (36,42–49). We show here that the region has a universal role in miRNA processing, so an additional, interesting mechanism might be that some of the proteins could act by inducing conformational changes in the RNAs to satisfy or dissatisfy the structural requirements of the general miRNA processing enzymes.
FUNDING
The Minnesota Medical Foundation (3674-9227-06, 3842-9201-08); U.S. Department of Defense (W81XWH-07-1-0183); National Institute of Drug Abuse (P50 DA 011806). Funding for open access charge: Departmental start-up fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 3.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Denli AM, Tops B, Plasterk RHA, Ketting RF, Hannon GJ. Processing of pri-microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 9.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 19.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Hutvágner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 22.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RHA. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 25.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Y, Cullen BR. Sequence requirements for microRNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Y, Cullen BR. Single-stranded extensions on primary microRNA transcripts are required for Drosha cleavage. J. Biol. Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 35.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 36.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods Mol. Biol. 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 43.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 46.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska E. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 49.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]