Abstract
All tRNAHis possess an essential extra G–1 guanosine residue at their 5′ end. In eukaryotes after standard processing by RNase P, G–1 is added by a tRNAHis guanylyl transferase. In prokaryotes, G–1 is genome-encoded and retained during maturation. In plant mitochondria, although trnH genes possess a G–1 we find here that both maturation pathways can be used. Indeed, tRNAHis with or without a G–1 are found in a plant mitochondrial tRNA fraction. Furthermore, a recombinant Arabidopsis mitochondrial RNase P can cleave tRNAHis precursors at both positions G+1 and G–1. The G–1 is essential for recognition by plant mitochondrial histidyl-tRNA synthetase. Whether, as shown in prokaryotes and eukaryotes, the presence of uncharged tRNAHis without G–1 has a function or not in plant mitochondrial gene regulation is an open question. We find that when a mutated version of a plant mitochondrial trnH gene containing no encoded extra G is introduced and expressed into isolated potato mitochondria, mature tRNAHis with a G–1 are recovered. This shows that a previously unreported tRNAHis guanylyltransferase activity is present in plant mitochondria.
INTRODUCTION
With the exception of a few α-proteobacterial tRNAHis (1), all known tRNAHis molecules have an unusual 5′-end consisting of an additional guanylate residue called G–1. The resulting extra base-pair in the acceptor stem is a fundamental determinant for tRNAHis aminoacylation by histidyl-tRNA synthetases both in prokaryotes and in eukaryotes (2,3).
This extra guanylate residue can be generated via two different processes. In Escherichia coli and in chloroplasts, the G–1 is genome-encoded and retained during tRNA maturation because of an unusual cleavage of the pre-tRNAHis at the −1 position by RNase P (4,5). In Saccharomyces cerevisiae as well as in Drosophila melanogaster the G–1 is not genome-encoded and must be post-transcriptionally added at the 5′ terminus of the nuclear-encoded tRNAHis by a specific tRNAHis guanylyltransferase (6,7). In chicken mitochondria, the extra G is also post-transcriptionally added (8) and a mitochondrial tRNAHis guanylyltransferase must be present in this organism. Indeed tRNAHis guanylyltransferases with putative mitochondrial targeting sequences were retrieved from databases by Gu et al. (9).
In plants, all mitochondrial-encoded trnH genes sequenced so far show a potential extra guanylate residue at their 5′ ends. In the liverwort Marchantia polymorpha and in larch, a gymnosperm, the ‘native’ trnH gene encodes an extra G at position −1 (10,11). In angiosperms, a ‘chloroplast-like’ trnH gene is expressed on the mitochondrial DNA (i.e. ref. 12). As on plastidial DNA, this mitochondrial gene also potentially encodes a G–1. Thus, it was suggested that, as in chloroplasts, the extra G is mitochondrially encoded, although no experimental evidence was brought so far in plant mitochondria. Here, we provide evidence that, even though plant mitochondrial trnH genes potentially encode the G–1, this residue can also be added post-transcriptionally by a guanylyltransferase enzyme. First, we show that plant mitochondrial RNase P cleaves tRNAHis precursor at both positions −1 and +1 in vivo and in vitro. Second, using a direct DNA uptake methodology, we show that, during tRNA expression and processing in potato mitochondria, a guanylate residue is added to a mutated larch mitochondrial tRNAHis precursor containing no encoded extra G. Finally, using a biochemical approach, a tRNAHis-dependent guanylyltransferase activity was found in potato mitochondria. Allover, our data show that there is an apparent flexibility in the processing of tRNAHis in plant mitochondria and strikingly that the two possible routes can be used to generate a functional tRNA for translation.
MATERIALS AND METHODS
In vitro synthesis of tRNA transcripts
Larch mitochondrial pre-tRNAHis and mature tRNAHis transcripts were synthesized from clones described in (11,13). Oligonucleotide-directed mutagenesis used to generate a larch mitochondrial trnH gene having no G–1 was performed using the QuickChangeTM site-directed mutagenesis kit (Stratagene). Potato tRNAHis precursor construct was amplified by polymerase chain reaction (PCR) using relevant pairs of primers and cloned into pGEM-T Easy (Promega). The construct containing Arabidopsis thaliana cytosolic trnA gene sequence was obtained previously (14). The Riboprobe Kit (Promega) was used to synthesize in vitro transcripts in the presence of either Sp6 or T7 polymerases.
Vaccinia guanylyltransferase labeling of potato mitochondrial tRNA fraction and Southern analysis
A potato mitochondrial tRNA fraction prepared as described in ref. (15) was labeled using commercial vaccinia guanylyltransferase in the presence of 50 µCi of [α-32P]GTP (800 Ci/mmol; 10 mCi/ml) according to manufacturer's recommendation (Ambion Inc.). This labeled tRNA fraction was then hybridized to a Southern blot carrying 1 µg per lane of PCR-generated DNA corresponding to either larch mitochondrial tRNAHis potato mitochondrial tRNACys or tRNAHis sequences using conditions described elsewhere (13).
Potato mitochondria isolation, DNA uptake experiment and mitochondrial transcription of imported DNA
Mitochondria were isolated from potato (Solanum tuberosum) tubers by differential centrifugation and purification on Percoll gradient as described in ref. (16). As previously shown, the mitochondrial fraction is free of any cytosolic and plastidial contamination. Mitochondrial import of DNA followed by mitochondrial transcription of imported DNA sequences was mostly carried out as described in ref. (13). The gene construct corresponding to the potato mitochondrial 18S rRNA gene promoter sequence fused to larch mitochondrial trnH sequence with its 5′- and 3′-flanquing sequences (13) was used as a substrate for oligonucleotide-directed mutagenesis to a generate a construct containing no G–1 and G–2 upstream of the trnH gene. Oligonucleotide-directed mutagenesis was performed using the QuickChangeTM site-directed mutagenesis kit (Stratagene).
Circular reverse transcription-PCR
Circular reverse transcription (RT)-PCR (cRT-PCR) was used to determine 5′- and 3′-termini of RNA substrates. DNase-treated RNA extracted from mitochondria was incubated with 40 U of T4 RNA ligase (New England Biolabs) in the supplied buffer supplemented with 2 U of RNase inhibitor and in a total volume of 25 µl. Following circularization, all steps of RT-PCR were carried out essentially as described in ref. (13).
Mitochondrial RNase P activity assays
Arabidopsis thaliana mitochondrial RNase P (called PRORP1) was overexpressed and purified according to ref. (17). Cleavage assays were carried out with 500 ng of pre-tRNA and 100 ng recombinant PRORP1 for 15 min in the reaction buffer previously described (17). The cleavage products were loaded in an 8% (v/v) acryl gel. The band corresponding to the 5′-matured precursors were cut and the matured tRNA was eluted and resuspended in water for subsequent cRT-PCR procedure.
Aminoacylation assays
An enzymatic extract was prepared from highly purified potato mitochondria as already described in ref. (15). Aminoacylation was conducted under optimal conditions in the presence of 8 µg of mitochondrial enzymatic extract, 50 µM [3H]-His (52 Ci/mmol) and 20 µM of tRNA transcript.
Assay for tRNAHis guanylyltransferase activity
The potato mitochondrial enzymatic extract prepared for aminoacylation assays was used to test for the presence of a guanylyltransferase activity. Guanylylation assay of 20 µM of tRNA transcript was done in the presence of 8 µg of mitochondrial enzymatic extract and 2.5 µCi of [α-32P]GTP (800 Ci/mmol) in a buffer containing 25 mM HEPES pH 7.3, 10 mM MgCl2, 100 mM KCl, 6 mM adenosine triphosphate (ATP), 3 mM DTT. Upon a 2-h incubation at 37°C, RNAs were phenol-extracted. After centrifugation, the nucleic acids recovered in the aqueous phase were purified by gel-filtration through a Sephadex G-50 spin column and resolved on a 15% (v/v) polyacrylamide gel containing 8 M urea.
Cloning of constructs
The Arabidopsis At2g31580 and At2g32320 genes encode a tandem duplication of the S. cerevisiae Thg1p homolog [(9) and Supplementary Data Figure SI]. The 5′ cDNA copy of each of the Arabidopsis genes was amplified by RT-PCR using total Arabidopsis RNA as template. PCR products were inserted in pB7FWG2 gateway expression vector according to manufacturer's instruction (Invitrogen).
In vivo localization analysis
Transient expression in Nicotiana benthamiana protoplasts and visualization of green fluorescent protein (GFP) fluorescence by confocal mitcroscopy was performed as described in (18).
Miscellaneous
Oligonucleotides were synthesized by Sigma-Aldrich. Oligonucleotide sequences used in this study are available upon request.
RESULTS
Different populations of tRNAHis exist in potato mitochondria
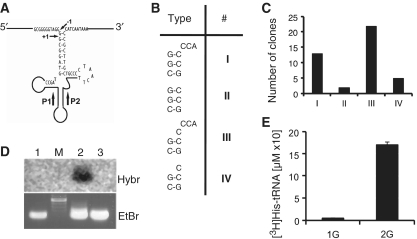
The 5′- and 3′-termini of potato mitochondrial tRNAHis (Figure 1A) were determined by cRT-PCR. For this purpose, a potato mitochondrial tRNA fraction was first circularized by T4 RNA ligase and then used for RT-PCR analysis. A PCR product of about 70 bp was amplified, cloned and sequenced. Out of 42 sequences (Figure 1B and C), only seven revealed tRNAHis sequences without the 3′-CCA triplet. Among them, two possess the extra G at position −1 (type II), whereas for five sequences the G–1 is not present (type IV). In 35 clones, the extra 3′-CCA sequence was found and among them, 13 clones possess the G–1 (type I), whereas in 22 sequences the G–1 is not present (type III). These results show the presence of various 5′ tRNAHis maturation intermediates in potato mitochondria with either one or two G at their 5′ends. Interestingly, the presence or not of a G at position −1 seems to be independent from the CCA addition. While the presence of tRNAHis molecules containing an extra G was expected as a result of an unusual cleavage of RNase P, the presence of tRNAHis molecules showing no extra G at position G–1 is more surprising. A capping experiment was performed on a potato mitochondrial tRNA fraction in the presence of vaccinia guanylyltransferase and [α-32P]GTP. The labeled RNA fraction was then probed against larch mitochondrial trnH and potato mitochondrial trnC and trnH. Noteworthy, the ‘native’ larch mitochondrial trnH (11) and the ‘chloroplast-like’ potato mitochondrial trnH sequences (19) differ greatly (28 different nucleotides out of 75). A hybridization signal was obtained only with the potato mitochondrial tRNAHis gene, whereas, as expected, no signal was observed with the two other tRNA genes (Figure 1D). As already shown for the chicken mitochondrial tRNAHis (8), our results indicate that a proportion of tRNAHis molecules present in potato mitochondria is a good substrate for the vaccinia guanylyltransferase and does not likely contain a G–1. In agreement with these data, it is worth to mention that, when larch mitochondrial tRNAHis precursor was processed in vitro using a potato mitochondrial processing extract, two 5′ termini were observed by primer extension (11). At first glance, it can be hypothesized that, in plant mitochondria, the presence of fully processed tRNAHis molecules with a G–*1 are obtained by an unusual cleavage by RNase P. However, the presence in vivo of tRNAHis molecules without G–1 suggests that the usual cleavage by RNase P can exist. Therefore, the post-transcriptional addition of an extra G by a plant mitochondrial guanylyltransferase to generate a functional tRNAHis could also be possible. In agreement with this hypothesis, we showed by using a potato mitochondrial enzymatic extract in an in vitro aminoacylation assay that no histidine can be charged on potato mitochondrial tRNAHis transcript lacking G–1, whereas tRNAHis transcript with a G–1 is a good substrate for histidylation (Figure 1E). Thus, likewise other organisms (2,3), the additional guanylate residue at position −1 is essential for histidylation of tRNAHis in plant mitochondria.
Figure 1.
Potato mitochondria contain a heterogeneous population of tRNAHis molecules. (A) Schematic cloverleaf structure of potato mitochondrial tRNAHis precursor molecule (EMBL accession number X93577). The primers P1 and P2 used for cRT-PCR analysis are depicted by black arrows. G+1 and G–1 are indicated by thin arrows. (B) Analysis by cRT-PCR of the 5′ and 3′ ends of the tRNAHis population present in a potato mitochondrial tRNA fraction. Four types of extremities (I, II, III and IV) were obtained. (C) The number of different clones found for each type of extremities is given. (D) [α-32P]GTP labeled potato mitochondrial tRNAs were probed against PCR fragments corresponding to (1) potato mitochondrial tRNACys, (2) potato mitochondrial tRNAHis and (3) larch mitochondrial tRNAHis. Hybr, autoradiogram after hybridization. EtBr, corresponding image of the ethidium bromide-stained gel of PCR products. (E) In vitro histidylation of potato mitochondrial tRNAHis transcripts with or without a G at position –1. The mean value of three independent experiments is presented. Error bars are given. 1G and 2G, potato mitochondrial tRNAHis transcripts without or with a G–1 respectively.
A guanylyl-transferase activity exists in potato mitochondria
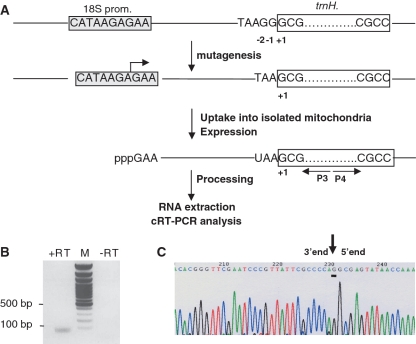
Using direct DNA uptake into potato mitochondria, we investigated the in organello maturation of a larch mitochondrial tRNA precursor transcript containing no extra G at position −1 in potato mitochondria. On the larch mitochondrial genome, three G residues are found at positions G+1, G–1 and G–2 on the trnH gene [(11) and Figure 2A]. First, using PCR mutagenesis, G–1 and G–2 were deleted from the previously used gene construct (13). In this construct, two consecutive A residues precede G+1 (Figure 2A). Then, following DNA uptake and expression of this mutated version into isolated potato mitochondria, mitochondrial RNAs were extracted, circularized by T4 RNA ligase, and used for cRT-PCR analysis. A specific reverse transcriptase-dependent product of about 70 nt was amplified (Figure 2B), cloned and sequenced. The size of this PCR product corresponds to the expected size of the product if the precursor RNA expressed by the transgene was correctly processed in organello. Out of 22 clones (Supplementary Data Figure S1), six sequences correspond to either processing intermediates or degradation products. Eight sequences showed the presence of the 3′-CCA sequence and terminate at G+1 and eight other sequences (see a representative sequence on Figure 2C) showed the presence of the 3′-CCA sequence and an additional G at position −1. As this G is not gene-encoded, this residue is likely to have been added post-transcriptionally by a potato mitochondrial guanylyl transferase activity consecutively to a normal cleavage by RNase P at G+1.
Figure 2.
Analysis of the fate of the larch mitochondrial tRNAHis precursor transcript having no G–1 in potato mitochondria. (A) Schematic representation of the strategy. First, G–1 and G–2 encoded by the larch mitochondrial trnH gene are deleted. Then, upon DNA uptake into potato mitochondria, larch mitochondrial tRNAHis is transcribed from the potato 18S rRNA promoter sequence (gray box). Total nucleic acids were analyzed by cRT-PCR. (B) Image of the ethidium bromide-stained gel of PCR product amplified using primers P3 and P4. The presence (+RT) or absence (–RT) of reverse transcriptase during the cDNA synthesis in the presence of primer P3 is indicated. The lane marked M shows the migration of the DNA ladder. (C) The 70-bp PCR product shown in (B) was cloned and 21 clones were sequenced. A sequence showing the junction (black vertical arrow) between 5′- and 3′- termini is presented. This sequence shows that the CCA triplet and the G–1 (underlined) have been post-transcriptionally added.
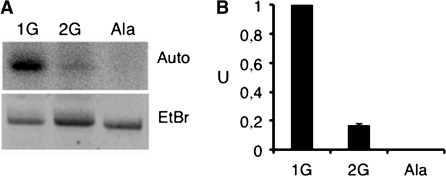
To confirm this result, we incubated similar amount of potato mitochondrial tRNAHis transcript with or without extra G–1 or of plant cytosolic tRNAAla transcript in the presence of [α-32P]GTP and a potato mitochondrial enzymatic extract. Upon incubation, the corresponding transcripts were resolved on a denaturing polyacrylamide gel. As expected, no radiolabeled guanosine triphosphates (GTP) was incorporated in the tRNAAla transcript, whereas a strong signal was obtained for the potato mitochondrial tRNAHis having no extra G (Figure 3). It must be noted that a weak incorporation of radioactivity was observed for the tRNAHis containing a G–1. This is very likely due to a heterogeneous population of tRNA molecules synthesized during in vitro transcription with T7 RNA polymerase.
Figure 3.
A tRNAHis-dependent guanylyl transferase activity is present in potato mitochondria. Larch mitochondrial tRNAHis without (1G) or with (2G) a G–1 and plant cytosolic tRNAAla (Ala) transcripts incubated with a potato mitochondrial enzymatic extract in the presence of [α-32P]GTP were analyzed on a polyacrylamide gel electrophoresis. (A) EtBr, image of the ethidium bromide-stained gel of RNA transcripts. Auto, autoradiography of the same gel. (B) Quantification of the GTP incorporation in the three tRNA transcripts. The amount of labeled tRNA was quantified using the ImageGauge software (Fuji). One unit (U) represents the amount of labeled larch mitochondrial tRNAHis transcript without G–1 (1G).
The two A. thaliana Thg1p homologs are predominantly localized to the nucleoplasm
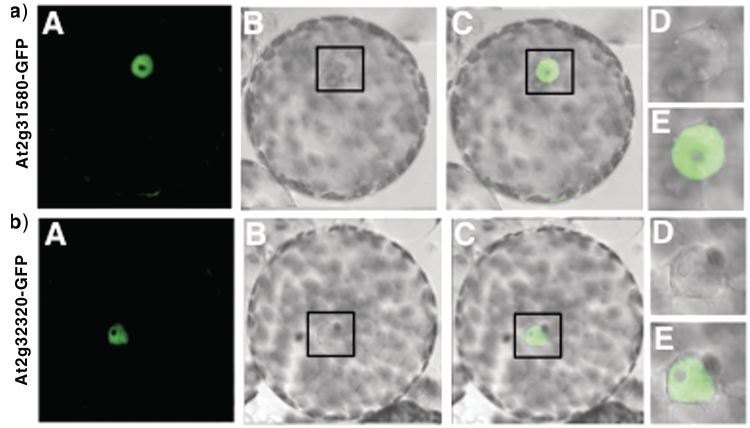
A tRNAHis-dependent guanylyl transferase activity is present in plant mitochondria. Which component is responsible for this activity? In eubacteria, a tRNAHis-dependent guanylyl transferase is not necessary and such an enzyme has not been identified. In eukaryotes, the nuclear enzyme responsible for this activity has been well characterized in S. cerevisiae and is called Thg1p (7,9,20,21). From the complete Arabidopsis nuclear genome, two genes (At2g32320 and At2g31580) that encode Thg1p homologs can be retrieved [(9) and Supplementary Data Figure S2]. The corresponding proteins share 83.3% identity and both correspond to a tandem duplication of two copies of the Thg1p homolog. Although none of them contains any predictable N-terminal mitochondrial targeting sequence, the protein encoded by the At2g31580 gene still presents an N-terminal extension as compared to the second protein. To address the potential involvement of these proteins in mitochondrial tRNAHis biogenesis, their subcellular localization was investigated. The 5′ copy of each Arabidopsis protein homolog to Thg1p (see Supplementary Figure S2) was fused to the GFP and visualized in N. benthamiana protoplasts by confocal microscopy. While no GFP fluorescence was observed in chloroplasts and mitochondria, both At2g31580- and At2g31320-GFP proteins were localized in the nucleoplasm (Figure 4). They are thereby not involved in the addition of the G–1 on the plant mitochondrial tRNAHis but rather implicated in the addition of this residue on nuclear-encoded tRNAHis.
Figure 4.
Nuclear localization of the two Arabidopsis proteins homolog to Thg1p. Half of the coding sequences of At2g31580 (a) and At2g32320 (b) proteins were cloned in frame with GFP. The resulting fusion proteins were expressed in N. Benthamiana. The localization of the two fusion proteins (At2g31580-GFP and At2g31580-GFP) was analyzed in transformed protoplasts by laser confocal microscopy. (A) GFP fluorescence, (B) DIC image and (C) merged image of (A) and (B), (D) and (E) enlarged view of the framed area shown in (B) and (C), respectively.
The recombinant Arabidopsis mitochondrial RNase P, PRORP1, cleaves potato mitochondrial tRNAHis precursors at two positions in vitro
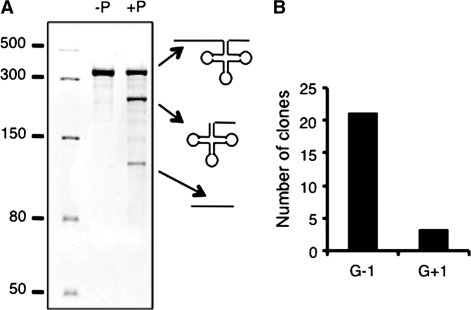
Our results suggest that, in vivo, plant mitochondrial RNase P can cleave tRNAHis precursors at two positions, thereby generating tRNAHis with or without a G–1. In order to verify this hypothesis, in vitro cleavage assays were performed with the purified recombinant Arabidopsis mitochondrial RNase P called PRORP1 (17) and precursor transcripts of potato mitochondrial tRNAHis. In the presence of PRORP1, tRNAHis precursor is submitted to an endonucleolytic cleavage and two RNA fragments of 240 and 125 nt length, respectively, are generated (Figure 5A). This is consistent with the expected cleavage at the 5′ end of tRNAHis resulting in the release of the 5′-leader sequence (125-nt-long RNA fragment) and a tRNAHis with a 3′-trailer (240-nt-long RNA fragment). To precisely map the 5′ endonucleolytic cleavage site, the 5′ matured tRNA precursor fragment was further analyzed by cRT-PCR, using the pair of primers P1 and P2 (Figure 1A). The amplicon obtained is ∼240 bp and could correspond to the expected size if the correctly processed 5′ end of the tRNAHis has been ligated to the 3′ end of tRNAHis precursor. The PCR product was cloned and sequenced. Out of 26 clones, 23 sequences showed that the 3′ terminus of the precursor transcript is ligated to G–1, whereas in three sequences, G–1 is missing and G+1 is ligated to the 3′-trailer sequence (Figure 5B). This result shows that, in vitro, although PRORP1 preferentially cleaves at the unusual cleavage site, this enzyme is also able to cleave at the classical cleavage site.
Figure 5.
In vitro cleavage of potato mitochondrial tRNAHis precursor transcripts in the presence of recombinant Arabidopsis mitochondrial RNase P, PRORP1. (A) The RNA molecules generated upon incubation of a 365-nt-long potato mitochondrial tRNAHis precursor transcript incubated (+P) or not (–P) with PRORP1 were analyzed on a polyacrylamide gel. Molecular mass markers given in nucleotides are indicated on the left. (B) The circularized 240-nt-long RNA product shown in (A) amplified by RT-PCR using primers P1 and P2 (Figure 1A) was cloned. Clones were sequenced, allowing the analysis of the 5′ extremity. The number of clones corresponding to a cleavage at position G–1 or at position G+1 is given.
DISCUSSION
Until now, the biogenesis of plant mitochondrial tRNAHis had never been studied. Because plant mitochondrial genomes encode the G–1, the direct pathway was thought to occur in these organelles (8). In this work, we report that, although unusual cleavage by RNase P can occur, the second pathway involving an RNase P cleavage between G+1 and G–1 and a guanylyl transferase activity also exists in plant mitochondria. In some archaea, a G–1 is encoded in the genome but a tRNAHis-dependent guanylyl transferase is active in vivo and the existence of the direct pathway has not been demonstrated so far in these organisms (22). Therefore, the data provided here in plant mitochondria are the first experimental evidence demonstrating that the two routes can coexist for the biogenesis of tRNAHis in a same compartment. This observation raises the puzzling questions of how and why the two pathways were maintained during evolution.
Very well conserved homologs of the S. cerevisiae guanylyl transferase Thg1p are widely distributed within eukaryotes (9). Among them, N-terminal mitochondrial targeting sequences were identified in metazoan Thg1p homologs. In chicken mitochondria, G–1 is not gene-encoded but rather post-transcriptionally added by a guanylyl transferase activity (8). Thus, the eukaryotic enzyme has likely been recruited to function in metazoan mitochondria. Here, a previously unreported guanylyl transferase activity was found in plant mitochondria but none of the two Arabidopsis Thg1p homologs is localized in the organelle. As no other class of tRNAHis-dependent guanylyl transferase can be predicted, there is no obvious protein candidate and biochemical and/or genetic approaches will be necessary to identify this new enzyme.
Until recently, the RNase P characterized in most organisms were shown to be ribonucleoproteins composed of a catalytic RNA subunit and several protein subunits (23,24). This ribozyme-type RNase P precisely cleaves tRNAHis precursors only at G–1 when the extra G is gene-encoded. By contrast, in plant mitochondria, the RNase P activity can be performed by a single protein called PRORP1, which, alone, is able to cleave the canonical structure of tRNAs (17). The question of plant mitochondrial tRNAHis precursor processing by a proteinaceous RNase P had not been addressed yet. According to the results obtained here, this proteinaceous RNase P seems to be less efficient than a ribozyme-type RNase P to precisely cleave tRNAHis precursors only at G–1 when the extra G is gene-encoded and the standard cleavage at G+1 is also performed by PRORP1. Consequently, a second factor with a tRNAHis-dependent guanylyl transferase activity presumably evolved or was maintained in plant organelles to repair the nonfunctional tRNAHis molecules. Due to the existence of the two distinct mechanisms, both plant mitochondrial tRNAHis with or without G–1 were found in vivo, while only tRNAHis with G–1 can be charged in vitro. In yeast when the tRNAHis and the histidyl-tRNA synthetase are overexpressed in a mutant strain lacking Thg1p (25), the strain is viable meaning that part of the tRNAHis can be charged in vivo. Therefore, a small proportion of plant mitochondrial tRNAHis without G–1 can conceivably be aminoacylated in vivo, although most of this tRNA species is very likely present as uncharged within mitochondria. The presence of tRNAHis without G–1 in the organelle can be the direct consequence of an inefficient repair system to add the extra G. Both in prokaryotes and in eukaryotes, uncharged tRNAs act as effector molecules to regulate gene expression, for example under adverse environmental conditions (e.g. ref. 26). In eukaryotes, under stress conditions, activation of the GCN2 kinase is mediated by uncharged tRNA (27,28) and plays an important role in regulating translation. So far, nothing is known about the presence and hypothetical function of uncharged tRNA in mitochondria or chloroplast. Here, the tRNAHis without G–1 is detectable in mitochondria under normal growth condition. Whether this uncharged tRNAHis has or not a function in the organelle will have to be addressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Centre National de la Recherche Scientifique (CNRS); the Agence National pour la Recherche (ANR); Fondi di Ateneo dell’Università degli studi di Bari. French Ministère Délégué à l’Enseignement Supérieur et à la Recherche, fellowship (to F.S.); A.G. is a fellow of the ANR; Università degli Studi di Bari ‘Aldo Moro’, 2-year research grant (to A.P.). Funding for open access charge: Institut de Biologie Moléculaire des Plantes (IBMP), CNRS.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wang C, Sobral BW, Williams KP. Loss of a universal tRNA feature. J. Bacteriol. 2007;189:1954–1962. doi: 10.1128/JB.01203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudinger J, Florentz C, Giegé R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues –1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22:5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen AE, Brooks BS, Guth E, Francklyn CS, Musier-Forsyth K. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA. 2006;12:1315–1322. doi: 10.1261/rna.78606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orellana O, Cooley L, Söll D. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol. 1986;6:525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkard U, Willis I, Söll D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J. Biol. Chem. 1988;263:2447–2451. [PubMed] [Google Scholar]
- 6.Cooley L, Appel B, Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc. Natl Acad. Sci. USA. 1982;79:6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell. Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L'Abbé D, Lang BF, Desjardins P, Morais R. Histidine tRNA from chicken mitochondria has an uncoded 5′-terminal guanylate residue. J. Biol. Chem. 1990;265:2988–2992. [PubMed] [Google Scholar]
- 9.Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Ohyama K. Transfer RNA genes in the mitochondrial genome from a liverwort, Marchantia-Polymorpha – the absence of chloroplast-like transfer RNAs. Nucleic Acids Res. 1992;20:3773–3777. doi: 10.1093/nar/20.14.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maréchal-Drouard L, Kumar R, Remacle C, Small I. RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res. 1996;24:3229–3234. doi: 10.1093/nar/24.16.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 13.Placido A, Gagliardi D, Gallerani R, Grienenberger JM, Maréchal-Drouard L. Fate of a larch unedited tRNA precursor expressed in potato mitochondria. J. Biol. Chem. 2005;280:33573–33579. doi: 10.1074/jbc.M505269200. [DOI] [PubMed] [Google Scholar]
- 14.Carneiro VTC, Dietrich A, Maréchal-Drouard L, Cosset A, Pelletier G, Small I. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol. Biol. 1994;26:1843–1853. doi: 10.1007/BF00019497. [DOI] [PubMed] [Google Scholar]
- 15.Maréchal-Drouard L, Small I, Weil JH, Dietrich A. Transfer RNA import into plant mitochondria. Methods Enzymol. 1995;260:310–327. doi: 10.1016/0076-6879(95)60148-1. [DOI] [PubMed] [Google Scholar]
- 16.Pujol C, Bailly M, Kern D, Maréchal-Drouard L, Becker H, Duchêne AM. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc. Natl Acad. Sci. USA. 2008;105:6481–6485. doi: 10.1073/pnas.0712299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 18.Berglund AK, Pujol C, Duchêne AM, Glaser E. Defining the determinants for dual targeting of amino acyl-tRNA synthetases to mitochondria and chloroplasts. J. Mol. Biol. 2009;393:803–814. doi: 10.1016/j.jmb.2009.08.072. [DOI] [PubMed] [Google Scholar]
- 19.Fey J, Dietrich A, Cosset A, Desprez T, Maréchal-Drouard L. Evolutionary aspects of “chloroplast-like” trnN and trnH expression in higher-plant mitochondria. Curr. Genet. 1997;32:358–360. doi: 10.1007/s002940050288. [DOI] [PubMed] [Google Scholar]
- 20.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12:1007–1014. doi: 10.1261/rna.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackman JE, Phizicky EM. Identification of critical residues for G-1 addition and substrate recognition by tRNA(His) guanylyltransferase. Biochemistry. 2008;47:4817–4825. doi: 10.1021/bi702517q. [DOI] [PubMed] [Google Scholar]
- 22.Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Söll D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc. Natl Acad. Sci. USA. 2009;106:21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preston MA, Phizicky EM. The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA. 16:1068–1077. doi: 10.1261/rna.2087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zhou H. tRNAs as regulators in gene expression. Sci. China Ser. C- Life Sci. 2009;52:245–252. doi: 10.1007/s11427-009-0039-y. [DOI] [PubMed] [Google Scholar]
- 27.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas C, Edwards-Ingram LC, Zeef L, Shenton D, Ashe MP, Grant CM. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.