Abstract
SARS coronavirus encodes non-structural protein 13 (nsP13), a nucleic acid helicase/NTPase belonging to superfamily 1 helicase, which efficiently unwinds both partial-duplex RNA and DNA. In this study, unwinding of DNA substrates that had different duplex lengths and 5′-overhangs was examined under single-turnover reaction conditions in the presence of excess enzyme. The amount of DNA unwound decreased significantly as the length of the duplex increased, indicating a poor in vitro processivity. However, the quantity of duplex DNA unwound increased as the length of the single-stranded 5′-tail increased for the 50-bp duplex. This enhanced processivity was also observed for duplex DNA that had a longer single-stranded gap in between. These results demonstrate that nsP13 requires the presence of a long 5′-overhang to unwind longer DNA duplexes. In addition, enhanced DNA unwinding was observed for gapped DNA substrates that had a 5′-overhang, indicating that the translocated nsP13 molecules pile up and the preceding helicase facilitate DNA unwinding. Together with the propensity of oligomer formation of nsP13 molecules, we propose that the cooperative translocation by the functionally interacting oligomers of the helicase molecules loaded onto the 5′-overhang account for the observed enhanced processivity of DNA unwinding.
INTRODUCTION
Severe acute respiratory syndrome (SARS) that claimed almost 800 deaths in southern China within a few months between 2002 and 2003 was caused by a novel coronavirus, SARS coronavirus (SCV) (1). SARS continues to be a serious concern as long as there is no vaccine or effective drug. SCV is a single-stranded (ss) RNA positive-strand virus with a genome of 29 727 nt (2,3). A single 21.2-kb replicase gene at the 5′-end region of the SCV genome is translated into two large replicative polyproteins, pp1ab (∼790 kDa) and pp1a (∼490 kDa), which is generated with and without ribosomal frameshifting (−1), respectively (4,5). These two translational products are subsequently processed by the viral main protease termed Mpro or 3CLpro, generating a number of non-structural proteins (nsPs). These nsPs include the RNA-dependent RNA polymerase (nsP12) and the NTPase/helicase (nsP13), which primarily constitute the membrane-bound viral replicase complex (6,7). The viral replicase synthesizes the entire viral genome (replication) as well as eight subgenomic mRNAs (transcription) (8,9).
Since the viral helicase has been identified as a potential target for therapy in other viruses due to its indispensability in viral genome replication (10–12), SCV NTPase/Helicase (nsP13), which was recently purified and characterized, was suggested as an attractive target for the development of anti-SCV agents (7). Thus, a lot of efforts have been made to identify and test small molecule inhibitors of the SCV helicase as drug candidates (13–16). RNA and DNA aptamers against SCV helicase were also reported to have an inhibitory effect against nucleic acids unwinding (17,18). Although the tertiary structure of the SCV NTPase/Helicase has not been experimentally verified, the structure prediction of the protein was recently reported (19,20). SCV nsP13 has been shown to contain two separate domains, i.e. the helicase domain (Hel) and a metal-binding domain (MBD), which consists of several conserved Cys/His residues at the N-terminal (6,19). The SCV helicase Hel domain has been categorized into the Superfamily I helicase based on the conserved sequence motifs as shown in the Escherichia coli Rep helicase (21).
Helicases are molecular motor proteins that translocate along the nucleic acid and separate double-stranded (ds) nucleic acid into two ss nucleic acids using the energy generated from nucleoside triphosphate (NTP) hydrolysis (22–24). In choosing duplex nucleic acids as substrates for unwinding, most known helicases unwind only one type of nucleic acid, either the DNA or RNA duplex. However, SCV helicase nsP13 has been shown to display activity on different types of nucleic acid substrates. The helicase nsP13 can unwind both RNA and DNA duplexes with a 5′- to 3′- polarity, in which the presence of 5′ ss overhang is absolutely required to unwind DNA duplexes (6,7). Furthermore, it has been observed that both ss RNA and DNA stimulated the ATPase activity of the helicase to the same degree (7,25). Thus, it is highly important to understand the duplex nucleic acid unwinding mechanism of the nsP13 helicase with respect to putative controlling factor(s) in nucleic acid unwinding.
In this study, the DNA duplex was chosen as a model substrate, and the unwinding of DNA substrates with different duplex lengths and 5′-overhang by this helicase was kinetically investigated. We demonstrated that the length of the 5′-overhang, which was used as the loading strand for helicase oligomers, was an important factor dictating the processivity of the DNA unwinding process. Thus, we propose that ‘functional interaction’ of nsP13 helicase oligomers loaded onto the 5′-overhang accounts for the observed cooperative translocation of the helicase in DNA unwinding and processivity, which is similar to the mechanism previously suggested for hepatitis C virus NS3 helicase (26).
MATERIALS AND METHODS
Protein and nucleic acids
SCV helicase nsP13 was overexpressed in E. coli and purified as described earlier (18). DNA substrates were purchased from Integrated DNA Technologies (Coralville, IA, USA), and purified by a denaturing (8 M urea) polyacrylamide gel electrophoresis (PAGE). The DNA concentration was determined by 260 nm absorbance and its extinction coefficient. To produce duplex or triplex DNA substrates, the 32P-labeled DNA strand was mixed with a 2-fold excess of complementary DNA and then annealed by heating to 95°C followed by slowly cooling. The DNA substrates used in this study are listed in Table 1.
Table 1.
DNA substrates for unwinding by nsP13 helicase
 |
DNA unwinding assays
The nsP13 (200 nM) and 32P -labeled DNA substrates (5 nM) were mixed in buffer MixA [50 mM Tris–Cl (pH 6.8), 50 mM NaCl, 2 mM ATP, 5 mM EDTA and 10% glycerol]. The mixture was preincubated for 5 min at 22°C and the unwinding reactions were initiated by adding an equal volume of MixB [2 mM ATP, 13 mM MgCl2 and 3 μM trap DNA (unlabeled bottom strand)]. After various times, the reactions were quenched by adding an equal volume of the quenching solution (100 mM EDTA, 0.4% SDS, 20% glycerol and 0.1% bromophenol blue). The ds and ssDNAs were resolved by native (urea-free) PAGE. Size markers for the unwound products were produced by heating the duplex substrates with 600-fold trap DNA at 95°C. The radioactivity was quantified using a Cyclone (PerkinElmer) and analyzed by OptiQuant/Cyclone software (Packard Instrument Company). The ratio of unwound products was calculated as described earlier (27) and the data were fit to a single-exponential equation [Equation (1)].
| (1) |
Where F(t) is fraction unwound at time t, A is the amplitude, k1 is the observed rate constant of the burst phase.
Chemical cross-linking
The chemical cross-linking reagent dimethylsuberimidate (DMS) was used to study the oligomerization of nsP13. A 300 mg/ml DMS stock solution was prepared immediately before use in ice-cold triethanolamine (TEA) buffer (TEA/HCl, 0.15 M, pH 8.2) and the pH was readjusted to 8.2 with 1 N NaOH. The nsP13 (4 µM) was preincubated in 50 mM TEA buffer (pH 8.2) containing 50 mM NaCl and 5 mM EDTA for 10 min at 22°C and cross-linking was initiated by adding DMS (10 mg/ml). After incubation for 1 min at 22°C, the cross-linking reactions were quenched by adding an equal volume of 1 M glycine. The quenched samples were analyzed by SDS–PAGE (4–12% gradient) and proteins were detected by Coomassie Brilliant Blue R-250 staining.
RESULTS AND DISCUSSION
SCV helicase nsP13 has been shown to have dsDNA unwinding activity and was shown to belong to the family of SF1 helicases (28). Like many SF1 helicases, the SCV helicase requires an exposed ss DNA or RNA for binding to the nucleic acid complex and the helicase translocates along the nucleic acids by hydrolyzing ATP. We have previously shown that ss DNA or RNA stimulates the ATP hydrolysis activity, but dsDNA does not (25). Although the crystal structures of a few typical SF1 helicases complexed with DNA were revealed (29,30), e.g. Rep and PcrA, only a computational 3D model has been proposed for the SCV helicase nsP13 (20). Therefore, details about the nucleic acid unwinding mechanism of SCV helicase nsP13 have to be deduced from biochemical studies only. Hence, we investigated how the mechanistic efficiency of dsDNA unwinding by nsP13 is affected by the structural features of the dsDNAs, such as duplex length, ssDNA tail length and artificial ss gap between the two DNA duplexes.
Single-turnover kinetics of duplex DNA unwinding by nsP13
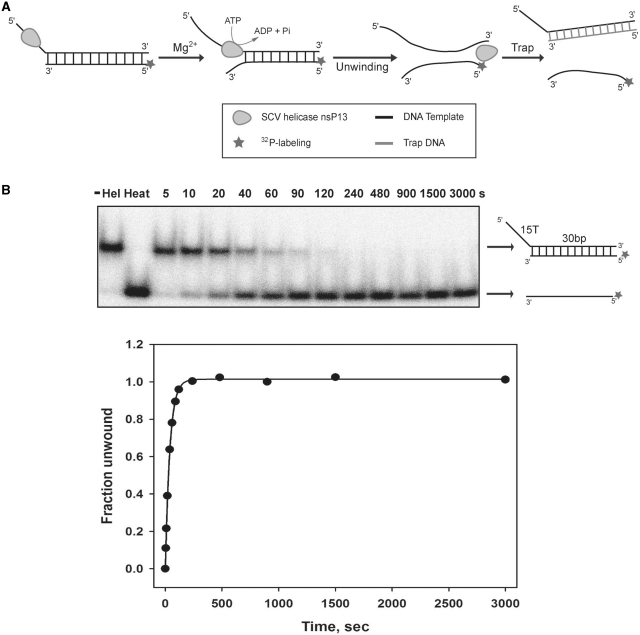
Because nsP13 has been shown to unwind both DNA and RNA duplexes that have a 5′-tail (6,7), we designed DNA duplexes with a 5′-tail at one side as the unwinding substrates to investigate the single-turnover kinetics of DNA unwinding by the helicase. Previously, analysis of nucleic acids unwinding by the coronavirus helicase was performed using a multiple turnover kinetics experiment, in which the helicase may rebind to the substrates after completion of a cycle of unwinding (6,7). In contrast, our experimental setup of the single-turnover condition enabled us to obtain quantitative aspects of the unwinding kinetics, especially the processivity of unwinding by the helicase. The reaction scheme is shown in Figure 1A, in which the helicase is initially allowed to bind to the substrate without ATP hydrolysis. In the absence of ATP hydrolysis, unwinding was not observed (data not shown). However, the addition of magnesium ions to the reaction allowed the helicase to unwind the DNA duplexes (Figure 1A).
Figure 1.
Single-turnover reaction of DNA unwinding by nsP13. (A) Schematic drawing of the unwinding of duplex DNA by nsP13. (B) Shown is the representative native gel shift assay of nsP13 unwinding in the presence of the 15T30bp dsDNA substrate. Unwinding products were resolved on an 8% native-PAGE and visualized using a phosphorimager Cyclone.
The duplex DNA unwinding reaction was initiated by adding a solution of MgCl2 (13 mM) and a large excess (3.0 µM) of trap oligonucleotides (unlabeled bottom strand) to a mixture of the pre-incubated reaction containing duplex DNA substrates (5 nM) and helicase (200 nM). The trap oligonucleotides served to prevent the re-initiation of unwinding by trapping free helicases and any helicases that dissociated from the substrates during unwinding (Supplementary Data). The trap oligonucleotides also prevented reannealing of the displaced bottom strand (Supplementary Data). Under the single-turnover reaction condition, the nsP13 helicase unwinded the dsDNA substrates and generated ssDNA products that were resolved by non-denaturing PAGE (Figure 1B). The kinetic time-course of ssDNA accumulation was plotted and fitted to an exponential function to obtain the reaction amplitudes and unwinding rates of only the helicases that were initially bound to the DNA substrates.
Processivity of duplex DNA unwinding
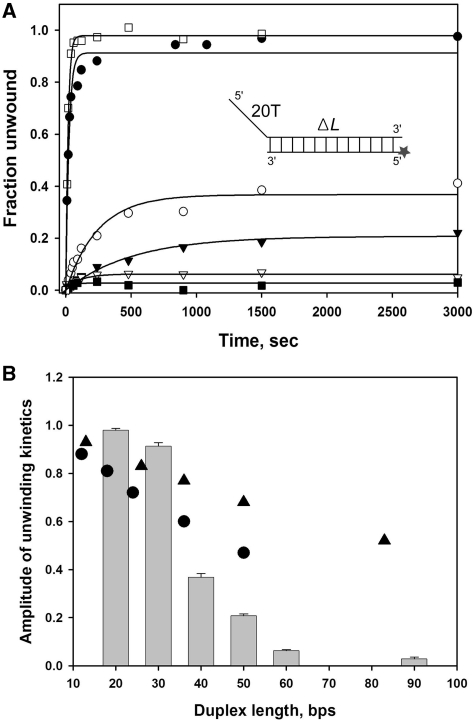
The helicase may dissociate from the duplex DNA before it is completely unwound. Therefore, to investigate the processivity of nsP13, kinetic studies of nsP13-catalyzed DNA unwinding were performed with DNA substrates containing different duplex lengths (shown in Table 1) under the single-turnover reaction condition. Measurements of the reaction amplitude of the fraction of dsDNA molecules unwound allows one to calculate the processivity of the helicase (31,32). As shown in Figure 2, the amplitude decreased as the length of the DNA duplex increased from 20 to 90 bp. This decrease in amplitude with an increase in duplex length may have occurred partly due to the reannealing of DNA strands behind the helicase and to the helicase dissociation from the DNA during unwinding. Nevertheless, the large excess of trap DNA that is included in the reaction rapidly anneals to the strand that is displaced from the duplex substrates during unwinding. A decrease in the amplitude of DNA unwound was observed in the absence of trap DNA at a given duplex DNA length (data not shown), indicating that the trap DNA prevented reannealing of displaced strands after completion of unwinding.
Figure 2.
Unwinding of duplex DNA substrates with varying length. (A) Single-turnover unwinding of duplex DNA substrates with different lengths by nsP13. dsDNA substrates contained 20 T tail and 20 bp (open square), 30 bp (filled circle), 40 bp (open circle), 50 bp (filled inverted triangle), 60 bp (open inverted triangle), 90 bp (filled square). The amplitudes are as follows: 20 bp = 0.98 ± 0.0077; 30 bp = 0.91 ± 0.020; 40 bp = 0.37 ± 0.016; 50 bp = 0.21 ± 0.0082; 60 bp = 0.062 ± 0.0045 and 90 bp = 0.028 ± 0.0074. (B) Amplitudes of nP13 are shown in the bar graph against duplex length and amplitudes of NPH (filled triangle) and NS3(filled circle) are marked on the same graph for comparison.
A plot of the final reaction amplitude versus duplex length showed that processivity of the nsP13 helicase in unwinding of DNA with a 5′-tail of 20 nt is very low (Figure 2B) compared with other related helicases, such as NPH-II RNA helicase and HCV NS3 DNA helicase (33,34). The processivity of a helicase is defined as the probability of unwinding a base pair versus dissociation of the helicase from the DNA at that position (24). At present, it is hard to calculate the exact processivity of the nsP13 helicase due to the absence of ‘kinetic step size’ information of active nsP13 (31,35). The monomeric form of most SF1 and SF2 helicases shows that they are rapid and processive at translocating along ssDNA, but not processive in terms of dsDNA unwinding (36–38). This raises the possibility that nsP13 may function as an oligomer in its active form like other SF1 and SF2 helicases, despite the fact that the helicase is non-processive by itself. Considering the hexameric ring-shaped helicases such as T7 bacteriophage helicase, the formation of ring was expected to result in high processivity due to DNA passage through a central channel. However, single-turnover kinetic analysis showed that the T7 helicase by itself has a low processivity for unwinding dsDNA (∼15 bp), but had a 9-fold higher helicase activity (∼130 bp) in the presence of polymerase and thioredoxin (31,39). The fact that SARS coronavirus has a polymerase and many non-structural proteins of unknown functions led to the hypothesis that additional proteins may be involved in the helicase activity of nsP13 (40).
An alternative but not exclusive explanation for the low processivity of nsP13 is that the DNA may not be the proper substrate for nsP13. Despite the fact that the SARS nsP13 helicase is a RNA helicase, little is known about its interaction with RNA. Although we have previously investigated the ATP hydrolysis activity of nsP13 in the presence of ssDNA or ssRNA (25), more rigorous experiments to monitor its affinities toward DNA or RNA are required to explain this low processivity. Whether nsP13 has different affinities for various types of DNAs or RNAs might be a determining factor for the processivity of nsP13 in duplex unwinding.
Enhancement of duplex DNA unwinding by increasing the length of 5′-overhang
Partial duplex DNA substrates containing 50 bp with a 5′-ssDNA overhang of varying lengths (Tn50bp in Table 1) were designed in order to investigate whether DNA unwinding is dependent on the length of the loading strand. Although nsP13 has been shown to have stimulated ATPase activity in the presence of polynucleotides, detailed binding affinity experiments have not been conducted. More importantly, the extent of stimulation of ATPase activity with ss nucleic acids was previously shown to be proportional to the length of ssDNA or ssRNA at a given concentration (25). These observations may indicate that nsP13 binds to the ss nucleic acids with a defined binding site size. Thus, we can speculate that as the length of the loading strand increases, more nsP13 helicases would be bound to the substrate DNA under the single-turnover reaction condition, in which the helicase outnumbers the amount of substrate DNAs.
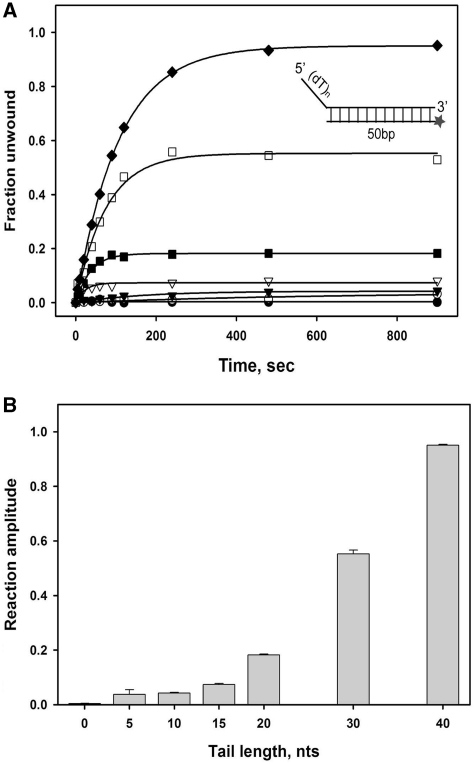
Time-courses of duplex DNA unwinding by 100 nM nsP13 with 2.5 nM of each duplex DNA substrate with various 5′-tail lengths (0, 5, 10, 15, 20, 30 and 40 nt) are shown in Figure 3. The quantity of unwound duplex DNA increased as the length of the ss 5′-tail increased. These results suggest that more ssDNA products were formed from duplex DNA substrates that could potentially bind more than one helicase molecule on the 5′-tail, implying that multiple nsP13 molecules might exhibit a higher processivity. Alternatively, a tighter binding affinity of the helicase monomer or oligomers to longer ss tails might also explain this increase in DNA unwinding. Based on these two explanations, we hypothesized that the helicase may occupy the defined length of the ssDNA overhang as a minimal binding site size and the stronger affinity is caused by cooperative binding of multiple helicases on a longer ssDNA. A tail length of 20 nt enabled nsP13 to marginally unwind 50 bp duplex DNA substrates, and a tail length of 40 nt allowed nsP13 to maximally unwind more duplex DNA (>95%) that was 50 bp in length. We postulate that the functional nsP13 monomer or oligomer requires a ssDNA loading strand that is at least 20 nt to translocate and unwind duplex DNA, which is analogous to a putative binding site size.
Figure 3.
Unwinding of DNA substrates with varying length of 5′-overhang by nsP13. (A) Single-turnover unwinding of 50 bp duplex DNA substrates with 5′-overhangs of varying lengths by 0 T (filled circle), 5 T (open circle), 10 T (filled inverted triangle), 15 T (open inverted triangle), 20 T (filled square), 30 T (open square) and 40 T (filled diamond). Data were fit to a single-exponential. The amplitudes were as follows: 0 T = 0.0035 ± 0.0013, 5 T = 0.038 ± 0.017, 10 T = 0.042 ± 0.0028, 15 T = 0.074 ± 0.0040, 20 T = 0.18 ± 0.0031, 30 T = 0.55 ± 0.013 and 40 T = 0.95 ± 0.0039. (B) The bar graph shows the reaction amplitudes of nsP13-catalyzed dsDNA unwinding as the length of the 5′-overhang increased from 0 to 40 nt.
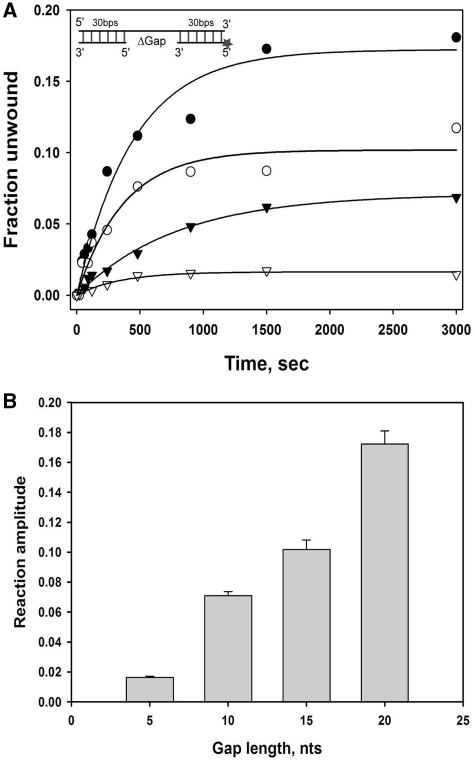
To further examine the effect of the length of the loading stand on nsP13 DNA duplex unwinding kinetics, we designed a series of duplex DNA substrates containing a DNA gap on the bottom strand (Table 1 and Figure 4). The gapped DNA duplex substrates did not have a 5′-tail at either end, which is available as loading strand for the helicase. Instead, the nsP13 helicase may recognize the internal ssDNA portion in the gapped DNA as the loading strand. The gap size was varied from 0 to 20 nt and unwinding at the bottom of the DNA strand (30 nt) annealed to the 3′-side of the top strand was monitored. Because entry of the helicase is not available at both blunt ends of the gapped DNA, the only available entry point of the helicase would be the ssDNA gap. In these experiments, the bottom strand (30 nt) annealed to the 5′-side of the top strand was displaced only when an overhang was present at the 5′-end of the gapped DNA duplex (data not shown).
Figure 4.
Unwinding of gapped DNA substrates by nsP13. (A) Unwinding of DNA substrates containing gaps of varying length: 20 bases gap (filled circle), 15 bases gap (open circle), 10 bases gap (filled inverted triangle), and 5 bases gap (open inverted triangle). The substrates consisted of two ds regions in the 5′-side (30 bp) and 3′ side (30 bp) of the upper strand. The gap was located between the two ds regions. Reaction amplitudes of gapped DNA substrates were as follows: 20 bases gap = 0.17 ± 0.0088, 15 bases gap = 0.10 ± 0.0063, 10 bases gap = 0.071 ± 0.0027 and 5 bases gap = 0.016 ± 0.0008. (B) The bar graph shows the amplitudes of nsP13-catalyzed unwinding with triple-stranded DNA substrates containing gaps of different lengths.
When blunt ended duplex DNA with an internal gap of 0 nt (nicked DNA) was reacted with the nsP13 helicase, no ssDNA product was observed, suggesting that the presence of ssDNA of a certain length is required for unwinding by nsP13. Increasing the gap size allowed nsP13 to bind to ssDNA and to unwind the bottom strand DNA annealed to the 3′ side of the top strand. When gaps of 5 or 10 nt were present in the bottom strand, the nsP13 helicase unwound the substrate with a very low processivity; <10% of substrates were unwound (Figure 4B). However, more ssDNA was formed when substrates contained gaps of 15 or 20 nt in the middle of the duplex DNA (Figure 4B). This result indicates that the longer loading strand facilitated more binding of nsP13 molecules to the ssDNA gap, resulting in a higher processivity in duplex (30 bp) unwinding. An increase in the amplitude of ssDNA product formation for DNA substrates containing 5–20 nt gaps was observed, indicating that DNA unwinding processivity was enhanced by an increase in the ssDNA gap. Interestingly, the amplitude obtained for 30-bp duplex DNA substrates containing a 20-nt gap was not as high as when the substrate contained a 20-nt 5′-tail overhang (>0.9 in Figure 2). This result suggests that the state of the nsP13 helicases bound to the 20-nt ssDNA gap were different from the one bound to the 20-nt 5′-tail.
The results of this study clearly show that increasing the ssDNA tail length enhances the unwinding amplitude. This result was also previously observed for SF1 and SF2 helicase (26,41); longer ssDNA tails provide more chances for helicase molecules to bind to ssDNA and multiple binding leads to an enhanced unwinding efficiency. In the present experimental setup, where the enzyme concentration was greater than the substrate concentration, all ssDNA tails were assumed to be initially bound by nsP13. The stacked oligomers and forward movement of nsP13 caused by the binding of multiple nsP13s on the ssDNA tail are believed to be the main reasons for the enhanced unwinding on substrates with a longer ssDNA loading strand. Taken together, when nsP13 concentration was in excess, the amplitude of ssDNA formation increased as the length of either the ssDNA overhang or gap increased. These results suggest that multiple nsP13 molecules may bind to the ssDNA loading strand and unwind longer duplex DNA with a higher processivity.
Cooperative translocation by oligomer of nsP13 helicases in duplex unwinding
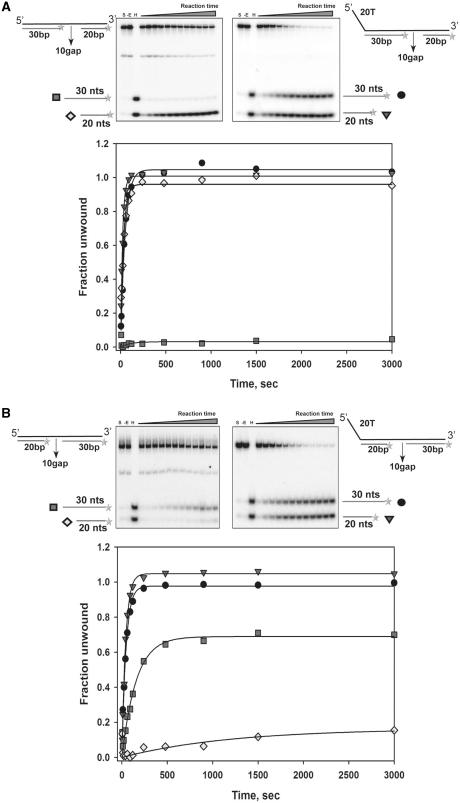
To further investigate whether multiple nsP13s can track through the loading strand and participate in the unwinding of the DNA duplex, we prepared gapped DNA substrates with or without a 5′-ssDNA tail (Table 1). Gapped substrates with a 5′-ssDNA tail, which contained a ssDNA gap that was 10 nt in length between the 30 bp of the lead duplex and 20 bp of the trailing duplex (20T30bp10G20bp in Table 1), was readily unwound by nsP13, resulting in displacement of the two duplexes (Figure 5A). However, for the gapped substrate that did not have the 5′-ssDNA tail (0T30bp10G20bp in Table 1), nsP13 was able to unwind only the 20 bp of the trailing duplex and not the 30 bp of the leading duplex (Figure 5A). This result indicates that the two gapped DNA substrates are recognized differently by nsP13 and the 5′-ssDNA loading strand is of primary importance for recognition and unwinding of the DNA duplex. Notably, a 10-nt gap was sufficient for the internal initiation site to completely unwind the 20 bp of the trailing DNA duplex, which was not observed for the unwinding of the longer 30-bp DNA duplex with a 10 nt gap (substrate ‘10 Gap’ in Figure 4). This indicates that nsP13 requires the presence of a longer 5′-ssDNA track to unwind a longer duplex DNA.
Figure 5.
Unwinding of gapped DNA substrates by nsP13 with or without a 5′-overhang. S indicates substrates that were incubated in the absence of nsP13 for 3000 s. –E indicates the absence of the enzyme (nsP13). H indicates substrates that were heated in the presence of excess trap DNA. Each substrate is described in the graph. (A) The amplitudes of 0T30bp10G20bp were as follows: 30b (filled square) = 0.031 ± 0.011, 20b (filled diamond) = 0.96 ± 0.025. The amplitudes of 20T30bp10G20bp were as follows: 30b (filled circle) = 1.05 ± 0.013, 20b (filled inverted triangle) = 1.01 ± 0.014. (B) The amplitudes of 0T20bp10G30bp were as follows: 30b (filled square) = 0.69 ± 0.22, 20b (filled diamond) = 0.16 ± 0.074. The amplitudes of 20T20bp10G30bp were as follows: 30b (filled circle) = 0.98 ± 0.012, 20b (filled inverted triangle) = 1.05 ± 0.0088.
It was of interest to determine whether nsP13 might traverse from the 5′-ssDNA tail to the internal gap and facilitate unwinding of the incoming duplex DNA, like a train on a railroad track pushing another train in the lead. To this end, we prepared DNA substrates with a longer trailing duplex (30 bp) and 10-nt gap (substrates in Figure 5B). As stated earlier, the 10-nt gap was not sufficient for nsP13 to completely unwind the 30-bp DNA duplex on the 3′-side, which was unwound when the gapped DNA substrate without the 5′-ssDNA overhang was used (0T20bp10G30bp in Figure 5B). Note that a blunt-ended DNA substrate (the lead 20-bp duplex of 0T20bp10G30bp) slightly unwound under this experimental setup. A portion of the blunt end duplex might have a 5′-ssDNA at the end due to breathing of the DNA duplex, which supports unwinding of the 20-bp DNA duplex. However, this effect was not sufficient for the unwinding of the longer 30-bp DNA duplex (square symbols in Figure 5A). Most importantly, nsP13 was able to completely unwind the 30 bp of the trailing duplex in the 10-nt gapped DNA substrate containing a 5′-ssDNA tail (20T20bp10G30bp in Figure 5B). This result suggests that the translocated nsP13 molecules pile up and facilitate unwinding of the 3′-side of the DNA duplex by the helicases that are internally bound at the gap. This may be the reason why the 30-bp DNA duplex containing the 10-nt gap and 5′-overhang (20T20bp10G30bp) unwound more efficiently than the gapped DNA substrate containing the longer lead DNA duplex (30 bp) with a blunt end (‘10 Gap’ shown in Figure 4). A portion of the nsP13 helicases may stack up after the 5′-side of the DNA duplex is unwound and may then unwind the 3′-side of the DNA duplex. Therefore, the nsP13 helicase can track through the loading strand and facilitate unwinding of the DNA duplex as long as the translocated helicases stay on the track.
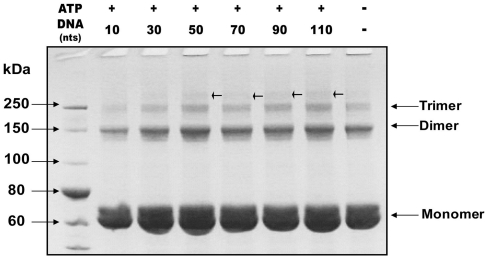
In order to investigate whether oligomerization of the nsP13 monomer is facilitated in the presence of ssDNA and ATP, we used a chemical cross-linking assay to examine the oligomeric state of nsP13 in the presence or absence of ligands. Chemical cross-linking with DMS showed that the degree of cross-linking was slightly altered in the presence of ligands; a higher order oligomers were observed in the presence of longer ssDNA (arrowheads in Figure 6). Interestingly, the nsP13 protein itself readily formed a dimer and trimer in the absence of any ligands. Thus, oligomerization of nsP13 is an intrinsic property of the protein, which is facilitated in the presence of long ssDNA and ATP. However, we could not determine the definite oligomeric state of the helicase in this study, suggesting that a dynamic oligomeric status of the helicase might exist in the presence of ss DNA.
Figure 6.
Chemical cross-linking using DMS indicates nsP13 oligomerization. Coomassie blue staining of a SDS–PAGE (4–12%) gel showing cross-linking of nsP13 either in the absence or presence of ATP (2 mM) and ssDNA (125 nM; 10, 30, 50, 70, 90, 110 nt). See the text for detailed experiment.
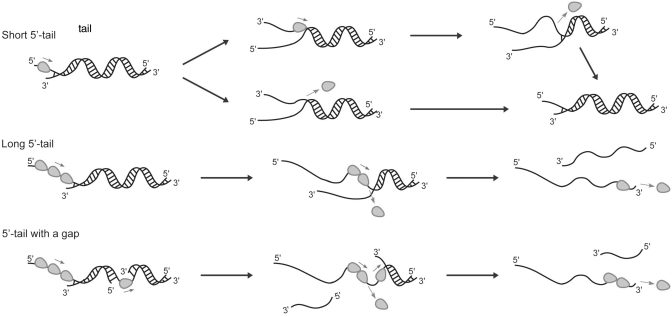
Based on the results obtained when the length of the 5′-ssDNA tail and internal ssDNA gap in duplex DNA was varied, together with the observation that nsP13 oligomerization occurs in the presence of ssDNA, we believe that nsP13 more efficiently unwinds partial-duplex nucleic acid substrates containing ss regions at the 5′-side that are long enough to allow the binding of multiple monomers (Figure 7). The proposed model, schematically shown in Figure 7, is very similar to that proposed for the unwinding of DNA by the HCV NS3 helicase and T4 Dda helicase (26,41). The proposed model for DNA unwinding by those helicases does not require protein oligomerization per se to obtain greater processivity. The increase in the helicase activity was explained by functional cooperativity rather than structural contacts between helicase monomers. Although multiple nsP13 helicases do not necessarily need to form a functional oligomeric form, multiple molecules of nsP13 are more efficient at unwinding longer substrates under single-turnover conditions. As depicted in Figure 7, multiple molecules of nsP13 loaded on the leading strand are more likely to complete the unwinding process before leading helicase monomers disassociate from the substrate. This cooperative translocation is demonstrated by our results on the unwinding of the gapped duplex DNAs with or without the 5′-ssDNA overhang (Figure 5). Unwinding of the 3′-side of the DNA duplex with a short gap size was impeded when fewer nsP13 molecules translocated from the 5′-side of the DNA, suggesting that multiple nsP13 monomers loaded on the same ssDNA track function together to produce more displaced ssDNA than a single nsP13 monomer can do. Taken together, the model presented in Figure 7 is a summary of how the non-processive nsP13 belonging to SF 1 helicase accomplishes substantial processivity in unwinding duplex nucleic acids in vitro. It should be noted that nsP13 may interact with other hosts and/or viral proteins when unwinding its RNA genome in infected cells and it may exhibit different properties during RNA unwinding.
Figure 7.
Proposed mechanism of DNA unwinding by the nsP13 helicase with different processivity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Korean Government, National Research Foundation of Korea (KRF-2008-313-C00531) and a grant (A090410) of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (to Y.-J.J.); National Research Foundation of Korea through the WCU Project (R33-10128) and a grant (20080401034026) from the BioGreen 21 Program, Rural Development Administration, Republic of Korea (to D.-E.K.). Funding for open access charge: Grant (2008-0062074) from the Ministry of Education, Science and Technology, the Korean government.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.World Health Organization report. Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS) 2003 http://www.who.int/csr/sars/country/en (1 July 2010, date last accessed) [Google Scholar]
- 2.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 3.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 5.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner JA, Watt RM, Chai YB, Lu LY, Lin MC, Peiris JS, Poon LL, Kung HF, Huang JD. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 9.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowski P, Schalinski S, Schmitz H. Nucleotide triphosphatase/helicase of hepatitis C virus as a target for antiviral therapy. Antiviral Res. 2002;55:397–412. doi: 10.1016/s0166-3542(02)00096-7. [DOI] [PubMed] [Google Scholar]
- 11.Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 12.Kleymann G, Fischer R, Betz UA, Hendrix M, Bender W, Schneider U, Handke G, Eckenberg P, Hewlett G, Pevzner V, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 13.Tanner JA, Zheng BJ, Zhou J, Watt RM, Jiang JQ, Wong KL, Lin YP, Lu LY, He ML, Kung HF, et al. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12:303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C, Lee JM, Lee NR, Jin BS, Jang KJ, Kim DE, Jeong YJ, Chong Y. Aryl diketoacids (ADK) selectively inhibit duplex DNA-unwinding activity of SARS coronavirus NTPase/helicase. Bioorg. Med. Chem. Lett. 2009;19:1636–1638. doi: 10.1016/j.bmcl.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang N, Tanner JA, Wang Z, Huang JD, Zheng BJ, Zhu N, Sun H. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem. Commun. 2007;14:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- 16.Yang N, Tanner JA, Zheng BJ, Watt RM, He ML, Lu LY, Jiang JQ, Shum KT, Lin YP, Wong KL, et al. Bismuth complexes inhibit the SARS coronavirus. Angew. Chem. Int. Ed. Engl. 2007;46:6464–6468. doi: 10.1002/anie.200701021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shum KT, Tanner JA. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem . 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang KJ, Lee NR, Yeo WS, Jeong YJ, Kim DE. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernini A, Spiga O, Venditti V, Prischi F, Bracci L, Huang J, Tanner JA, Niccolai N. Tertiary structure prediction of SARS coronavirus helicase. Biochem. Biophys. Res. Commun. 2006;343:1101–1104. doi: 10.1016/j.bbrc.2006.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann M, Eitner K, von Grotthuss M, Rychlewski L, Banachowicz E, Grabarkiewicz T, Szkoda T, Kolinski A. Three dimensional model of severe acute respiratory syndrome coronavirus helicase ATPase catalytic domain and molecular design of severe acute respiratory syndrome coronavirus helicase inhibitors. J. Comput. Aided Mol. Des. 2006;20:305–319. doi: 10.1007/s10822-006-9057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 23.Patel SS, Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 24.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Ann. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 25.Lee NR, Lee AR, Lee B, Kim DE, Jeong YJ. ATP hydrolysis analysis of severe acute respiratory syndrome (SARS) coronavirus helicase. Bull. Kor. Chem. Soc. 2009;30:1724–1728. [Google Scholar]
- 26.Levin MK, Wang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- 27.Ahnert P, Patel SS. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 28.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug. Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 30.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 31.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl Acad. Sci. USA. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 33.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 34.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 36.Cheng W, Hsieh J, Brendza KM, Lohman TM. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 37.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 39.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, Klenk HD, Rappuoli R. SARS: beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.